Abstract
The ubiquitin proteasome system (UPS) plays a crucial role in biological processes integral to the development of the cardiovascular system and cardiovascular diseases. The UPS prototypically recognizes specific protein substrates and places polyubiquitin chains on them for subsequent destruction by the proteasome. This system is in place to degrade not only misfolded and damaged proteins, but is essential also in regulating a host of cell signaling pathways involved in proliferation, adaptation to stress, regulation of cell size, and cell death. During the development of the cardiovascular system, the UPS regulates cell signaling by modifying transcription factors, receptors, and structural proteins. Later, in the event of cardiovascular diseases as diverse as atherosclerosis, cardiac hypertrophy, and ischemia reperfusion injury, ubiquitin ligases and the proteasome are implicated in protecting and exacerbating clinical outcomes. However, when misfolded and damaged proteins are ubiquitinated by the UPS, their destruction by the proteasome is not always possible due to their aggregated confirmations. Recent studies have discovered how these ubiquitinated misfolded proteins can be destroyed by alternative “specific” mechanisms. The cytosolic receptors p62, NBR, and HDAC6 recognize aggregated ubiquitinated proteins and target them for autophagy in the process of “selective autophagy”. Even the ubiquitination of multiple proteins within whole organelles that drive the more general macro-autophagy may be due, in part, to similar ubiquitin-driven mechanisms. In summary, the cross-talk between the UPS and autophagy highlight the pivotal and diverse roles the UPS plays in maintaining protein quality control and regulating cardiovascular development and disease.
Keywords: Cardiovascular, development, ubiquitin, proteasome, signaling, autophagy
Introduction
A growing number of studies implicate post-translational modifications by the ubiquitin proteasome system (UPS) in regulating the complex cell signaling processes fundamental to cardiovascular development and disease. More recently though, our understanding of the role of the UPS in protein quality control has expanded with new studies delineating its role in the endoplasmic reticulum and its cross talk with the process of autophagy. In this review, we present recent evidence that suggests that the UPS plays an essential role not only in cardiovascular development, but also in the dynamic pathophysiology of cardiovascular diseases (Table 1).
Table 1.
Proteins involved in the UPS regulation of cardiovascular development and pathophysiology of cardiovascular diseases
| Proteins: Vascular Development | Function | Role |
|---|---|---|
| 1. Numb | Notch antagonist | Ubiquitinated by LNX, preventing internal sequestration of Notch |
| 2. Itch | Ubiquitin ligase | Polyubiquitinates Notch in absence of a ligand, which promotes endocytosis and inhibition of Notch |
| 3. FBW7/Sel-10 | Ubiquitin ligase | Targets Notch for proteasomal degradation during vascular development |
| 4. MIB (Mind Bomb) family | Ubiquitin ligase | Regulates Notch ligand signaling (MIB1 developing embryo; MIB2 adult tissues) |
| 5. Nedd-4 | Ubiquitin ligase | Targets VEGF-receptor 2 for proteasomal degradation |
| 6. VHL (von Hippel-Lindau) | Ubiquitin ligase | Regulates hypoxia inducible factor 1 (HIF1) to adapt to low oxygen concentrations |
| 7. CHIP (C terminus of Hsc70-interacting protein) | Ubiquitin ligase | Targets myocardin, FOXO1; mediates smooth muscle cell differentiation and proliferation |
| 8. HIF1α(Hypoxia inducible factor 1) | Ubiquitin ligase/transcription factor | During hypoxia transcribes pro-angiogenic factors e.g. VEGF, TGF-β3 |
| 9. FIH (Factor inhibiting HIF) | Hydroxylation factor | Binds HIF1 to make it transcriptionally inactive |
| 10. Siah2 | Ubiquitin ligase | Promotes HIF1 activity at specific oxygen concentrations |
| 11. VDU2 | VHL de-ubiquitinase | Stabilizes HIF1 by deubiquitination |
| 12. ASB4 (Ankyrin repeat SOCS box protein 4) | Ubiquitin ligase | Interacts directly with FIH; high levels expressed with drastic increases in oxygen tension |
| 13. FBW7, Cul 7 | Substrate recognition protein/SCF (Skp1, Cul1, F-box protein) type ubiquitin ligase | Required for cardiovascular development; implicated in cancers (targets destruction of oncogenes e.g. myc, c-Jun, Notch, cyclin E) |
| 14. Cullin | SCF type ubiquitin ligase | Deletion results in hemorrhagic vasculature and abnormal placental endothelial differentiation |
| Proteins: Atherosclerosis | Function | Role |
| 1. Proteasome inhibitors | Reduce endotoxin-induced gene expression; prevent LPS-induced inflammatory responses; upregulate nitric oxide synthase in endothelial cells | |
| 2. TNFa, NF-kB | Inflammatory mediator/transcription factor | Correlated with protein ubiquitination and 20S proteasome activity |
| 3. CYLD | Deubiquitinating enzyme | Inhibits TNFa-induced NF-kB activation and expression of Cyclin D1 through deubiquitination of TNFR-associated factor 2 (TRAF2) and Bcl-3 |
| 4. PAF (Platelet Activating Factor) | Inflammatory mediator | Downregulated by lysosomal- and ubiquitin-dependent proteasomal-mediated receptor degradation |
| 5. CRP | Acute phase protein | Transgenic expression of human CRP reduced atherosclerotic lesion size and was associated with increased plaque expression of 26S proteasome subunits |
| 6. LIG (Lipoprotein-inducible gene) | Human homologue of bovine ubiquitin- conjugating (E2) enzyme | Increased during aggregated LDL challenge of macrophages |
| 7. HMGCoA | Cholesterol synthesis regulatory enzyme (hepatocyte) | Turnover mediated by ubiquitination and activity of 26S proteasome |
| 8. Idol | Ubiquitin ligase | Regulates LDL receptor |
| 9. COP9 signalosome | Mediates ABCA1 (ATP-binding cassette protein A1) degradation | |
| Proteins: Cardiovascular Biology | Function | Role |
| 1. cIAP, XIAP | Degrade caspases | Regulation of apoptosis |
| 2. MDM2, COP1, Pirh2, ARF-BP1, CHIP | Degrade p53 | Regulation of apoptosis |
| 3. MuRF1, CHIP, MAFBx/Atrogin-1, MDM2 | Ubiquitin ligases | Pathophysiology of various cardiac diseases |
| Proteins: ERAD, ER Stress, Unfolded Protein Response (UPR) | Function | Role |
| 1. PERK, ATF6, IRE-1 | ER transmembrane proteins | Mechanisms by which ER senses stress (Unfolded Protein Response) |
| 2. GRP78 | ER chaperone | In normal times, binds to internal surfaces of PERK, ATF6, IRE-1 and blocks downstream signaling |
| 3. GRP94 | SR chaperone | Overexpression protects against Ca2+ overload, ischemia-induced cell death |
| 4. Puma (p53-upregulated modulator of apoptosis) | Modulates apoptosis | Upregulated during induced ER stress and UPR |
| Proteins: Cardiac Development | Function | Role |
| 1. N-recognins: UBR1, UBR2 | Ubiquitin ligases | Mediate the N-end rule pathway by recognizing N-degrons |
| Proteins: Cardiac Disease | Function | Role |
| 1. MuRF1, MuRF2, MuRF3, MAFBx/Atrogin-1, CHIP, MDM2 | Ubiquitin ligases | Mechanistically characterized in hypertrophy and ischemia reperfusion injury |
| Proteins: Selective Autophagy | Function | Role |
| 1. HSP-E3 Complex | Protein quality control | Promotes folding of misfolded proteins, enhances ubiquitination, and targets misfolded proteins for proteasome degradation |
| 2. Atg12-Atg5; LC3(Atg8)-PE | Autophagy pathways | Conjugate to Atg proteins forming complexes essential for recruitment of LC3 and formation of autophagosome membranes |
| 3. p62 | Intracellular receptor, adaptor protein | Recognizes ubiquitin chains on targeted proteins and delivers them to autophagosomes; also regulates clearance of proteins by UPS |
| 4. NBR1 | Intracellular receptor, adaptor protein | Mediates cross-linking of ubiquitinated proteins; interacts w/p62 to clear misfolded proteins |
| 5. HDAC6 | Intracellular receptor, adaptor protein | Recognizes ubiquitinated/misfolded proteins shuttling them into aggresomes; delivers for autophagy via microtubule organizing centers (MTOC) |
| 6. BAG3 | Hsc/Hsp10 co-chaperone | Mediates autophagic degradation of ubiquitinated proteins in aging cells; co-localizes with p62-positive aggregated proteins |
| 7. BAG1 | Mediates proteasome-dependent degradation of ubiquitinated proteins w/CHIP |
Overview of the Ubiquitin Proteasome System
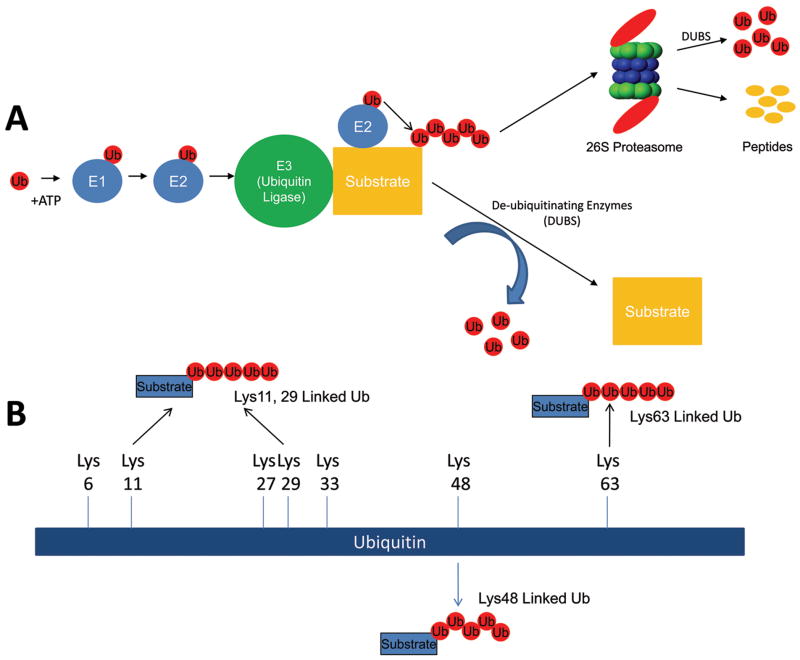
The destruction of proteins at the cellular level is a dynamic process regulated primarily by the ubiquitin proteasome system (UPS). The UPS is a cascade of carefully regulated enzymes which consist of E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase) enzymes (see Figure 1A) which target proteins for destruction by the proteasome. Each of these enzymes (E1, E2, and E3) plays a unique role in the post-translational modification of specific proteins. The E1 uses ATP to generate a high energy thioester bond with ubiquitin between catalytic cysteine residues within E1 and the c-terminal glycine residue of ubiquitin. This “activated” ubiquitin is then available to be transferred to one of the ubiquitin conjugating enzymes (E2). There are dozens of these E2 enzymes to which the charged ubiquitin is added. These E2 enzymes then interact with of one of the hundreds of ubiquitin ligases (E3) to transfer the activated ubiquitin to the epsilon-amino group of a lysine residue in the target protein. The ubiquitin on the target protein then serves as an “acceptor” on which additional ubiquitins can be added. Several cycles of this ubiquitin ligase activity results in a poly-ubiquitin chain being formed on the target protein.
Figure 1. The ubiquitin proteasome system at a glance.
The ubiquitin proteasome is a system of enzymes that places ubiquitin (chains) on specific protein substrates to target them for degradation, change their localization, and/or enhance their activity. (A) Free monoubiquitin is activated by the E1 enzyme in an ATP-dependent manner and transferred to the E2 enzyme. The specificity of the system is in the E3 (ubiquitin ligase) that mediates the transfer of one or more ubiquitin moieties sequentially to form ubiquitin chains on the substrate. The canonical lysine chains linked by their lysine48 (Lys48) are recognized by the 26S proteasome which degrades the protein into constituent peptides and free ubiquitin. (B) The role of noncanonical polyubiquitination (i.e. Lys63-linked ubiquitin chains) is increasingly being described in the cardiovascular system as a way to regulate protein (i.e. transcription factors) activity. A total of seven lysine moieties exist in ubiquitin which can be used for chain formation (Lys6, Lys11, Lys27, Lys29, Lys 33, Lys48, Lys63). A number of studies discussed in this review reveal a role for more linear Lys63 chains, in addition to canonical Lys48 linkages in the cardiovascular system. However, the significance of Lys6, Lys11, Lys 27, Lys29, Lys33 and branching/complex ubiquitin chains has not been elucidated in general.
Ubiquitin is a 76 amino acid moiety with multiple lysines capable of making isopeptide linkages to form polyubiquitin chains (Figure 1B). This allows a diversity of polubiquitin chain configurations that can drive different fates for the proteins to which they are attached. The most commonly identified polyubiquitin linkages occurs on the lysine at amino acid 48 (Lys48). Polyubiquitin chains that link through its Lys48 are called canonical ubiquitin chains and target the ubiquitinated substrate for destruction. However, there are a total of seven lysine moieties in ubiquitin which can be used for chain formation (Lys6, Lys11, Lys27, Lys29, Lys 33, Lys48, Lys63)1. Conformationally, polyubiquitin chains formed through Lys63 have been identified to be more linear than chain made through Lys48 as illustrated in Figure 1B. Lys63 polyubiquitin chains can modify the activity of the target proteins 2 and have been implicated in DNA repair mechanisms3 and in the regulation of physiologic cardiac hypertrophy2. The significance of the addition of polyubiquitin chains formed through the noncanonical lysines (Lys6, Lys11, Lys 27, Lys29, Lys33) has not been completely elucidated. The addition of a single ubiquitin on a protein, called mono-ubiquitination, regulates DNA repair, nuclear export, and histone regulation. In the heart, several signaling processes are regulated by monoubiquitination in the heart, including receptors involved in signaling pathways (EGF)4, cell-to-cell electrical coupling (connexin 43)5, 6, apoptosis (caspase 3 and caspase 7) 7, and calcium regulation (via calmodulin)8. The post-translational modification of a protein by poly- or mono-ubiquitination may determine its fate and regulate its activity. For example, polyubiquitin chains may target misfolded proteins for degradation while monoubiquitination may tag these same proteins for delivery to other cellular compartments effectively inhibiting their activity9. De-ubiquitinating enzymes (DUBs) counteract the ubiquitination process (Figure 1A). The nearly 100 described DUBs cleave ubiquitin and help recycle ubiquitin removed at the 26S proteasome as ubiquitinated proteins are degraded 10. Not only can DUBs reverse the modification of ubiquitinated proteins, they play a role in the remodeling of polyubiquitin chains10.
The UPS regulates cell signaling in vascular development
Development of the vascular system is one of the earliest and most pivotal events that occur during embryogenesis. The development of blood vessels de novo occurs when mesodermally derived angioblasts differentiate to form primitive blood vessels (vasculogenesis). Sprouting and bridging of this primary plexus occurs through angiogenesis, where endothelial cell outlines are covered by smooth muscle cells in the large vessels. Arteriogenesis, the process of remodeling existing capillaries in response to increased flow demand, is also fundamentally involved in vascular development. These processes all regulate precursor cells in the developing embryo as well as in the adult through common signaling pathways such as Notch, VEGF, and HIF1α among others11, 12. In turn, each of these signaling pathways can be and are regulated by the UPS.
UPS regulation of Notch signaling
The UPS regulates Notch signaling by its interaction with and regulation of the Notch antagonist Numb. The ubiquitin ligase LNX can ubiquitinate Numb, preventing internal sequestration of Notch, resulting in enhanced downstream Notch signaling 13. The ubiquitin ligase Itch can polyubiquitinate Notch in the absence of a ligand, promoting endocytosis and inhibition of Notch 14. Furthermore, during vascular development the ubiquitin ligase FBW7/Sel-10 can target Notch to the proteasome for degradation 15. Adding to the complexity of UPS-mediated regulation of Notch, is the fact that Notch ligands are also targets of proteasomal degradation. Two studies have demonstrated that the Mind Bomb (MIB) family of ubiquitin ligases serves a regulatory role in Notch ligand signaling, with individual family members temporally restricted in expression to either revascularizing adult tissue (MIB2) or the developing embryo (MIB1) 16, 17.
UPS regulation of VEGF signaling
Perhaps the most widely studied component of vascular development is VEGF signaling. VEGF signaling is crucial for angiogenesis, vasculogenesis, cell migration, proliferation, and cell survival18. Mice lacking VEGF die at ~ E8.5 and have significant impairments in angiogenesis and blood-island formation 19. On the opposite end of the spectrum, even modest increases in VEGF disrupt vascular development 20. Improper regulation of VEGF has also been implicated in the pathophysiology of pulmonary inflammatory disease, cancer proliferation, diabetic retinopathy, and rheumatoid arthritis. The VEGF-receptor 2 can be ubiquitinated by Nedd-4, targeting it to the proteasome for degradation21. However, this Nedd-4 mediated regulation can itself be regulated by its association with Grb10 21. VEGF is also regulated by oxygen sensing mechanisms; the ubiquitin ligase VHL regulates hypoxia inducible factor 1 (HIF1) to adapt to low oxygen concentrations.
UPS regulation of smooth muscle cell development
Another major component of vascular development involves the development of vascular smooth muscle cells (SMCs). The precise coordination of proliferation and differentiation of SMCs is required for proper vasculature. Recent studies indicate that the ubiquitin ligase CHIP (C terminus of Hsc70-interacting protein) mediates both SMC differentiation and proliferation through ubiquitination and proteasomal destruction of specific substrates. CHIP’s targeting of myocardin, a key co-transcription factor of SRF, decreases in SMC differentiation 22. However, when CHIP targets FOXO1, a repressor of SMC differentiation, the subsequent FOXO1 repression is ameliorated, thus mitigating apoptosis and enhancing SMC growth 23. Other ubiquitin ligases (i.e. Skp2, MDM2) have been described in SMC biology, but their role in differentiation has yet to be explored 24, 25.
HIF1α as a prototypic transcription factor regulated by the UPS in vascular development
The best characterized ubiquitin ligase that regulates vascular development is hypoxia inducible factor 1 (HIF1). The HIF1 signaling cascade mediates the necessary adaptations the vasculature needs to make in the presence of low oxygen concentrations, including the formation of vessels in both embryos and adults. HIF1 is a dimeric transcription factor composed of an α andβ subunit. Under normoxic conditions, HIF1α subunits are rapidly degraded by the UPS. However, in response to decreased oxygen levels, HIF1α becomes stabilized and transcribes an array of pro-angiogenic factors, such as VEGF, TGF-β3, and various components of glucose transport and glycolysis, which are generally thought to overcome vascular insufficiency 26–28. While regulation of HIF1 can occur at the mRNA level29, it is widely believed its primary regulation is through post-translational modification and degradation of HIF1α via the UPS.
The von Hippel-Lindau tumor suppressor protein (pVHL) is an essential part of the ubiquitin ligase complex that negatively regulates HIF1α. During periods of normoxia, prolyl hydroxylase (PHD) catalyzes the hydroxylation of the HIF1α subunits on conserved proline residues, converting them into hydroxyproline. This in turn is recognized by the pVHL ubiquitin ligase complex resulting in HIF1α ubiquitination and subsequent degradation by the proteasome. A second form of HIF1 inhibition occurs through its interaction with the factor inhibiting HIF (FIH). FIH binds HIF1 and hydroxylates an asparagine residue on the C-terminal transactivation domain (C-TAD) of HIF1. This hydroxylation prevents the coactivator p300/CBP from associating with the C-TAD of HIF1, rendering it transcriptionally inactive. While apparently redundant, it has recently become clear that FIH and PHD-pVHL modes of HIF1 inhibition operate in segregation as well, depending on the precise oxygen gradient, as evidenced by mathematical and biological predictions30. Though this canonical view of HIF1 regulation by pVHL and FIH is generally believed to be the predominant modulator in oxygen-mediated vascular development, other elements exist adding further layers of complexity. For example, by negatively regulating the activity of PHD enzymes, the ubiquitin ligase Siah2 promotes HIF1 activity at specific O2 concentrations 31. Additionally, the VHL de-ubiquitinase VDU2 is able to stabilize HIF1α by de-ubiquitinating it, resulting in the enhancement of HIF1 activity32.
Another crucial factor involved in vascular differentiation and development regulated by FIH is the ankyrin repeat SOCS box protein 4 (ASB4), originally identified as a ubiquitin ligase that is differentially expressed in vasculature lineages in embryoid bodies 33. ASB4 interacts directly with FIH and is itself a substrate of FIH-mediated hydroxylation, the results of which may promote binding to and degradation of substrates of ASB4 33. High levels of ASB4 are expressed in the embryonic vasculature at times when drastic increases in oxygen tension occur (E8.5-E9.5) 33. Additionally, in situ mRNA distribution analysis reveals that ASB4 is spatially compartmentalized to the developing capillary plexi, intersomitic vessels, and placenta 33. As the vasculature matures and oxygen levels are stabilized, ASB4 is expression is downregulated, further implicating its role in differentiation. ASB4 makes up a ubiquitin ligase complex with elongin B/elongin C/cullin/Roc and serves to give the complex its specificity 33. Overexpression of ASB4 promotes differentiation of vascular precursors into the vascular lineage in an oxygen-dependent manner, strengthening its proposed role in vascular development 33.
Other UPS components linked to signaling in vascular development: FBW7 and Cul7
The substrate recognition protein FBW7 (F-box and WD repeat domain-containing 7) is part of a conserved Skp1, Cul1, and F-box protein (SCF) type ubiquitin ligase complex. FBW7 is required for cardiovascular development, as FBW7 null mice die at embryonic day 11 from impaired cardiac and vascular development 15, 34. Conditional knockouts in hematopoietic stem cells results in a decrease in all lineages of blood cells (pancytopenia)35 and deletion of FBW7 in mouse T-cells, disrupting their cell-cycle exit36. Conditional FBW7 knockouts can also develop leukemia. This is not surprising since FBW7 is implicated in many human cancers, targeting the destruction of oncogenes such as myc, c-Jun, Notch, and cyclin E35. Cullin is another component of the SCF ubiquitin ligase complex that has been linked to the developing vasculature. Arai, et al. identified that the deletion of the Cul1 homologue p185 (Cul7) results in neonate lethality from respiratory distress37. They further show that embryos exhibit hemorrhagic vasculature, and abnormal placental endothelial differentiation and vascular structure. Interestingly, in the same report Cul7 was also shown to form a SCF-like complex that includes glomulin (aka FAP48 and FAP68), which has been suggested as the loss-of-function component in familial glomuvenous malformation. This hereditary syndrome is characterized by abnormalities in vascular morphology and morphogenesis 38.
The role of the UPS in the pathophysiology of atherosclerosis
Recent studies have identified the contribution the UPS makes to the development of atherosclerosis by regulating vascular inflammation, oxidative stress, apoptosis, and cholesterol metabolism. These studies, outlined below, provide evidence that implicates the UPS in many diverse mechanisms that contribute to the pathogenesis of atherosclerosis.
The UPS regulates vascular inflammation and oxidative stress responses
Current dogma states that atherosclerosis is, in large part, an inflammatory disease of the vessel wall. Several reports provide insight into how the UPS may affect specific components of the inflammatory response and may therefore modulate the atherosclerotic process. The UPS plays a role in modifying the function of inflammatory and vascular cells. Regulatory T cell (Treg) function plays a role in attenuating the inflammatory/immune component of atherogenesis. Meier, et al. have shown that treatment of Treg cells with oxidized LDL or uremic serum causes a reduction in proteasomal activity that leads to cell cycle arrest and apoptosis39. They postulate that the resulting immune dysfunction exacerbates inflammation and atherogenesis in patients, especially those with end stage renal disease 39. The macrophage, central to the development of inflammation and atherosclerosis, is dependent in part upon its proteasome activity to function. Proteasome inhibitors reduce endotoxin-induced gene expression including the Toll-like receptor 2, and can prevent LPS-induced inflammatory responses 40. Thus, regulation of the macrophage proteasome activity can modulate the function of these cells and may therefore alter the atherogenic process. Endothelial cell function is also intimately involved in atherogenesis, and regulation of endothelial proteasome activity can modulate the inflammatory phenotype. For example, proteasome inhibition causes the upregulation of endothelial nitric oxide synthase in endothelial cells and thereby enhances endothelial-dependent vasorelaxation of rat aortic rings 41. Thus, proteasome function may be necessary to induce the endothelial cell dysfunction that contributes to atherosclerosis, other vascular diseases, and systemic diseases characterized in part by vascular dysfunction.
The UPS can also modify the function of various inflammatory mediators. TNFa stimulates inflammation and immune responses in part through activation of NF-kB, a nuclear transcription factor that plays an important and central role in the generation of inflammation, apoptosis and cell proliferation. In carotid artery plaques from patients with various clinical characteristics, protein ubiquitination and 20S proteasome activity is correlated with the presence of TNFa and NF-kB 42–44. Plaques from patients with morning surges in blood pressure have increased protein ubiquitination and 20S proteasome activity which is associated with increases in NF-kB, TNFa, inflammatory cell number, markers of oxidative stress, and matrix metalloproteinase 9, but decreases in collagen content and IkB levels compared to controls43. Similar findings have been reported in plaques from post-menopausal women not receiving hormone replacement therapy compared to those who are 44, and in plaques from diabetics not treated with rosiglitazone compared to those who were treated42. These findings demonstrate that protein ubiquitination and 20S proteasome activity is associated with inflammation, oxidative stress, and histologic changes leading to an unstable plaque phenotype. TNFa also stimulates an increase in the expression of the de-ubiquitinating enzyme cylindromatosis (CYLD) in endothelial and smooth muscle cells. CYLD inhibits TNFa-induced NF-kB activation and expression of Cyclin D1 through de-ubiquitination of TNFR-associated factor 2 (TRAF2) and Bcl-3, respectively. Overexpression of CYLD inhibits cell viability and neointima formation in a rat model of carotid artery injury 45. A recent study assessing the role of the UPS in atherosclerosis in rabits investigated how inhibiting the 20S proteasome by aspirin (ASA) affected atherosclerosis progression. Rabbits were fed a high fat diet and some were additionally treated with ASA. In ASA-treated rabbits, atherosclerotic lesions were less apparent and more ubiquitinated proteins were present. Signaling through NF-κB was inhibited by ASA, as determined by a number of measures, suggesting that the therapeutic effect of ASA may be due, in part, to the inhibition of the proteasome and subsequent degradation of IkB46.
Platelet Activating Factor (PAF) is a potent mediator of inflammation thought to be important in atherogenesis 47. However, its receptor-stimulated activity is characterized by rapid desensitization due to receptor down-regulation. This down-regulation is due to lysosomal- and ubiquitin-dependent proteasomal-mediated receptor degradation 48, suggesting that inhibition of the UPS may exacerbate the effects of PAF on vascular cells and be pro-atherogenic. Finally, C-reactive protein (CRP), an acute phase protein increased in inflammation, has been postulated to contribute to atherogenesis 49. A recent study in a strain of atherosclerosis-prone mice that are low density lipoprotein (LDL) receptor-deficient and express ApoB100 demonstrated that transgenic expression of human CRP reduces lesion size and is associated with increased aortic plaque expression of several subunits of the 26S proteasome. These include genes for the 20S subunit (PSMA7, PSMB7, PSMB9), the 19S cap unit (PSMC6), and the 11S cap unit (PSME2) 50. These studies provide evidence that the UPS may modulate the activity and/or function of many of the inflammatory mediators and cell types that participate in mechanisms of atherogenesis. Given the large number of possible targets and variation of effects the UPS system has on substrates, predicting the overall role the UPS has on atherosclerosis will be difficult. This may explain why therapeutic proteasome inhibition has made atherosclerosis both better and worse, depending on the model42–44, 51–54.
UPS regulation of vascular cell apoptosis
Certain aspects of lipoprotein metabolism are important in atherosclerosis and may be modulated by the UPS. It is well accepted that modified forms of low density lipoprotein (LDL) are risk factors for atherosclerosis. Studies of the effects of aggregated LDL (agLDL) on macrophages have shown that challenge with agLDL triggers polyubiquitination of intracellular proteins and ubiquitin-dependent degradation of the apoptosis inducer p53. There is a concomitant increase in expression of low density lipoprotein-inducible gene (LIG), a human homologue of the bovine ubiquitin-conjugating (E2) enzyme E2-25K. Inhibiting the proteasome blocks this anti-apoptotic effect of agLDL, and increases the half-life of p53. Thus, agLDL may protect macrophages from apoptosis in a LIG- and proteasome-dependent manner 55. Other studies have found a role for the UPS in oxidized LDL (oxLDL)-induced apoptosis of vascular cells. OxLDL-induced ubiquitination of cellular proteins induces early activation but late depression of proteolysis. Proteasome inhibition exacerbates the toxicity of oxLDL, demonstrating that the UPS may be involved in oxLDL-induced apoptosis 56. OxLDL also downregulates insulin-like growth factor-1 receptor, leading to smooth muscle cell apoptosis 57. Higashi et al. 58 has demonstrated that this downregulation is dependent on enhanced Nedd4-dependent receptor ubiquitination, but that degradation of the receptor is independent of the proteasome pathway. In contrast to earlier studies, these later reports suggest that ubiquitination and proteasomal activity may enhance apoptosis of certain vascular cell types. Thus, the role of the UPS in cellular apoptosis and its contribution to atherogenesis may vary depending on the mechanism of apoptosis and target cell type.
UPS modulation of cholesterol metabolism
Marfella et al. recently summarized the available evidence demonstrating that the UPS may regulate key processes in the pathogenesis of atherosclerosis 59. They identified that the UPS regulates: 1) insulin resistance in vascular tissues, 2) hyperglycemia-induced endothelial activation and dysfunction, and 3) plaque destabilization. Cholesterol metabolism and transport is central to the development of atherosclerosis, and there is evidence that the UPS may play a role in these processes. HMGCoA is a key regulatory enzyme of cholesterol synthesis in the liver, and pharmacologic inhibition of HMGCoA has been very successful in lowering blood cholesterol concentrations. Studies show that sterol-stimulated turnover of HMGCoA is mediated by its ubiquitination as well as activity of the 26S proteasome 60, implicating the UPS in (dys)regulation of cholesterol production for peripheral tissues. The LDL receptor itself has recently been shown to be regulated by ubiquitin-dependent degradation by the ubiquitin ligase Idol (inducible degrader of the LDLR), which is transcriptionally regulated by nuclear receptors to maintain cholesterol homeostasis 61. Overexpressing Idol promotes LDLR degradation and elevates plasma LDL levels demonstrating its potential anti-atherogenic role 61. Reverse cholesterol transport from peripheral tissues back to the liver is mediated by high density lipoprotein (HDL), and loading of HDL with cholesterol is mediated by ATP-binding cassette protein A1 (ABCA1). Work by Azuma, et al. 62 suggests that ABCA1 degradation is mediated by the COP9 signalosome and is a key controller of ubiquitination. Therefore, the UPS may modulate atherogenesis, in part, through these several mechanisms involving the regulation of cholesterol metabolism.
UPS and cardiovascular biology
The UPS regulates a wide array of biological processes in the cardiovascular system (see recent reviews63–65). For example, the UPS plays a role in the regulation of voltage-gated channels, including the hERG1 channel linked to familial long QT syndromes. Many of the 200+ mutations in this gene lead to misfolded proteins that are rapidly degraded by the endoplasmic reticulum-associated degradation (ERAD) pathway (discussed next) 66, 67. The UPS also regulates a number of signal transduction pathways and transcription factors. The UPS has a significant role in attenuating MAPK activation 68, signaling through NFAT via calcineurin 69, 70, and in regulating NF-κB signaling71. The UPS also regulates apoptosis directly through the degradation of caspases (by cIAP, XIAP) and p53 (by MDM2, COP1, Pirh2, ARF-BP1, CHIP) 63. Additional ubiquitin ligases specifically regulate the pathophysiology of cardiac diseases, including MuRF1, CHIP, MAFBx/Atrogin-1, and MDM2. The number and diversity of processes the UPS regulates in the cardiovascular system continues to increase, such that the role of UPS is now recognized as intimately associated with ER stress, ERAD, and autophagy in the cardiovascular system.
ERAD, ER Stress, and the unfolded protein response in the cardiovascular system
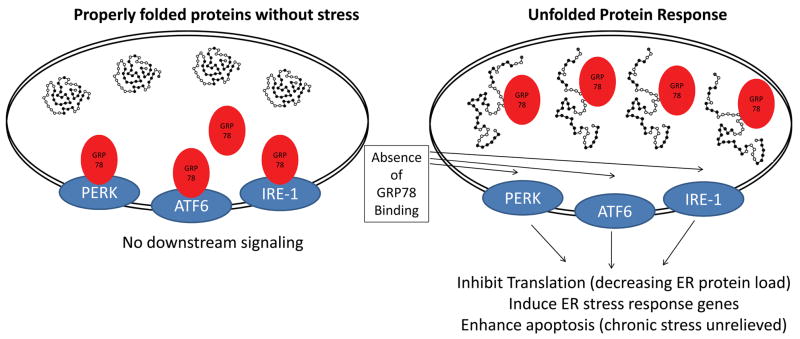
The unfolded protein response (UPR) is a signal transduction system activated in response to stresses that affect the ability of the ER to properly fold proteins. This system is activated in response to increased protein misfolding (Figure 2). Several factors are necessary to maintain efficient protein folding in the ER including the maintenance of the redox state, maintenance of the glycosylation machinery, as well as chaperones to optimally fold newly formed proteins in the ER. The ER senses stress by at least 3 transmembrane proteins: 1) the protein kinase R-like ER kinase (PERK); 2) activating transcription factor-6 (ATF6); and 3) the inositol-requiring enzyme-1 (IRE-1)72–78. With normal functioning, the ER chaperone GRP78 (glucose-regulated protein 78) bind to the internal ER surfaces of PERK, ATF6, and IRE-1 effectively blocking downstream signaling through these receptors (Figure 2). When normal protein folding is disrupted, GRP78 accumulates with the misfolded proteins in the ER in an apparent attempt to refold the proteins (Figure 2) 72–76. Without GRP78 to bind the UPR receptors in the ER, the three ER receptors mediate downstream signaling in response to the increase in ER stress. The distal effectors of these three UPR receptors then mediate an arrest in protein biosynthesis (translation), increased expression in ER responsive genes (including chaperones, calcium binding proteins, and disulfide isomerases), and effectors of apoptosis (through CHOP and JNK/Caspase-12) as recently reviewed 79, 80. Through these mechanisms, the UPR can exert both positive and negative influcences on cell survival.
Figure 2. Endoplasmic reticulum stress and the unfolded protein response.
In response to stress, proteins synthesized in the rough endoplasmic reticulum are refolded by resident molecular chaperones GP78 (glucose-regulated protein-78). Upon refolding, GRP78 looses its association with the luminal domains of the PKR-like ER kinase (PERK), activating transcription factor-6 (ATF6), and inositol-required enzyme-1 (IRE-1). This leads to PERK, ATF6, and IRE-1 activation and downstream activation of ER stress response genes which help stabilize the misfolding of proteins, including ER-targeted chaperones. If the unfolded proteins are not adequately removed after the activation of the UPR, signaling pathways for apoptosis can be activated. Adapted from: Glembotski et al., 200780 and Glembotski et al,. 200879.
ER stress signaling pathways in the heart: UPR-mediated protection
The induction of the UPR (or components thereof) protect against ischemic challenge in cardiomyocytes. Hearts from transgenic mice with cardiac-specific ATF6 overexpression are protected against ischemia/reperfusion injury, suggesting that ATF6-mediated signaling upregulates proteins that protect against cell death 81. In cultured cardiomyocytes, increasing the expression of GRP78 during preconditioning imparts protection 82, 83. Similarly, increasing the sarcoplasmic reticulum chaperone GRP94 in C2C12 myocytes or H9C2 cardiomyocytes protects against Ca2+ overload or ischemia-induced cell death determined by propidium iodide uptake 84. GRP94 overexpression in neonatal cardiomyocytes also results in protection against simulated ischemia. The UPR in cultured cells can be induced experimentally using the antibiotic tunicamycin. Tunicamycin-evoked UPR results in an increase in GRP78 in H9C2 cardiomyocytes, and subsequent protection against simulated ischemia/reperfusion 85.
ER stress signaling pathways in the heart: UPR-mediated apoptosis
The UPR also plays a critical role in mediating apoptosis and cell death in response to cardiac ischemia reperfusion injury. Activation of the UPR can induce apoptosis, while inhibiting it may protect against ischemia reperfusion-induced cell death. For example, tunicamycin challenge in cultured cardiomyocytes induces the rapid translocation of deltaPKC and subsequent cell death, determined by assays for necrosis (LDH release) and apoptosis (caspase activation, TUNEL staining) 86. However, if deltaPKC activation is inhibited, tunicamycin-induced cell death is reduced, as are other specific indicators of UPR, such as GRP78 expression and JNK phosphorylation 86. Similarly, blockade of deltaPKC activation resulting from ischemia reperfusion injury-induced UPR protects against induced cell death 86. Since the protein Puma (p53-upregulated modulator of apoptosis) is required for the induction of cardiomyocyte cell death in ischemia reperfusion injury87, recent studies have investigated its role in the UPR response. Upregulation of Puma and increased apoptosis are seen in rat and mouse neonatal cardiomyocytes that have been treated with thapsigargin or tunicamycin to induce ER stress and the UPR88. Inhibiting Puma significantly protects these cardiomyocytes from ER-stress induced apoptosis, suggesting Puma’s critical role in mediating ER-induced cardiomyocyte death 87.
Temporal regulation of the ER stress response
The ER stress response may be temporally controlled according to recent studies. When neonatal cardiomyocytes undergo apoptosis in response to simulated ischemia or serum/glucose/oxygen deprivation, activation of the UPR precedes caspase activation. The initial response of cardiomyocytes to these stresses is the activation of the UPR, as assessed by increases in GRP78, XBP1, and eIF2alpha phosphorylation 89. At later time points, the UPR activation transitions to the activation of CHOP and pro-caspase 12 processing 89. These studies suggest that UPR effectors initially try to maintain protein quality by increasing the folding capacity (e.g. increasing the GRP78 chaperone), and later induce apoptosis if they are unable to overcome the stress. The downstream signaling processes in the cardiomyocyte UPR are obviously complex given the number of receptors and effectors involved (Figure 2). The cardiac UPR is also activated by the induction of diabetes by streptozocin 90 and the use of proteasome inhibitors 91. With the growing prevalence of diabetes and the increased use of proteasome inhibitors, understanding the ER stress response in the heart may be increasingly relevant to cardiac health in diseases not primarily of cardiac origin.
The role of ubiquitin ligase N-recognins in cardiac development
Recent studies have identified ubiquitin ligases that recognize structural motifs or degradation signals (“degrons”) within target proteins present within the substrate structure. A degron is defined as the minimal part of a protein sufficient for recognition and degradation. There are three components of the N-degron signal in eukaryotic proteins: 1) a destabilizing N-terminal residue; 2) its internal lysine residue(s) where the polyubiquitin chain forms; and 3) the conformational flexibility of areas around these determinants 92–95. Recent studies have identified that a family of ubiquitin ligases, called N-recognins, mediate the N-end rule pathway 92, 96, 97. Mammalian N-recognins have been identified that recognize N-degrons: UBR1 and UBR2 (ubiquitin protein ligase E3 component n-recognin 1 and 2) 98, 99. These components of the N-end rule pathway are essential for proper cardiac development as evideicned by the wide range of cardiovascular abnormalities seen in UBR1 and UBR2 deficient mice.
The ubiquitin ligases UBR1 and UBR2 are necessary for cardiac development
UBR1 and UBR2 have indistinguishable patterns of binding to N-degrons 98; however, their in vivo roles do not clearly overlap 100. Mice lacking UBR1 (UBR1 −/−) are viable and fertile, exhibiting only a mild hypoglycemia, disturbed fatty acid synthase activity and exocrine pancreatic insufficiency 101, 102. Mice lacking UBR2 (UBR2 −/−) exhibit gender specific defects: males are viable but infertile while females die as embryos 98. The differences in the apparent role of UBR1 and UBR2 in development may be due to differential expression patterns in cell types and tissues, although this has yet to be specifically tested. A better understanding of the role of UBR1 and UBR2 in development has been achieved by the creation of UBR1 −/−//UBR2 −/−double null mice 100. These mice die at midgestation with defects in cardiovascular development. Unlike single UBR1 −/−or UBR2 −/− mice, double null mice had severe cardiovascular defects characterized by local hemorrhages and a swollen pericardial sac 100. Development of the atria and ventricles is arrested by E10.5 and disorganization of the myocardial wall and ventricular atrophy is observed 100. Subsequent studies using synthesized small-molecules that competitively inhibit the recognition of N-recognins have been performed to determine the specific effects on the heart 103. In both mouse and rat cardiomyocytes, these studies demonstrate that the N-end rule pathway functions to regulate cardiac proliferation and hypertrophy, which further supports a role for degrons in cardiac development and the degradation of important cardiac regulators 103.
The role of ubiquitin ligases in cardiac disease
Approximately 500 ubiquitin ligases are estimated to exist in the human genome. At least 9 have been described in the heart, with 6 of these (MuRF1, MuRF2, MuRF3, MAFBx/Atrogin-1, CHIP, MDM2) being mechanistically characterized in cardiac hypertrophy and ischemia reperfusion injury. In particular, exciting new studies have been published recently implicating cardiac ubiquitin ligases in cardiac atrophy104, cardiac metabolism 105, and cardiac ischemia reperfusion injury70. Several recent reviews cover the role of these cardiac ubiquitin ligases in detail, including the 3 additional ubiquitin ligases (Cbl, E6AP, cIAP) which have distinct functions described in other model systems 63, 64, 106–108 and so this topic will not be explored further in this review.
The role of ubiquitin in selective autophagy
Ubiquitination, the proteasome, and autophagy
Misfolding is a constant threat to proteins and results from the presence of oxidative stress, mutations, and external stresses such as heat shock. To protect against these stresses, the cell has constitutive and inducible molecular chaperones of the heat shock family to assist in refolding. Misfolded proteins have exposed hydrophobic residues that, when properly folded, are not present. Heat shock proteins bind these hydrophobic residues in misfolded proteins and help to refold them (Figure 3A). A cell’s first option when encountering misfolded proteins is to refold them. However, if refolding isn’t possible, the protein is slated for degradation by the proteasome. Evidence demonstrates that ubiquitin ligases interact with heat shock proteins, illustrating the close relationship within the cell of the processes involved in monitoring protein quality. These heat shock protein-E3 complexes promote folding and, when this is not possible, enhance ubiquitination of recognized substrates, targeting misfolded proteins for proteasome degradation 109.
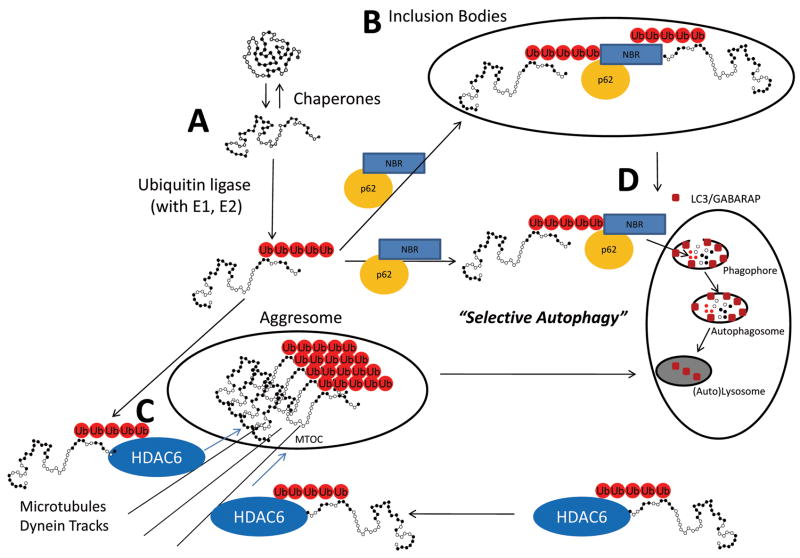
Figure 3. Selective Autophagy through recognition of misfolded and ubiquitinated proteins through NRB1 and p62.
(A) Stress-induced misfolding of proteins is a constant threat to the well-being of the cell. Chaperones continually refold proteins by recognizing the hydrophobic regions of the protein exposed during stress. Heat shock protein-ubiquitin ligase complexes promote folding, but if this is not possible they enhance the ubiquitination of recognized substances effectively targeting proteins for either proteasome degradation (see Figure 1) or by selective autophagy. (B) Misfolded ubiquitinated proteins can polymerize to form inclusion bodies and (C) aggresomes which form from the transport of aggregated ubiquitinated proteins that are transported via dynein on microtubule tracks. (D) “Selective autophagy” is the catabolism of macromolecule and organelles based on the recognition of ubiquitination chains on proteins which plays an important role in maintaining protein quality control in the cell. MTOC, Microtubule organizing centers. Adapted from: Kirkin, et al., 2009130 and Larmark et al., 2009 144.
Misfolded proteins unable to be resolved by chaperone-mediated refolding or proteasome degradation, form aggregates. Aggregates of misfolded proteins may then polymerize to form structures microscopically recognized as inclusion bodies (Figure 3B) and aggresomes (Figure 3C) 110, 111. From these structures, bulky, misfolded proteins can be degraded via pathways that are independent from the proteasome degradation pathway. By shunting protein degradation to these alternative pathways, the accumulation of ubiquitinated misfolded proteins can be prevented. The accumulation of protein aggregates is a proximal trigger of cardiomyocyte autophagy which is the mechanism by which aggresomes can be cleared 112.
Autophagy occurs continuously at low levels in the normal heart. It is regulated by autophagy (Atg) proteins that make up 2 conjugation pathways that parallel the ubiquitin ligation pathway described in Figure 1: 1) the Atg12-Atg5 pathway; and 2) the LC3(Atg8)-PE (light chain 3-phosphatidylethanolamine) pathways (Recently reviewed by Gustafsson et al., 2009113). In this system, Atg12/Atg5 or Atg8 (LC3) are conjugated to Atg7, Atg10, Atg5, or Atg3 via lysine residues, forming complexes essential for the recruitment of LC3 and the formation of the membranes needed to form the autophagosomes (Figure 3D) 114, 115. During cardiac ischemia or cardiac loading, autophagy increases as a means to adapt to the significant amount of remodeling that accompanies these processes 113, 116.
Selective autophagy occurs through receptors that recognize ubiquitinated proteins
While the 26S proteasome degrades most intracellular proteins, it is limited in its capacity to degrade misfolded proteins which become aggregated. Since the proteasome is unable to degrade these proteins, parallel systems have evolved to help remove proteins that cannot be degraded by the proteasome. This is done by intracellular receptors which recognize ubiquitinated protein aggregates, and target these proteins for destruction by the autophagosome 117, 118. Autophagy is a general term to describe several processes in which lysosomes engulf cytosolic proteins for degradation. Recently, a group of loosely associated receptors have been described that recognize ubiquitin chains covalently attached to proteins and are capable of delivering ubiquitinated proteins to autophagosomes. Two of these receptors, p62 and NBR1, target ubiquitinated proteins not cleared by the proteasome for autophagic clearance. This selective uptake of cellular organelles has previously been described for mitochondria, aggregations of protein, and bacteria 119. Identifying the receptors that recognize ubiquitin led to advancements in understanding the underlying mechanisms of the apparently “specific” (selective) autophagy. There is increasing appreciation that ubiquitin plays a role in autophagy, and that the uptake of damaged proteins and organelles occurs in a much more specific way than previously realized.
Selective autophagy: Targeting ubiquitinated proteins for autophagy through p62, NBR1, HDAC6, and BAG1/BAG3
The multi-functional ubiquitin receptor p62 has recently been identified as part of the autophagic apparatus 120. p62 contains a zinc-finger domain and ubiquitin-binding UBA domain in its C-terminal region. The UBA domain is able to bind Lys48-linked and Lys63-linked ubiquitin chains, with higher affinity for Lys63 (Figure 3) 121–123. The p62-associated clearance of aggregated proteins by autophagy was first suggested by the discovery of colocalization of ubiquitin-positive inclusion bodies with p62 124, 125. Further evidence for the relationship between p62 and LC3 comes from mouse studies 126, 127. Mice with deficiencies in autophagy (Atg7 −/− mice) display accumulation of p62 in ubiquitin positive inclusion bodies 127. Mice lacking both Atg7 and p62 (Atg7 −/−//p62−/− mice) have reduced numbers of protein aggregates 127. The role of p62 in the formation of autophagosomes is further supported by similar findings in Drosophila 128. Recent studies have also found that p62 regulates the clearance of proteins cleared by the ubiquitin proteasome system. In models where autophagy is inhibited, p62 accumulation is seen as expected 129. Unexpectedly, however, there is also decreased clearance of proteins normally removed by the ubiquitin proteasome system, such as p53, in addition to the accumulation of aggregation prone proteins129.
NBR1 binds ubiquitin by its UBA domain, favoring Lys63-linked poly-ubiquitinated chains 130. The recruitment of ubiquitin-linked cargo to lysosomes is dependent on both p62 and NBR1. The cross-linking of ubiquitinated misfolded proteins is mediated by NBR1, and like p62, is necessary for protein aggregation and inclusion body formation following autophagy inhibition 130. NBR1 associates with itself through its coiled-coiled domain to clear ubiquitinated misfolded proteins, or it can interact with oligomeric p62 and ubiquitinated mis-folded proteins 130. In muscle cells, NBR1 interacts directly with p62 and has been implicated as a part of a signaling complex of the giant protein titin kinase where mechanical stretch-inducing titin kinase activity is associated with the regulation of the ubiquitin ligase MuRF2 131. An additional role for NBR1 in autophagic degradation of ubiquitinated targets is illustrated by NBR1’s ability to bind directly to the autophagosome-specific ATG8/LC3/GABARP in the presence or absence of p62 (Figure 3D) 130.
Histone deacetylase 6 (HDAC6) is another adaptor protein that recognizes ubiquitinated and misfolded proteins, shuttling them into aggresomes where they are sequestered within the cell 132–134. The aggresomes are then targeted for degradation by the autophagic pathway after HDAC delivers them via the microtubule organizing centers (MTOC) (Figure 3C). When aggresomes are experimentally induced with ubiquitinated proteins, they contain HDAC6. Inhibition of HDAC6 using siRNA reportedly compromises aggresome formation, a process that can be rescued with HDAC constructs containing the ubiquitin binding region 135. Like NBR1, HDAC6 binds Lys63-linked ubiquitin chains 133. HDAC6 interacts with dynein motors necessary for the transport of the aggresome via the microtubules (Figure 3C) 135.
Other proteins that do not have ubiquitin binding domains can be associated with ubiquitinated proteins and autophagosomal markers, including BAG1 and BAG3. The Hsc/Hsp70 co-chaperone BAG3 has been proposed to mediate autophagic degradation of ubiquitinated proteins in aging cells 135–137. BAG3 co-localizes with p62-postive aggregated proteins, but is not itself degraded by autophagy 136. In contrast to BAG3 (found primarily in older cells), BAG1 mediates the proteasome-dependent degradation of ubiquitinated proteins in coordination with the CHIP ubiquitin ligase 136, 138. These findings suggest that the ratio of BAG1/BAG3 changes the mechanism by which aging cells regulate how ubiquitinated proteins are disposed. The activation of the senescence program, including the increase in the BAG3/BAG1 ratio, enhances the shuttling of ubiquitinated proteins to autophagy pathways with advancing age, whereas younger cells with the reverse ratio tend to shuttle ubiquitinated proteins for proteasomal degradation 136.
Ubiquitination may play a role in selectively degrading whole organelles, such as the mitochondria139, peroxisomes140, ribosomes141, and bacteria 140. This is most readily seen in the starvation response. The role of ubiquitin in clearing specific organelles is not fully understood. Mitochondrial degradation has been reported to be ubiquitin dependent 142, while autophagy of the peroxisomes is partially dependent on p62 and associated with mono-ubiquitinated proteins 143. There is much work to be done to understand how complex organelles are targeted for selective autophagy through ubiquitination and recognition by adaptor proteins such as p62 and NBR1.
Conclusion
A picture of the UPS as a simple isolated system no longer adequately describes the myriad of functions it has in the cardiovascular system. The UPS plays a fundamental role in the development of the vascular system through its regulation of key signaling pathways including Notch, VEGF, and HIF1. In the mature vasculature, the UPS regulates inflammation, oxidative stress, and apoptosis in addition to cholesterol metabolism in ways that may affect the development and severity of atherosclerosis. In the stressed heart, the UPS maintains protein quality control of nascent proteins in the endoplasmic reticulum by the unfolded protein response (including signaling through PERK, ATF6, IRE-1) and ER-associated degradation of misfolded proteins. Despite increasing appreciation of the UPS in regulating a myriad of biological processes, some of the newest and most fascinating findings implicate autophagy (“selective autophagy”) as an alternative to the proteasome to get rid of unfolded, damaged, and aggregated proteins. The cytosolic receptors p62, NBR, and HDAC6 play a fundamental role in targeting damaged proteins to this extra-proteasomal “selective autophagy” form of destruction. This emphasizes the fundamental role of the UPS in maintaining protein quality by the selective destruction of worn and damaged proteins. While our appreciation of the UPS in maintaining protein quality control continues to grow, so does our realization of the ubiquitous nature of its control over cell signaling pathways by harnessing the same destructive power.
Acknowledgments
Sources of Funding
This work was supported by the American Heart Association Scientist Development Grant (to M.W.), the pre-doctoral Training Program in Integrative Vascular Biology (T32 HL069768 to W.H.D.T.T.), National Institutes of Health R01 HL090823 (to J.H.), and the National Institutes of Health R01HL065619 (to C.P.), R01HL061656 (to C.P), and R01GM061728 (C.P.).
Non-standard Abbreviations and Acronyms
- agLDL
aggregated low density lipoprotein
- ASB4
ankyrin repeat and SOCS box-containing 4
- ASA
aspirin
- ATF6
activating transcription factor 6
- CHIP
c-terminus of Hsp70-interacting protein (CHIP)
- CHOP
cytosine-cytosine-adenine-adenine-thymine enhancer-binding protein (C/EBP) homologous protein
- COP1
constitutive photomorphic 1 protein
- CRP
c-reactive protein
- C-TAD
C-terminal transactivation domain
- Cul7
cullin 7 ubiquitin ligase
- CYLD
cylindromatosis
- DUBS
de-ubiquitinating enzymes
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated death
- FIH
factor inhibiting HIF
- GRP78
glucose-regulated protein 78
- HDL
high density lipoprotein
- HIF1
hypoxia inducible factor 1
- HMGCoA
3-hydroxy-3-methylglutaryl-Coenzyme A synthase
- I/R
ischemia reperfusion
- JNK
c-Jun NH2-terminal kinase
- c-Jun
jun oncogene
- LDL
low density lipoprotein
- LNX
ligand of numb-protein X 1
- LPS
lipopolysaccharide
- MAFBx
muscle atrophy F-box
- MDM2
murine double minute 2
- MIB1, 2
mind bomb1/2
- MTOC
Microtubule organizing centers
- MuRF1, 2
Muscle ring finger-1, -2
- NF-KB
nuclear factor kappa light chain enhancer of activated B cells
- oxLDL
oxidized low density lipoprotein
- PAF
platelet activating factor
- PERK
protein kinase R-like ER kinase
- PHD
prolyl hydroxylase
- Pirh2
p53-indiced protein with a RING-H2 domain
- PKC
protein kinase C
- SCF
Skp1Cul1-F-box-protein
- SRF
serum response factor
- TGF-β
transforming growth factor beta
- TNF-α
tumor necrosis factors-alpha
- TRAF2
TNF receptor-associated factor 2
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labeling
- UPR
unfolded protein response
- UBR1,2
ubiquitin protein ligase E3 component n-recognin 1,2
- UPS
ubiquitin proteasome system
- VEGF
vascular endothelial growth factor
- pVHL
von-Hippel Lindau tumor suppressor
Footnotes
Disclosures
None (M.W., D.T., E.K., J.H., C.P.)
References
- 1.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8(6):610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, Patterson C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest. 2007;117(11):3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15(3):1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haglund K, Shimokawa N, Szymkiewicz I, Dikic I. Cbl-directed monoubiquitination of CIN85 is involved in regulation of ligand-induced degradation of EGF receptors. Proc Natl Acad Sci U S A. 2002;99(19):12191–12196. doi: 10.1073/pnas.192462299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leithe E, Rivedal E. Ubiquitination and down-regulation of gap junction protein connexin-43 in response to 12-O-tetradecanoylphorbol 13-acetate treatment. J Biol Chem. 2004;279(48):50089–50096. doi: 10.1074/jbc.M402006200. [DOI] [PubMed] [Google Scholar]
- 6.Leithe E, Rivedal E. Epidermal growth factor regulates ubiquitination, internalization and proteasome-dependent degradation of connexin43. J Cell Sci. 2004;117(Pt 7):1211–1220. doi: 10.1242/jcs.00951. [DOI] [PubMed] [Google Scholar]
- 7.Huang H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J Biol Chem. 2000;275(35):26661–26664. doi: 10.1074/jbc.C000199200. [DOI] [PubMed] [Google Scholar]
- 8.Jennissen HP, Laub M. Ubiquitin-calmodulin conjugating activity from cardiac muscle. Biol Chem Hoppe Seyler. 1988;369(12):1325–1330. doi: 10.1515/bchm3.1988.369.2.1325. [DOI] [PubMed] [Google Scholar]
- 9.Torres MP, Lee MJ, Ding F, Purbeck C, Kuhlman B, Dokholyan NV, Dohlman HG. G Protein Mono-ubiquitination by the Rsp5 Ubiquitin Ligase. J Biol Chem. 2009;284(13):8940–8950. doi: 10.1074/jbc.M809058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moser M, Yu Q, Bode C, Xiong JW, Patterson C. BMPER is a conserved regulator of hematopoietic and vascular development in zebrafish. J Mol Cell Cardiol. 2007;43(3):243–253. doi: 10.1016/j.yjmcc.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23(4):543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- 13.Nie J, McGill MA, Dermer M, Dho SE, Wolting CD, McGlade CJ. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. Embo J. 2002;21(1–2):93–102. doi: 10.1093/emboj/21.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chastagner P, Israel A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 2008;3(7):e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279(10):9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- 16.Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, Lee J, Chitnis AB, Kim CH, Kong YY. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132(15):3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- 17.Koo BK, Yoon KJ, Yoo KW, Lim HS, Song R, So JH, Kim CH, Kong YY. Mind bomb-2 is an E3 ligase for Notch ligand. J Biol Chem. 2005;280(23):22335–22342. doi: 10.1074/jbc.M501631200. [DOI] [PubMed] [Google Scholar]
- 18.Cross MJ, Dixelius J, Matsumoto T, Claesson-Welsh L. VEGF-receptor signal transduction. Trends Biochem Sci. 2003;28(9):488–494. doi: 10.1016/S0968-0004(03)00193-2. [DOI] [PubMed] [Google Scholar]
- 19.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O’Shea KS, Powell-Braxton L, Hillan KJ, Moore MW. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380(6573):439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 20.Miquerol L, Langille BL, Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127(18):3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- 21.Murdaca J, Treins C, Monthouel-Kartmann MN, Pontier-Bres R, Kumar S, Van Obberghen E, Giorgetti-Peraldi S. Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J Biol Chem. 2004;279(25):26754–26761. doi: 10.1074/jbc.M311802200. [DOI] [PubMed] [Google Scholar]
- 22.Xie P, Fan Y, Zhang H, Zhang Y, She M, Gu D, Patterson C, Li H. CHIP represses myocardin-induced smooth muscle cell differentiation via ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2009;29(9):2398–2408. doi: 10.1128/MCB.01737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Xie P, Fan Y, Zhang H, Zheng L, Gu D, Patterson C, Li H. C Terminus of Hsc70-interacting Protein Promotes Smooth Muscle Cell Proliferation and Survival through Ubiquitin-mediated Degradation of FoxO1. J Biol Chem. 2009;284(30):20090–20098. doi: 10.1074/jbc.M109.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang X, Austin PF, Niederhoff RA, Manson SR, Riehm JJ, Cook BL, Pengue G, Chitaley K, Nakayama K, Nakayama KI, Weintraub SJ. The mechanoregulation of proliferation. Mol Cell Biol. 2009 doi: 10.1128/MCB.00465-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boesten LS, Zadelaar SM, De Clercq S, Francoz S, van Nieuwkoop A, Biessen EA, Hofmann F, Feil S, Feil R, Jochemsen AG, Zurcher C, Havekes LM, van Vlijmen BJ, Marine JC. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ. 2006;13(12):2089–2098. doi: 10.1038/sj.cdd.4401973. [DOI] [PubMed] [Google Scholar]
- 26.Maxwell PH, Ratcliffe PJ. Oxygen sensors and angiogenesis. Semin Cell Dev Biol. 2002;13(1):29–37. doi: 10.1006/scdb.2001.0287. [DOI] [PubMed] [Google Scholar]
- 27.Semenza GL. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev. 1998;8(5):588–594. doi: 10.1016/s0959-437x(98)80016-6. [DOI] [PubMed] [Google Scholar]
- 28.Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105(2):659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 29.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dayan F, Monticelli M, Pouyssegur J, Pecou E. Gene regulation in response to graded hypoxia: the non-redundant roles of the oxygen sensors PHD and FIH in the HIF pathway. J Theor Biol. 2009;259(2):304–316. doi: 10.1016/j.jtbi.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, Bowtell DD, Ronai Z. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117(7):941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6(4):373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson JE, 3rd, Wu Y, Smith K, Charles P, Powers K, Wang H, Patterson C. ASB4 is a hydroxylation substrate of FIH and promotes vascular differentiation via an oxygen-dependent mechanism. Mol Cell Biol. 2007;27(18):6407–6419. doi: 10.1128/MCB.00511-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tetzlaff MT, Yu W, Li M, Zhang P, Finegold M, Mahon K, Harper JW, Schwartz RJ, Elledge SJ. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc Natl Acad Sci U S A. 2004;101(10):3338–3345. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 36.Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alboran IM, Nakayama K, Nakayama KI. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204(12):2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arai T, Kasper JS, Skaar JR, Ali SH, Takahashi C, DeCaprio JA. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc Natl Acad Sci U S A. 2003;100(17):9855–9860. doi: 10.1073/pnas.1733908100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brouillard P, Boon LM, Mulliken JB, Enjolras O, Ghassibe M, Warman ML, Tan OT, Olsen BR, Vikkula M. Mutations in a novel factor, glomulin, are responsible for glomuvenous malformations (“glomangiomas”) Am J Hum Genet. 2002;70(4):866–874. doi: 10.1086/339492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meier P, Golshayan D, Blanc E, Pascual M, Burnier M. Oxidized LDL modulates apoptosis of regulatory T cells in patients with ESRD. J Am Soc Nephrol. 2009;20(6):1368–1384. doi: 10.1681/ASN.2008070734. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Qureshi N, Vogel SN, Van Way C, 3rd, Papasian CJ, Qureshi AA, Morrison DC. The proteasome: a central regulator of inflammation and macrophage function. Immunologic research. 2005;31(3):243–260. doi: 10.1385/IR:31:3:243. [DOI] [PubMed] [Google Scholar]
- 41.Stangl V, Lorenz M, Meiners S, Ludwig A, Bartsch C, Moobed M, Vietzke A, Kinkel HT, Baumann G, Stangl K. Long-term up-regulation of eNOS and improvement of endothelial function by inhibition of the ubiquitin-proteasome pathway. FASEB J. 2004;18(2):272–279. doi: 10.1096/fj.03-0054com. [DOI] [PubMed] [Google Scholar]
- 42.Marfella R, D’Amico M, Di Filippo C, Baldi A, Siniscalchi M, Sasso FC, Portoghese M, Carbonara O, Crescenzi B, Sangiuolo P, Nicoletti GF, Rossiello R, Ferraraccio F, Cacciapuoti F, Verza M, Coppola L, Rossi F, Paolisso G. Increased activity of the ubiquitin-proteasome system in patients with symptomatic carotid disease is associated with enhanced inflammation and may destabilize the atherosclerotic plaque: effects of rosiglitazone treatment. J Am Coll Cardiol. 2006;47(12):2444–2455. doi: 10.1016/j.jacc.2006.01.073. [DOI] [PubMed] [Google Scholar]
- 43.Marfella R, Siniscalchi M, Portoghese M, Di Filippo C, Ferraraccio F, Schiattarella C, Crescenzi B, Sangiuolo P, Ferraro G, Siciliano S, Cinone F, Mazzarella G, Martis S, Verza M, Coppola L, Rossi F, D’Amico M, Paolisso G. Morning blood pressure surge as a destabilizing factor of atherosclerotic plaque: role of ubiquitin-proteasome activity. Hypertension. 2007;49(4):784–791. doi: 10.1161/01.HYP.0000259739.64834.d4. [DOI] [PubMed] [Google Scholar]
- 44.Marfella R, Di Filippo C, Portoghese M, Ferraraccio F, Crescenzi B, Siniscalchi M, Barbieri M, Bologna C, Rizzo MR, Rossi F, D’Amico M, Paolisso G. Proteasome activity as a target of hormone replacement therapy-dependent plaque stabilization in postmenopausal women. Hypertension. 2008;51(4):1135–1141. doi: 10.1161/HYPERTENSIONAHA.107.105239. [DOI] [PubMed] [Google Scholar]
- 45.Takami Y, Nakagami H, Morishita R, Katsuya T, Hayashi H, Mori M, Koriyama H, Baba Y, Yasuda O, Rakugi H, Ogihara T, Kaneda Y. Potential role of CYLD (Cylindromatosis) as a deubiquitinating enzyme in vascular cells. Am J Pathol. 2008;172(3):818–829. doi: 10.2353/ajpath.2008.070312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan C, Li Y, Tan X, Pan H, Huang W. Inhibition of the ubiquitin-proteasome system: a new avenue for atherosclerosis. Clin Chem Lab Med. 2006;44(10):1218–1225. doi: 10.1515/CCLM.2006.209. [DOI] [PubMed] [Google Scholar]
- 47.Ghesquiere SA, Hofker MH, de Winther MP. The role of phospholipases in lipid modification and atherosclerosis. Cardiovasc Toxicol. 2005;5(2):161–182. doi: 10.1385/ct:5:2:161. [DOI] [PubMed] [Google Scholar]
- 48.Dupre DJ, Chen Z, Le Gouill C, Theriault C, Parent JL, Rola-Pleszczynski M, Stankova J. Trafficking, ubiquitination, and down-regulation of the human platelet-activating factor receptor. J Biol Chem. 2003;278(48):48228–48235. doi: 10.1074/jbc.M304082200. [DOI] [PubMed] [Google Scholar]
- 49.Wilson AM, Ryan MC, Boyle AJ. The novel role of C-reactive protein in cardiovascular disease: risk marker or pathogen. International journal of cardiology. 2006;106(3):291–297. doi: 10.1016/j.ijcard.2005.01.068. [DOI] [PubMed] [Google Scholar]
- 50.Kovacs A, Tornvall P, Nilsson R, Tegner J, Hamsten A, Bjorkegren J. Human C-reactive protein slows atherosclerosis development in a mouse model with human-like hypercholesterolemia. Proc Natl Acad Sci U S A. 2007;104(34):13768–13773. doi: 10.1073/pnas.0706027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marfella R, Di Filippo C, Laieta MT, Vestini R, Barbieri M, Sangiulo P, Crescenzi B, Ferraraccio F, Rossi F, D’Amico M, Paolisso G. Effects of ubiquitin-proteasome system deregulation on the vascular senescence and atherosclerosis process in elderly patients. J Gerontol A Biol Sci Med Sci. 2008;63(2):200–203. doi: 10.1093/gerona/63.2.200. [DOI] [PubMed] [Google Scholar]
- 52.Herrmann J, Edwards WD, Holmes DR, Jr, Shogren KL, Lerman LO, Ciechanover A, Lerman A. Increased ubiquitin immunoreactivity in unstable atherosclerotic plaques associated with acute coronary syndromes. J Am Coll Cardiol. 2002;40(11):1919–1927. doi: 10.1016/s0735-1097(02)02564-0. [DOI] [PubMed] [Google Scholar]
- 53.Herrmann J, Soares SM, Lerman LO, Lerman A. Potential role of the ubiquitin-proteasome system in atherosclerosis aspects of a protein quality disease. J Am Coll Cardiol. 2008;51(21):2003–2010. doi: 10.1016/j.jacc.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 54.Herrmann J, Saguner AM, Versari D, Peterson TE, Chade A, Olson M, Lerman LO, Lerman A. Chronic proteasome inhibition contributes to coronary atherosclerosis. Circ Res. 2007;101(9):865–874. doi: 10.1161/CIRCRESAHA.107.152959. [DOI] [PubMed] [Google Scholar]
- 55.Kikuchi J, Furukawa Y, Kubo N, Tokura A, Hayashi N, Nakamura M, Matsuda M, Sakurabayashi I. Induction of ubiquitin-conjugating enzyme by aggregated low density lipoprotein in human macrophages and its implications for atherosclerosis. Arterioscler Thromb Vasc Biol. 2000;20(1):128–134. doi: 10.1161/01.atv.20.1.128. [DOI] [PubMed] [Google Scholar]
- 56.Vieira O, Escargueil-Blanc I, Jurgens G, Borner C, Almeida L, Salvayre R, Negre-Salvayre A. Oxidized LDLs alter the activity of the ubiquitin-proteasome pathway: potential role in oxidized LDL-induced apoptosis. FASEB J. 2000;14(3):532–542. doi: 10.1096/fasebj.14.3.532. [DOI] [PubMed] [Google Scholar]
- 57.Scheidegger KJ, James RW, Delafontaine P. Differential effects of low density lipoproteins on insulin-like growth factor-1 (IGF-1) and IGF-1 receptor expression in vascular smooth muscle cells. J Biol Chem. 2000;275(35):26864–26869. doi: 10.1074/jbc.M002887200. [DOI] [PubMed] [Google Scholar]
- 58.Higashi Y, Sukhanov S, Parthasarathy S, Delafontaine P. The ubiquitin ligase Nedd4 mediates oxidized low-density lipoprotein-induced downregulation of insulin-like growth factor-1 receptor. Am J Physiol Heart Circ Physiol. 2008;295(4):H1684–1689. doi: 10.1152/ajpheart.00548.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marfella R, Filippo CD, Portoghese M, Siniscalchi M, Martis S, Ferraraccio F, Guastafierro S, Nicoletti G, Barbieri M, Coppola A, Rossi F, Paolisso G, D’Amico M. The ubiquitin-proteasome system contributes to the inflammatory injury in ischemic diabetic myocardium: the role of glycemic control. Cardiovasc Pathol. 2009 doi: 10.1016/j.carpath.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Ravid T, Doolman R, Avner R, Harats D, Roitelman J. The ubiquitin-proteasome pathway mediates the regulated degradation of mammalian 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2000;275(46):35840–35847. doi: 10.1074/jbc.M004793200. [DOI] [PubMed] [Google Scholar]
- 61.Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325(5936):100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Azuma Y, Takada M, Maeda M, Kioka N, Ueda K. The COP9 signalosome controls ubiquitinylation of ABCA1. Biochem Biophys Res Commun. 2009;382(1):145–148. doi: 10.1016/j.bbrc.2009.02.161. [DOI] [PubMed] [Google Scholar]
- 63.Willis MS, Patterson C. Into the heart: the emerging role of the ubiquitin-proteasome system. J Mol Cell Cardiol. 2006;41(4):567–579. doi: 10.1016/j.yjmcc.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 64.Willis MS, Schisler JC, Patterson C. Appetite for destruction: E3 ubiquitin-ligase protection in cardiac disease. Future Cardiol. 2008;4(1):65–75. doi: 10.2217/14796678.4.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Powell SR. The ubiquitin-proteasome system in cardiac physiology and pathology. Am J Physiol Heart Circ Physiol. 2006;291(1):H1–H19. doi: 10.1152/ajpheart.00062.2006. [DOI] [PubMed] [Google Scholar]
- 66.Ficker E, Dennis AT, Wang L, Brown AM. Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circ Res. 2003;92(12):e87–100. doi: 10.1161/01.RES.0000079028.31393.15. [DOI] [PubMed] [Google Scholar]
- 67.Gong Q, Keeney DR, Molinari M, Zhou Z. Degradation of trafficking-defective long QT syndrome type II mutant channels by the ubiquitin-proteasome pathway. J Biol Chem. 2005;280(19):19419–19425. doi: 10.1074/jbc.M502327200. [DOI] [PubMed] [Google Scholar]
- 68.Laine A, Ronai Z. Ubiquitin chains in the ladder of MAPK signaling. Sci STKE 2005. 2005;281:re5. doi: 10.1126/stke.2812005re5. [DOI] [PubMed] [Google Scholar]
- 69.Kishi T, Ikeda A, Nagao R, Koyama N. The SCFCdc4 ubiquitin ligase regulates calcineurin signaling through degradation of phosphorylated Rcn1, an inhibitor of calcineurin. Proc Natl Acad Sci U S A. 2007;104(44):17418–17423. doi: 10.1073/pnas.0704951104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, Patterson C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114(8):1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 72.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 73.Ma K, Vattem KM, Wek RC. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem. 2002;277(21):18728–18735. doi: 10.1074/jbc.M200903200. [DOI] [PubMed] [Google Scholar]
- 74.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15(12):3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 75.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90(6):1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 76.Shen J, Snapp EL, Lippincott-Schwartz J, Prywes R. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol. 2005;25(3):921–932. doi: 10.1128/MCB.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 78.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 79.Glembotski CC. The role of the unfolded protein response in the heart. J Mol Cell Cardiol. 2008;44(3):453–459. doi: 10.1016/j.yjmcc.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101(10):975–984. doi: 10.1161/CIRCRESAHA.107.161273. [DOI] [PubMed] [Google Scholar]
- 81.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, Glembotski CC. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98(9):1186–1193. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 82.Shintani-Ishida K, Nakajima M, Uemura K, Yoshida K. Ischemic preconditioning protects cardiomyocytes against ischemic injury by inducing GRP78. Biochem Biophys Res Commun. 2006;345(4):1600–1605. doi: 10.1016/j.bbrc.2006.05.077. [DOI] [PubMed] [Google Scholar]
- 83.Pan YX, Ren AJ, Zheng J, Rong WF, Chen H, Yan XH, Wu C, Yuan WJ, Lin L. Delayed cytoprotection induced by hypoxic preconditioning in cultured neonatal rat cardiomyocytes: role of GRP78. Life Sci. 2007;81(13):1042–1049. doi: 10.1016/j.lfs.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 84.Vitadello M, Penzo D, Petronilli V, Michieli G, Gomirato S, Menabo R, Di Lisa F, Gorza L. Overexpression of the stress protein Grp94 reduces cardiomyocyte necrosis due to calcium overload and simulated ischemia. FASEB J. 2003;17(8):923–925. doi: 10.1096/fj.02-0644fje. [DOI] [PubMed] [Google Scholar]
- 85.Zhang PL, Lun M, Teng J, Huang J, Blasick TM, Yin L, Herrera GA, Cheung JY. Preinduced molecular chaperones in the endoplasmic reticulum protect cardiomyocytes from lethal injury. Ann Clin Lab Sci. 2004;34(4):449–457. [PubMed] [Google Scholar]
- 86.Qi X, Vallentin A, Churchill E, Mochly-Rosen D. deltaPKC participates in the endoplasmic reticulum stress-induced response in cultured cardiac myocytes and ischemic heart. J Mol Cell Cardiol. 2007;43(4):420–428. doi: 10.1016/j.yjmcc.2007.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Toth A, Jeffers JR, Nickson P, Min JY, Morgan JP, Zambetti GP, Erhardt P. Targeted deletion of Puma attenuates cardiomyocyte death and improves cardiac function during ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2006;291(1):H52–60. doi: 10.1152/ajpheart.01046.2005. [DOI] [PubMed] [Google Scholar]
- 88.Nickson P, Toth A, Erhardt P. PUMA is critical for neonatal cardiomyocyte apoptosis induced by endoplasmic reticulum stress. Cardiovasc Res. 2007;73(1):48–56. doi: 10.1016/j.cardiores.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szegezdi E, Duffy A, O’Mahoney ME, Logue SE, Mylotte LA, O’Brien T, Samali A. ER stress contributes to ischemia-induced cardiomyocyte apoptosis. Biochem Biophys Res Commun. 2006;349(4):1406–1411. doi: 10.1016/j.bbrc.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 90.Xu J, Wang G, Wang Y, Liu Q, Xu W, Tan Y, Cai L. Diabetes- and angiotensin II-induced cardiac endoplasmic reticulum stress and cell death: Metallothionein protection. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fu HY, Minamino T, Tsukamoto O, Sawada T, Asai M, Kato H, Asano Y, Fujita M, Takashima S, Hori M, Kitakaze M. Overexpression of endoplasmic reticulum-resident chaperone attenuates cardiomyocyte death induced by proteasome inhibition. Cardiovasc Res. 2008;79(4):600–610. doi: 10.1093/cvr/cvn128. [DOI] [PubMed] [Google Scholar]
- 92.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci U S A. 1996;93(22):12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56(6):1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 94.Suzuki T, Varshavsky A. Degradation signals in the lysine-asparagine sequence space. EMBO J. 1999;18(21):6017–6026. doi: 10.1093/emboj/18.21.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11(9):830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 96.Du F, Navarro-Garcia F, Xia Z, Tasaki T, Varshavsky A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc Natl Acad Sci U S A. 2002;99(22):14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25(16):7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kwon YT, Xia Z, An JY, Tasaki T, Davydov IV, Seo JW, Sheng J, Xie Y, Varshavsky A. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23(22):8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Varshavsky A. ‘Spalog’ and ‘sequelog’: neutral terms for spatial and sequence similarity. Curr Biol. 2004;14(5):R181–183. doi: 10.1016/j.cub.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 100.An JY, Seo JW, Tasaki T, Lee MJ, Varshavsky A, Kwon YT. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci U S A. 2006;103(16):6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol Cell Biol. 2001;21(23):8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zenker M, Mayerle J, Lerch MM, Tagariello A, Zerres K, Durie PR, Beier M, Hulskamp G, Guzman C, Rehder H, Beemer FA, Hamel B, Vanlieferinghen P, Gershoni-Baruch R, Vieira MW, Dumic M, Auslender R, Gil-da-Silva-Lopes VL, Steinlicht S, Rauh M, Shalev SA, Thiel C, Ekici AB, Winterpacht A, Kwon YT, Varshavsky A, Reis A. Deficiency of UBR1, a ubiquitin ligase of the N-end rule pathway, causes pancreatic dysfunction, malformations and mental retardation (Johanson-Blizzard syndrome) Nat Genet. 2005;37(12):1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 103.Lee MJ, Pal K, Tasaki T, Roy S, Jiang Y, An JY, Banerjee R, Kwon YT. Synthetic heterovalent inhibitors targeting recognition E3 components of the N-end rule pathway. Proc Natl Acad Sci U S A. 2008;105(1):100–105. doi: 10.1073/pnas.0708465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Willis MS, Rojas M, Li L, Selzman CH, Tang RH, Stansfield WE, Rodriguez JE, Glass DJ, Patterson C. Muscle ring finger 1 mediates cardiac atrophy in vivo. Am J Physiol Heart Circ Physiol. 2009;296(4):H997–H1006. doi: 10.1152/ajpheart.00660.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Willis MS, Schisler JC, Li L, Rodriguez JE, Hilliard EG, Charles PC, Patterson C. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circ Res. 2009;105(1):80–88. doi: 10.1161/CIRCRESAHA.109.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodriguez JE, Schisler JC, Patterson C, Willis MS. Seek and Destroy: The Ubiquitin Proteasome System in Cardiac Disease. Curr Hypertens Rep. 2009 doi: 10.1007/s11906-009-0069-7. In press. [DOI] [PubMed] [Google Scholar]
- 107.Schisler JC, Willis MS, Patterson C. You spin me round: MaFBx/Atrogin-1 feeds forward on FOXO transcription factors (like a record) Cell Cycle. 2008;7(4):440–443. doi: 10.4161/cc.7.4.5451. [DOI] [PubMed] [Google Scholar]
- 108.Willis MS, Schisler JC, Portbury AL, Patterson C. Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovasc Res. 2009;81(3):439–448. doi: 10.1093/cvr/cvn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patterson C, Ike C, Willis PWt, Stouffer GA, Willis MS. The bitter end: the ubiquitin-proteasome system and cardiac dysfunction. Circulation. 2007;115(11):1456–1463. doi: 10.1161/CIRCULATIONAHA.106.649863. [DOI] [PubMed] [Google Scholar]
- 110.Markossian KA, Kurganov BI. Protein folding, misfolding, and aggregation. Formation of inclusion bodies and aggresomes. Biochemistry (Mosc) 2004;69(9):971–984. doi: 10.1023/b:biry.0000043539.07961.4c. [DOI] [PubMed] [Google Scholar]
- 111.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 112.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr, Rothermel BA, Hill JA. Intracellular Protein Aggregation Is a Proximal Trigger of Cardiomyocyte Autophagy. Circulation. 2008 doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gustafsson AB, Gottlieb RA. Autophagy in ischemic heart disease. Circ Res. 2009;104(2):150–158. doi: 10.1161/CIRCRESAHA.108.187427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116(Pt 9):1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 115.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152(4):657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circ Res. 2008;103(12):1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao J, Brault JJ, Schild A, Goldberg AL. Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy. 2008;4(3):378–380. doi: 10.4161/auto.5633. [DOI] [PubMed] [Google Scholar]
- 118.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 Coordinately Activates Protein Degradation by the Autophagic/Lysosomal and Proteasomal Pathways in Atrophying Muscle Cells. Cell Metab. 2007;6(6):472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 119.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 120.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32(2):95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 121.Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, Searle MS. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283(9):5427–5440. doi: 10.1074/jbc.M704973200. [DOI] [PubMed] [Google Scholar]
- 122.Tan JM, Wong ES, Dawson VL, Dawson TM, Lim KL. Lysine 63-linked polyubiquitin potentially partners with p62 to promote the clearance of protein inclusions by autophagy. Autophagy. 2007;4(2) [Google Scholar]
- 123.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283(11):6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 124.Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, Oyama F, Nukina N. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem. 2004;91(1):57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 125.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am J Pathol. 2002;160(1):255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 127.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 128.Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180(6):1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33(4):517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 131.Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edstrom L, Ehler E, Udd B, Gautel M. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308(5728):1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 132.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143(7):1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178(6):1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rodriguez-Gonzalez A, Lin T, Ikeda AK, Simms-Waldrip T, Fu C, Sakamoto KM. Role of the aggresome pathway in cancer: targeting histone deacetylase 6-dependent protein degradation. Cancer Res. 2008;68(8):2557–2560. doi: 10.1158/0008-5472.CAN-07-5989. [DOI] [PubMed] [Google Scholar]
- 135.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115(6):727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 136.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28(7):889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carra S, Seguin SJ, Landry J. HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy. 2008;4(2):237–239. doi: 10.4161/auto.5407. [DOI] [PubMed] [Google Scholar]
- 138.Berry NB, Fan M, Nephew KP. Estrogen receptor-alpha hinge-region lysines 302 and 303 regulate receptor degradation by the proteasome. Mol Endocrinol. 2008;22(7):1535–1551. doi: 10.1210/me.2007-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15(12):2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 140.Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1(2):75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 141.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 142.Rapoport S, Dubiel W, Muller M. Proteolysis of mitochondria in reticulocytes during maturation is ubiquitin-dependent and is accompanied by a high rate of ATP hydrolysis. FEBS Lett. 1985;180(2):249–252. doi: 10.1016/0014-5793(85)81080-2. [DOI] [PubMed] [Google Scholar]
- 143.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105(52):20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8(13):1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]