Abstract
It is known that CBFB-MYH11, the fusion gene generated by inversion of chromosome 16 in human acute myeloid leukemia, is causative for oncogenic transformation. However, the mechanism by which CBFB-MYH11 initiates leukemogenesis is not clear. Previously published reports showed that CBFB-MYH11 dominantly inhibits RUNX1 and CBFB, and such inhibition has been suggested as the mechanism for leukemogenesis. Here we show that Cbfb-MYH11 caused Cbfb/Runx1 repression–independent defects in both primitive and definitive hematopoiesis. During primitive hematopoiesis, Cbfb-MYH11 delayed differentiation characterized by sustained expression of Gata2, Il1rl1, and Csf2rb, a phenotype not found in Cbfb and Runx1 knockout mice. Expression of Cbfb-MYH11 in the bone marrow induced the accumulation of abnormal progenitor-like cells expressing Csf2rb in preleukemic mice. The expression of all 3 genes was detected in most human and murine CBFB-MYH11+ leukemia samples. Interestingly, Cbfb-MYH11+ preleukemic progenitors and leukemia-initiating cells did not express Csf2rb, although the majority of leukemia cells in our Cbfb-MYH11 knockin mice were Csf2rb+. Therefore Csf2rb can be used as a negative selection marker to enrich preleukemic progenitor cells and leukemia-initiating cells from Cbfb-MYH11 mice. These results suggest that Cbfb/Runx1 repression–independent activities contribute to leukemogenesis by Cbfb-MYH11.
Introduction
In humans, acute myeloid leukemia (AML) is often associated with specific, recurrent chromosomal rearrangements. Inversion of chromosome 16, inv(16)(p13;q22) or the related t(16;16)(p13;q22), is frequently found in patients with AML subtype M4 with eosinophilia1,2 and results in the fusion of the core-binding factor (CBF) beta gene CBFB with MYH11, the gene encoding smooth muscle myosin heavy chain (SMMHC).3 This fusion gene, CBFB-MYH11, encodes the chimeric protein CBFβ-SMMHC. It has been shown that CBFβ-SMMHC is necessary, but not sufficient for leukemogenesis; additional cooperating mutations are required.4 CBFβ-SMMHC is thought to impair differentiation of hematopoietic progenitor cells. The cooperating mutations may confer the increased proliferation required for oncogenic transformation.5
It has been proposed that CBFβ-SMMHC functions by inhibiting the normal activity of the CBF family members. This family of transcription factors is composed of 4 proteins, the 3 α subunits, Runt-related protein 1 (RUNX1; AML1), RUNX2, and RUNX3,6 and the single β subunit, CBFβ.7,8 The α subunits share the RUNT homology domain, which is responsible for binding to both their DNA targets and CBFβ. CBFβ does not bind DNA directly, but stabilizes the RUNX-DNA interaction allosterically.9 The CBFβ-SMMHC fusion protein retains the RUNX-binding domain in CBFβ and contains an additional RUNX-binding domain in the SMMHC tail, resulting in a higher binding affinity for the RUNX proteins than wild-type CBFβ.10 It has been proposed that this allows CBFβ-SMMHC to act as a dominant negative inhibitor of normal CBF family member activity.10–13
Studies in mice have shown that Cbfb and Runx1 are important for blood development. Hematopoiesis in mammals occurs in at least 2 waves: primitive and definitive. In mice, primitive hematopoiesis starts at embryonic day 7.5 (E7.5), and produces primarily large nucleated erythrocytes. Definitive hematopoiesis produces mainly enucleated red cells, and, as the embryo ages, all the cells of the myeloid and lymphoid lineages. In the mouse, the cells from definitive hematopoiesis enter circulation at E12.5, and are the predominant vehicle for gas exchange by E13.5.14 Cbfb and Runx1 are required for definitive hematopoiesis; mice homozygous for null alleles of either Cbfb (Cbfb−/−) or Runx1 (Runx1−/−) have a complete block in definitive hematopoiesis.15–20 However, little is known of their role in primitive hematopoiesis. No defects in primitive hematopoiesis have been reported in Cbfb−/− embryos. In Runx1−/− embryos, initial reports from 3 independent groups did not include any defects in primitive hematopoiesis,15,16,18 although subtle defects in Runx1−/− embryos were described in a more recent report.21
Previously, we generated mice in which the Cbfb-MYH11 fusion gene is knocked into the endogenous Cbfb locus.22 Embryos heterozygous for this allele (Cbfb+/MYH11) display a phenotype identical to both the Runx1- and Cbfb-null embryos, consistent with the hypothesis that Cbfb-MYH11 dominantly inhibits normal Runx/Cbfβ activity. Further, we observed delayed differentiation in the primitive hematopoietic cells of Cbfb+/MYH11 but not Cbfb−/− embryos. Based on this finding, we proposed that Cbfb-MYH11 has activities independent of Cbfb/Runx inhibition. In this report, we present evidence confirming this hypothesis. Moreover, we present data implying that this novel activity plays an important role in the initiation of leukemogenesis.
Methods
Animals
All animals used and the procedures performed in this study were approved by the National Human Genome Research Institute (NHGRI) Animal Care and Use Committee. Cbfb-MHY11 conventional (Cbfb+/MYH11) and conditional knockin (Cbfb+/56M), Cbfb knockout (Cbfb−/−), Runx1 knockout (Runx1−/−), and Mx1-Cre recombinase transgenic (Mx1-Cre+) mice have been described previously.4,16,17,23,24 All mice were genotyped as described previously by polymerase chain reaction (PCR) using tail-snip DNA. Mice at the age of 6 to 8 weeks were treated with 250 μg/dose of Polyinosine-polycytidylic acid (pI:pC) (InvivoGen) intraperitoneally to induce Cbfb-MYH11 expression. Unless otherwise noted, mice were given 3 doses of pI:pC on days 1, 3, and 5. To accelerate leukemia, mice were either treated with N-ethyl-N-nitrosourea (ENU) as described previously,23 or given a second round of 3 doses of pI:pC 10 weeks after the first round. Transplantations were performed using retro-orbital injection of 2.5 × 104 cells into sublethally irradiated (630 cGy), congenic (C57Bl6/129SvEv F1) mice.
Flow cytometry
Peripheral blood cells from embryos were washed with fluorescence activated cell-sorting (FACS) buffer (5% fetal calf serum in phosphate-buffered saline) before staining. Peripheral blood cells or spleen cells from adult animals were incubated in ACK lysing buffer (BioWhittaker) and then washed with FACS buffer before staining. Bone marrow cells were harvested from the femurs of adult mice at the indicated number of days after pI:pC injection. Lineage-negative cells were isolated using Easy-Sep Lineage Depletion Kit (StemCell Technologies) according to the manufacturer's instructions. Spleens were harvested from mice and cells were isolated after filtering through a 70-μm cell strainer (BD Biosciences) in the presence of FACS buffer. Cells were stained with fluorescein isothiocyanate–, phycoerythrin-, and allophycocyanin (APC)–conjugated antibodies to Ter119, Csf2rb (CD131), c-Kit, annexin V, and bromodeoxyuridine (BrdU; BD PharMingen); APC–cyanin 7–conjugated c-Kit, phycoerythrin–cyanin 7 FcγR II/III, APC-conjugated CD34 (eBioscience); fluorescein isothiocyanate–conjugated Il1rl1 (MD Biosciences); and Pacific blue–conjugated ScaI (Biolegend). Data were acquired using a LSRII (BD PharMingen) and analyzed using FlowJo software (TreeStar). Sorting was performed using a BDAria (BD PharMingen).
Apoptosis and proliferation assays
Apoptotic cells were analyzed by annexin V and 7-amino-actinomycin D (7AAD) staining (BD PharMingen) and cell proliferation was determined by BrdU incorporation using the BrdU Flow kit (BD PharMingen) according to the manufacturer's instructions. In brief, pregnant mice were injected intraperitoneally with 10 μg of BrdU per gram of body weight, and embryos were harvested 1 hour later. Cells were stained following the manufacturer's protocol and analyzed by flow cytometry.
Western blot analysis
Cells were lysed in 1× radioimmunoprecipitation assay buffer (Boston BioProducts) containing Complete Mini proteinase inhibitor tablets (Roche Diagnostics). Protein concentrations were determined using DC Protein Assay (Bio-Rad). Protein was run on Nu-PAGE 4% to 12% Bis (bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane)-Tris (tris(hydroxymethyl) aminomethane) gels (Invitrogen) and transferred to Invitrolon membranes (Invitrogen). Membranes were probed for Gata2 (anti-Gata2 [Cell Signaling Technologies], anti-rabbit–horseradish peroxidase (HRP) [Vector Laboratories]) or Cbfβ-SMMHC (anti-Cbfβ141,17 antimouse-HRP [Vector Laboratories]), stripped with Restore Western Blot Stripping Buffer (Pierce), and reprobed for α-tubulin (anti–α-tubulin [Abcam] and anti-mouse–HRP [Vector Laboratories]). Blots were visualized with ECL Western Blot Detection System (GE Healthcare) and Biomax light film (Kodak). Band intensity was determined using MultiGauge Version 3.0 (Fujifilm).
PCR for Cre excision efficiency
Excision of the Cbfb-56M allele was determined using primers that amplify the Cbfb allele and the excised Cbfb-56M allele, but not the unexcised Cbfb-56M allele, using DNA isolated from sorted cells with the DNeasy Blood & Tissue Kit (QIAGEN) according to the manufacturer's instructions. Band intensity was determined using MultiGauge Version 3.0. Primer sequences are available upon request.
Reverse transcription–PCR
RNA was extracted using RNA Stat-60 (Tel-Test Inc). cDNA was generated using Super-Script One Step (Invitrogen) and PCR amplified. Primer sequences are available upon request.
Histologic staining
Sorted cells were affixed to slides using a Shandon Cytospin (Thermo Fisher Scientific) and stained with Wright-Giemsa (Protocol Hema 3 system; Thermo Fisher Scientific) according to the manufacturer's protocol. Slides were imaged using either an Eclipse E800 (Nikon) with a 100×/1.4 oil objective, a CCD camera (Scanalytics) and IP Labs 3.5.5 (Scanalytics), or on an Axioscope 2 (Zeiss) with a 100×/1.4 oil objective (Hamamatsu) and iVision Mac 4.0.1 (BioVision Technologies).
Colony-forming assay
Equal numbers of sorted cells from mice 14 days after pI:pC treatment were suspended in Methocult 3434 (StemCell Technologies) according to the manufacturer's protocol. After 12 days in culture, plates were scored for colony type and number.
Microarray analysis
Peripheral blood from 8 to 10 embryos (E12.5) of the same genotype was pooled and RNA was extracted using RNA Stat-60. RNA was then labeled with One Cycle Target Labeling kit (Affymetrix) and hybridized to Affymetrix Genechip mouse microarray (430 2.0) chips. Four chips were used for both the Cbfb+/MYH11 and littermate control samples. Three chips were used for the Cbfb−/− and littermate control samples. The data were analyzed with GeneSifter software (VizX Laboratories) and log transformed and normalized using Robus Multichip Analysis algorithm.25 Student t tests were performed for every probe on the Affymetrix chip. P values were corrected by the Benjamini-Hochberg procedure. Functional analyses were generated through the use of Ingenuity Pathway Analysis (Ingenuity Systems, http://www.ingenuity.com). Expression in Cbfb+/MYH11 leukemic samples was determined using microarray data as described (Y.K., L.Z., M. Wunderlich, T. Corpora, R.K.H., T. Paul, M. Kundu, L. Garrett-Beal, S. C. G. Huang, L. Wolff, Y. Ito, J. Bushweller, J. C. Mulloy and P.P.L., submitted May 2009. A gene was considered expressed in the leukemic samples if it was called “present” using the Affymetrix software suite in 75% or more of the sample replicates. Expression in inv(16) patients was determined using the data from published studies deposited in National Center for Biotechnology Information Gene Expression Omnibus database.26–31 For studies using the Affymetrix array, a gene was considered expressed if it was called present using the Affymetrix software suite. In studies using other platforms, a gene was considered expressed if the signal intensity was in the 50th percentile or greater. All microarray data have been deposited in the Gene Expression Omnibus public database under accession number GSE19194.
Statistical analysis
To assess the significance in FACS staining between samples, and the difference in colony numbers, Student t tests were performed using Excel (Microsoft).
Results
Expression of Cbfb-MYH11 induced defects in differentiation, proliferation, and apoptosis of primitive hematopoietic cells
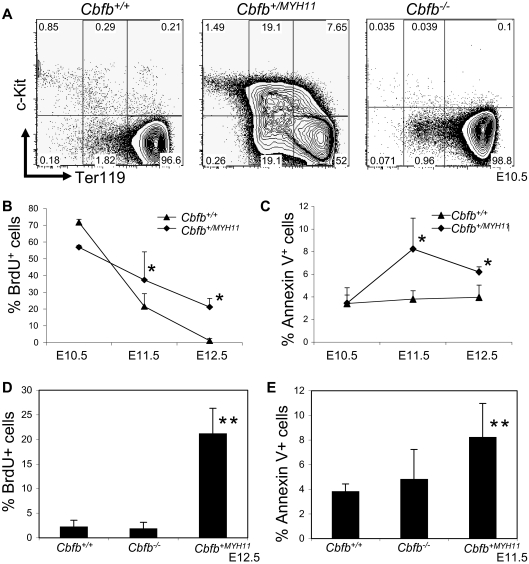
We showed previously that the peripheral blood from Cbfb+/MYH11 embryos has increased numbers of histologically immature primitive blood cells.22 The immature cells were found to be increased at E12.5, whereas no apparent morphologic differences were observed between Cbfb+/MYH11 and Cbfb+/+ littermate embryos at earlier time points, such as E10.5 and E11.5.22 To analyze the hematopoietic defects by a different assay, we stained the peripheral blood of Cbfb+/+ and Cbfb+/MYH11 embryos with markers of primitive blood differentiation: c-Kit and Ter119. In the peripheral blood of E10.5 Cbfb+/+ embryos, most of the primitive blood cells were c-Kit−, Ter119high cells. In contrast, the peripheral blood from E10.5 Cbfb+/MYH11 embryos showed significant populations of cells with delayed development, including cells with continued c-Kit expression (c-Kit+, Ter119− and c-Kit+, Ter119low), as well as c-Kit−, Ter119low cells (Figure 1A, supplemental Figure 1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). In E12.5 embryos, the differences in c-Kit and Ter-119 expression levels between Cbfb+/MYH11 and Cbfb+/+ littermate embryos were less severe, but still significant (supplemental Figure 1B). Therefore, the differentiation blockage in the Cbfb+/MYH11 embryos occurs earlier than previously observed morphologically.
Figure 1.
Cbfb-MYH11 causes Cbfb/Runx1 repression–independent defects during primitive hematopoiesis. (A) Representative FACS plots of Ter119 and c-Kit staining of the primitive blood from embryos of the indicated ages and genotypes. Percentage of cells in each gate is given. Line graphs of the percentage (%) of (B) BrdU-positive (+) and (C) annexin V+, 7AAD-low cells in the peripheral blood of littermate embryos of the indicated genotypes and ages. Bar graphs of the percentage of (D) BrdU+ and (E) annexin V+, 7AAD-low cells in the peripheral blood of Cbfb+/+ and Cbfb−/− littermate embryos of the indicated ages. Data from age-matched Cbfb+/MYH1 embryos was included for comparison. *Statistically significant difference (P < .05) compared with Cbfb+/+ embryos; **statistically significant difference (P < .05) compared with Cbfb+/+ and Cbfb−/− embryos. N ≥ 3 for all genotypes and ages. Error bars indicate SD within each genotype.
We also used BrdU incorporation and annexin V staining to measure proliferation and apoptosis in the blood of Cbfb+/+ and Cbfb+/MYH11 embryos. We found that the E11.5 and E12.5 Cbfb+/MYH11 embryos did not show the same decrease in BrdU+ blood cells found in the blood of their wild-type littermates, resulting in significantly higher percentages of BrdU+ cells in the blood (Figure 1B). We also found statistically significant increase of annexin V staining in the primitive blood of E11.5 and E12.5 Cbfb+/MYH11 embryos, even though the overall level of such staining was very low (Figure 1C). These effects on proliferation and apoptosis were likely secondary to the differentiation defect, as they occurred later during embryogenesis.
Lack of significant defects in Cbfb−/− and Runx1−/− primitive hematopoiesis
We tested whether Cbfb−/− and Runx1−/− embryos also had defects in primitive hematopoiesis. In the primitive blood of Cbfb−/− embryos, we saw no difference in c-Kit or Ter119 staining (Figure 1A and supplemental Figure 2A) compared with their Cbfb+/+ littermates. In the blood of Runx1−/− embryos, we observed a statistically significant delay (P < .01) in differentiation compared with their Runx1+/+ littermates (supplemental Figure 2B), consistent with the recent findings of Yokomizo et al.21 However, this delay lessened with age; by E11.5 the difference was not statistically significant (supplemental Figure 2C). In addition, these changes were significantly less severe than those observed in Cbfb+/MYH11 embryos (Runx1−/− vs Cbfb+/MYH11, P < .01). We also examined the rates of proliferation and apoptosis in Cbfb−/− and Runx1−/− embryos (Figure 1D-E, supplemental Figure 3). We found no statistically significant difference in the rates of BrdU incorporation and annexin V staining in Cbfb−/− and Runx1−/− embryos. Together these results indicate that loss of Cbfb or Runx1 activity is not sufficient to cause the defects in primitive hematopoiesis observed in Cbfb+/MYH11 embryos and that Cbfb-MYH11 has gain-of-function activities that are independent of Cbfb/Runx1 repression.
Expression of Cbfb-MYH11 and loss of Cbfb have different effects on gene expression
To determine whether Cbfb-MYH11 effects gene expression independent of Cbfb repression, we used microarray technology to assess gene expression profiles of blood cells from E12.5 Cbfb−/− and Cbfb+/MYH11 embryos, as well as their wild-type littermates. Hierarchic clustering analysis indicated that gene expression in the Cbfb+/+ and Cbfb−/− samples was more similar to each other than either was to the Cbfb+/MYH11 sample (supplemental Figure 4A). We also found that in the Cbfb+/MYH11 embryos, 658 genes showed at least a 2-fold change in expression level compared with the wild-type littermate controls (supplemental Figure 4B). The majority of these 658 genes (95%) showed increased expression in the Cbfb+/MYH11 embryos. In contrast, in the Cbfb−/− sample, only 174 genes showed at least a 2-fold change. Again, the majority of these genes (77%) showed increased expression. Only 71 genes were deregulated in both the Cbfb+/MYH11 and Cbfb−/− embryos. Pathway analysis of the 587 genes uniquely deregulated in the Cbfb+/MYH11 embryos indicated that this set of genes is involved in multiple cellular processes, including DNA replication, cell cycle, and cancer (supplemental Figure 4C). Specifically, we found genes associated with proliferation such as those that encode cyclins (CcnB1, CcnE2, CcnG1, CcnH) and cell division–associated proteins (Cdca4, Cdca5, Cdca8) were up-regulated, which is consistent with the increased proliferation seen only in the Cbfb+/MYH11 embryos (Figure 1B,D).
To determine whether any of the 587 genes found uniquely deregulated in Cbfb+/MYH11 embryos were also expressed during leukemogenesis, we examined the expression of these genes by microarray in leukemic cells from mice expressing Cbfb-MYH11.22 We found that most of the genes from this set (88%) were expressed in the mouse Cbfb-MYH11+ leukemic cells (Table 1 and data not shown). These findings indicate that Cbfb-MYH11 has similar effects on gene expression during leukemogenesis, implying that a Cbfb/Runx1 repression–independent mechanism may also be important during leukemogenesis.
Table 1.
Expression of the genes uniquely deregulated in Cbfb+/MYH11 embryos in mouse and human leukemias
| Gene symbol | Gene name | Fold change in Cbfb-MYH embryos | Expressed in Cbfb-MYH leukemic mice | Expression in inv(16) patients |
|---|---|---|---|---|
| Il1rl1 | Interleukin 1 receptor-like 1 | 62 | Yes | 8/60 |
| Csf2rb | Colony-stimulating factor 2 receptor, beta | 48 | Yes | 59/60 |
| Ms4a2 | Membrane-spanning 4-domains, subfamily A, member 2 | 36 | Yes | 11/60 |
| Npl | N-acetylneuraminate pyruvate lyase | 24 | Yes | 17/55 |
| Prnp | Prion protein | 20 | Yes | 46/46 |
| Flot2 | Flotilin 2 | 17 | Yes | 54/60 |
| Gata2 | GATA-binding protein 2 | 17 | Yes | 55/60 |
| Adcy7 | Adenylate cyclase 7 | 15 | Yes | 46/60 |
| Pfkp | Phosphofructokinase, platelet | 15 | Yes | 53/60 |
| Gpc4 | Glypican 4 | 14 | No | 4/60 |
| Tpd52l1 | Tumor protein D52-like 1 | 14 | No | 2/60 |
| Slc41a3 | Solute carrier family 41 member 3 | 13 | Yes | 29/55 |
| Chpt1 | Choline phosphotransferase 1 | 13 | No | 54/55 |
| Slc29a1 | Solute carrier family 29, member a1 | 13 | Yes | 45/60 |
| Col5a1 | Collagen, type V, alpha 1 | 12 | Yes | 1/60 |
| Wnt2 | Wingless-related MMTV integration site 2 | 11 | No | 4/60 |
| Fabp7 | Fatty acid–binding protein 7, brain | 11 | No | 0/60 |
| Padi3 | Peptidly arginine deiminase, type III | 9 | No | 0/41 |
| Spns2 | Spinster homolog 2 (Drosophila) | 9 | No | 6/51 |
| Rnf128 | Ring finger protein 128 | 9 | Yes | 0/55 |
The 20 genes with human homologs and the greatest fold change in expression by microarray analysis in the blood of Cbfb+/MYH11 but not Cbfb−/− embryos compared with blood from wild-type littermates are listed. These genes were also examined for expression in leukemic cells from mice expressing Cbfb-MYH11 as determined from microarray data. Expression in inv(16) AML patient samples was determined using published gene expression data.26–30 These data are presented as a ratio of the number of patient samples expressing a given gene over the total number of inv(16) AML samples with available expression data for that gene.
To determine whether any of the genes from this set are also expressed in patient inv(16) AML cells, we examined data from published gene expression studies.26–30 We used either the signal intensity for a given probe or the signal compared with a mismatch control probe to determine whether a gene was expressed. As a negative control, we examined the expression of 2 genes not expressed in any blood cells: rhodopsin and dystrophin. We found that these 2 genes were expressed in 1 of 60 samples, and 1 of 55 samples, indicating that this is a valid method for determining gene expression in these patient samples. We found that many of the genes that show high up-regulation uniquely in Cbfb+/MYH11 embryos were expressed in human AML samples (Table 1), implying that Cbfb-MYH11's Cbfb/Runx1 repression–independent activities are involved in leukemogenesis in mice and humans.
Csf2rb up-regulation in Cbfb-MYH11–expressing cells reflects delayed differentiation
Csf2rb was the second-most up-regulated gene in the Cbfb+/MYH11 embryos and was expressed in virtually all human inv(16)+ leukemia samples analyzed in this study (Table 1). Csf2rb (CD131) encodes the common β chain of the receptors for interleukin 3, interleukin 5, and granulocyte-macrophage colony stimulating factor.32–34 A role for the Csf2rb heterodimerization partner, IL-3Rα, has been described in specifically marking leukemia stem cells.35 We therefore decided to further characterize the expression of Csf2rb in Cbfb-MYH11+ cells.
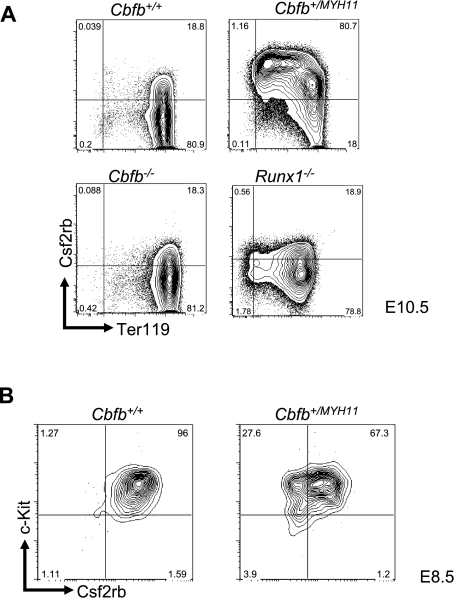
By FACS, we examined Csf2rb expression in the peripheral blood of E10.5 embryos and found that Csf2rb showed significantly increased expression at the protein level in the primitive blood cells of Cbfb+/MYH11 embryos compared with Cbfb+/+ littermates (Figure 2A), consistent with our microarray results. This high level of increased Csf2rb expression was not seen in either Cbfb−/− or Runx1−/− embryos (Figure 2A), confirming that Csf2rb expression is deregulated by Cbfb-MYH11 via a mechanism independent of Cbfb/Runx repression.
Figure 2.
Csf2rb expression is related to Cbfb-MYH11–induced differentiation defects. (A) Representative FACS plots of Csf2rb and Ter119 staining in primitive blood cells from E10.5 embryos of the indicated genotypes. Percentage of cells in each gate is given. N ≥ 3 for all genotypes. (B) Representative FACS plots of Csf2rb and c-Kit staining in the primitive blood of E8.5 embryos of the indicated genotypes. N ≥ 3 for all genotypes.
We next tested whether expression of Csf2rb in the primitive blood of Cbfb+/MYH11 embryos resulted from differentiation delays induced by Cbfb-MYH11. To test this, we stained the primitive blood from E8.5 Cbfb+/+ and Cbfb+/MYH11 embryos (Figure 2B). We found that Csf2rb was expressed at high levels in both Cbfb+/+ and Cbfb+/MYH11 embryos at this early time point, indicating that Csf2rb is predominantly expressed early in primitive hematopoietic differentiation, and the continued expression of Csf2rb in the E10.5 Cbfb+/MYH11 embryos is likely the result of delayed differentiation.
We next asked whether Csf2rb is expressed in analogous early progenitor populations during definitive hematopoiesis. We examined Csf2rb expression in the lineage-depleted (lin−) cells in the bone marrow (BM) of wild-type mice by FACS. In general, Csf2rb+ cells accounted for only 2% (± 0.6%, N = 3) of lin− cells. Interestingly, the lin−, c-Kit+, ScaI+ (LKS+) fraction contained a higher percentage (10.39% ± 0.39%) of Csf2rb+ cells than the lin−, c-Kit+, ScaI− (LKS−) compartment (0.97% ± 0.31%), with various levels in the common myeloid progenitor, granulomonocytic progenitor, and megakaryocyte/erythrocyte progenitor populations (supplemental Figure 5). Therefore, Csf2rb is expressed in a small fraction of hematopoietic stem cells and early progenitors in the adult BM, and its expression decreases as the hematopoietic cells become more lineage restricted.
Expression of Csf2rb, Il1rl1, and Gata2 proteins in mouse and human leukemia cells
Of the many genes that showed increased expression uniquely in Cbfb+/MYH11 embryos, 2 more genes were of particular interest: Il1rl1 (T1, St2), the gene encoding the receptor for Il-33,36,37 and the gene for the transcription factor Gata2.38 Both genes were among the most up-regulated in Cbfb+/MYH11 embryos. In addition, GATA2 is also expressed in essentially all patients with inv(16) AML (Table 1), implying that it has particular relevance to the disease in humans. We therefore examined the expression of these 2 genes, as well as Csf2rb, in mouse and human leukemia cells.
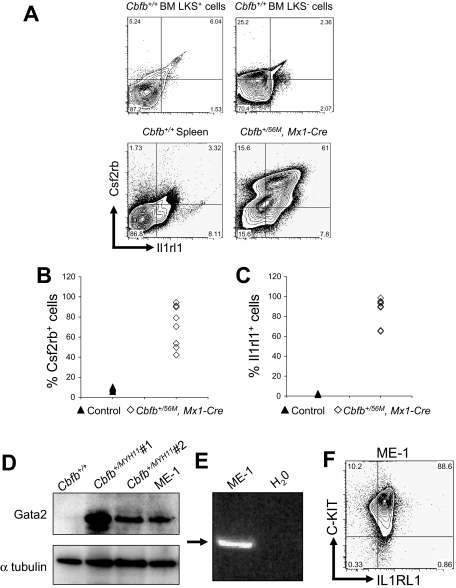
By FACS, we examined the expression of Csf2rb and Il1rl1 in leukemic cells, isolated from mice expressing a conditional allele of full-length Cbfb-MYH11 (Cbfb+/56M)23 paired with the Mx1-Cre recombinase (Mx1-Cre) transgene24 to effect inducible expression after treatment with pI:pC in all hematopoietic cells. To enhance the development of leukemia, these mice were also treated with the mutagen N-ethyl-N-nitrosourea (ENU), or given a second round of pI:pC. We found that both Csf2rb and Il1rl1 were expressed at high levels in most of the leukemic cells (Figure 3A-C), forming a Csf2rb/Il1rl1 double-positive population. This Csf2rb/Il1rl1 double expression appeared to be a specific signature of the leukemia cells because spleen cells from Cbfb+/+ mice did not express high levels of these 2 markers, nor did the LKS+ or LKS− cells from Cbfb+/+ mice (Figure 3A). By Western blot analysis, we found that Gata2 was also expressed at high levels in leukemic cells from ENU-treated, Cbfb-MYH11–expressing mice (Figure 3D). In addition, we found that all 3 of these genes were expressed in the inv(16) patient-derived cell line, ME-1 (Figure 3D-F). Collectively, these results show that these 3 genes were expressed at high levels in both mouse and human leukemia cells.
Figure 3.
Csf2rb, Il1rl1, and Gata2 are expressed in both mouse and human leukemic cells. (A) Representative FACS plots of Csf2rb and Il1rl1 staining in lineage-negative, c-Kit+ ScaI+ (LKS+) and c-Kit+, ScaI− (LKS−) bone marrow and spleen from nonleukemic Cbfb+/+ mice, and leukemic cells from the spleen of mice expressing a conditional allele of Cbfb-MYH11 (Cbfb+/56M, Mx1-Cre). N ≥ 3 for Cbfb+/56M, Mx1-Cre. Expression of Csf2rb (B) and Il1rl1 (C) in the peripheral blood of leukemic Cbfb+/56, Mx1 Cre+ and control littermate mice. The control group contained Cbfb+/+, Mx1-Cre−; Cbfb+/56M, Mx1-Cre−; and Cbfb+/+, Mx1-Cre+ mice. All mice were treated with pI:pC in the same way. (D) Western blot analysis of Gata2 expression in the peripheral blood of a nonleukemic Cbfb+/+ adult mouse, 2 different leukemic adult mice expressing Cbfb-MYH11, and the human inv(16) AML-derived ME-1 cells. The bottom panel shows α-tubulin expression using the same blot. (E) Reverse transcription–PCR analysis using primers specific for human CSF2RB in ME-1 cells. An arrow indicates the band of the expected size. (F) FACS plot of ME-1 cells stained for C-KIT and IL1RL1. Percentage of cells in each gate is given.
Induction of a Csf2rb+ cell population by Cbfb-MYH11 expression
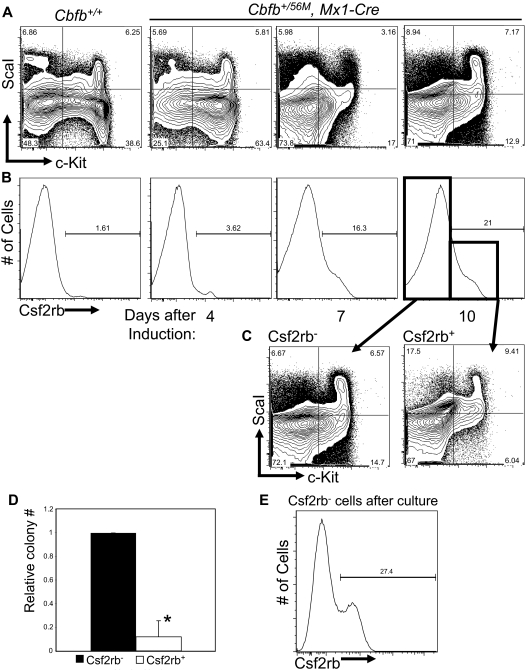
Based on the data described in Figures 2 and 3 we hypothesized that the expression of these genes marks an early step in leukemogenesis by Cbfb-MYH11. To test this hypothesis, we again used pI:pC-treated Cbfb+/56M, Mx1-Cre mice, but in this instance, the mice were not treated with ENU. At 4 days after the induction of Cbfb-MYH11, an accumulation of LKS− progenitors could be detected in the bone marrow (Figure 4A), similar to previously described findings.23 Interestingly, these changes were accompanied by a progressive increase in Csf2rb-expressing cells (Figure 4B). Staining of lin− bone marrow cells from Cbfb+/+ mice after pI:pC was indistinguishable from untreated mice at all time points (Figure 4 and data not shown), indicating that this effect is dependent on Cbfb-MYH11 expression. This defect was transient, however; by 10 weeks after Cbfb-MYH11 induction, the number of Csf2rb+ cells returned to wild-type levels (supplemental Figure 6A). In addition, at 10 weeks, we did not observe the block in myeloid differentiation previously described by Kuo et al23 (supplemental Figure 6B).
Figure 4.
Cbfb-MYH11 expression results in an abnormal Csf2rb+ population with reduced progenitor activity. Representative FACS plots of c-Kit and ScaI (A), and Csf2rb (B) staining in lineage-depleted (lin−) bone marrow from mice of Cbfb+/56/M, Mx1-Cre+ and Cbfb+/+ mice at the indicated number of days after treatment with pI:pC. Percentage of cells in each gate is given. N ≥ 3 for all genotypes. (C) Representative FACS plots of c-Kit and ScaI staining of the Csf2rb− and Csf2rb+ populations from lin− bone marrow from Cbfb+/56M, Mx1-Cre+ mice 10 days after Cbfb-MYH11 induction. (D) Bar graphs of the relative total numbers of colonies seen from lin− bone marrow cells from Cbfb+/56M, Mx1-Cre+ mice 10 days after induction of Cbfb-MYH11 expression, sorted for Csf2rb expression and grown in culture for 12 days. N ≥ 3. *Statistically significant difference (P < .01). (E) FACS staining for Csf2rb of colonies derived from the Csf2rb− cells described in panel C. Percentage of Csf2rb+ cells is indicated. Error bars indicate SD within the indicated cell population.
Although the accumulation of the LKS− and Csf2rb+ cells took place at the same time, these 2 populations were not identical to each other. Rather, both the Csf2rb− and Csf2rb+ cells contained LKS+ and LKS− cells (Figure 4C). These data imply that the Csf2rb+ cells in Cbfb-MYH11–expressing adult bone marrow contained cells with the immunophenotype of both hematopoietic stem cells (HSCs) and early progenitors.
We next asked whether the Csf2rb-expressing cells from adult bone marrow were arrested in differentiation at the stage of early progenitors. To test this, we sorted lin− bone marrow cells from nonleukemic mice 14 days after induction of Cbfb-MYH11 expression and performed colony-forming assays using equal numbers of lin−, Csf2rb− and lin−, Csf2rb+ cells. We found that the Csf2rb+ cells produced significantly fewer colonies than did the Csf2rb− cells (Figure 4D). Those colonies that did form from the Csf2rb+ cells were of similar size and cell type as seen from Csf2rb− cells (data no shown). From the colonies derived from Csf2rb− cells, we performed FACS staining for Csf2rb. We found that a significant portion of these cells had acquired Csf2rb expression (Figure 4E). These results indicate that the Csf2rb− population is enriched for earlier progenitor cell activity, and gives rise to the Csf2rb+ population.
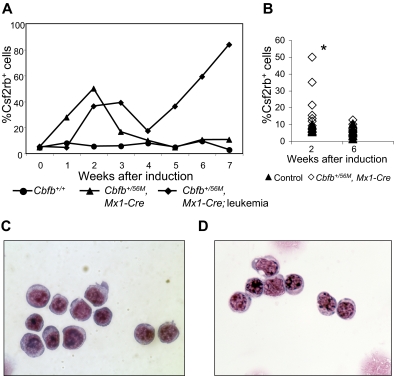
We also stained peripheral blood cells for Csf2rb expression after induction of Cbfb-MYH11. We found that, similar to bone marrow, Cbfb-MYH11 expression caused an increase of Csf2rb+ cells in the blood (Figure 5A-B). In nonleukemic mice, this population was transient and was not observable at 6 weeks (Figure 5B) and 10 weeks (supplemental Figure 6C) after Cbfb-MYH11 induction. In contrast, in mice that developed leukemia, there was a second wave of Csf2rb+ cells that was not transient, but continued expanding until the disease was fatal. Figure 5A shows a representative mouse that developed AML spontaneously (the line marked by diamonds) without treatment with ENU. To test whether all mice developed a similar, second wave of leukemic Csf2rb+ cells, we induced leukemia in a cohort of mice using 2 rounds of pI:pC. This protocol induced leukemia after a longer latency than combined treatment with ENU, allowing us to clearly separate Csf2rb+ cells from the initial wave after pI:pC treatment from those in the final leukemic wave. The percentage of Csf2rb+ cells in the initial, transient wave of cells is shown in Figure 5B, and the percentage of Csf2rb+ cells at the time of killing is shown in Figure 3B. Sorting of the Csf2rb+ cells from the initial wave of expression in nonleukemic mice showed that these cells had a blastlike morphology (Figure 5C). These cells are indistinguishable from the Csf2rb+ cells seen in the peripheral blood of leukemic mice (Figure 5D). These results indicate that, before oncogenic transformation, Cbfb-MYH11 expression caused a similar block in differentiation as seen in leukemia, but without additional mutations, these cells had a limited life span.
Figure 5.
A transient population of Csf2rb+ cells is seen in the peripheral blood after Cbfb-MYH11 induction. (A) Line graphs showing percentage (%) of Csf2rb+ cells in the peripheral blood at the indicated number of weeks after pI:pC treatment in 3 representative mice. The line marked with circles represents a wild-type mouse after pI:pC treatment. The line marked with triangles represents a conditional Cbfb-MYH11 knockin mouse after pI:pC treatment that did not develop leukemia within the observed time period (7 weeks). The line marked with diamonds represents a conditional Cbfb-MYH11 knockin mouse after pI:pC treatment that spontaneously developed AML (without ENU). (B) Percentage of Csf2rb+ cells in a larger cohort of mice at the indicated number of weeks after pI:pC treatments. *Statistically significant difference between Cbfb+/56M, Mx1-Cre and control mice (P < .02). Wright-Giemsa staining of sorted Csf2rb+ peripheral blood cells from (C) a preleukemic Cbfb+/56M, Mx1-Cre mouse 2 weeks after Cbfb-MYH11 induction and (D) a leukemic Cbfb+/56M, Mx1-Cre mouse. Magnification, 1000×.
Csf2rb is a useful marker for isolating preleukemic progenitors
Studies in humans have indicated that leukemia stem cells have HSC and early progenitor-like immunophenotypes.39,40 The analogous populations in mice are the LKS+ and LKS− cells, respectively. Previously, it was shown that Cbfb-MYH11–expressing cells from both populations could initiate AML.23 We found both of these populations in the Csf2rb− and Csf2rb+ fractions. Yet, as described in Figure 5, only the Csf2rb− population was enriched for progenitor cell activity.
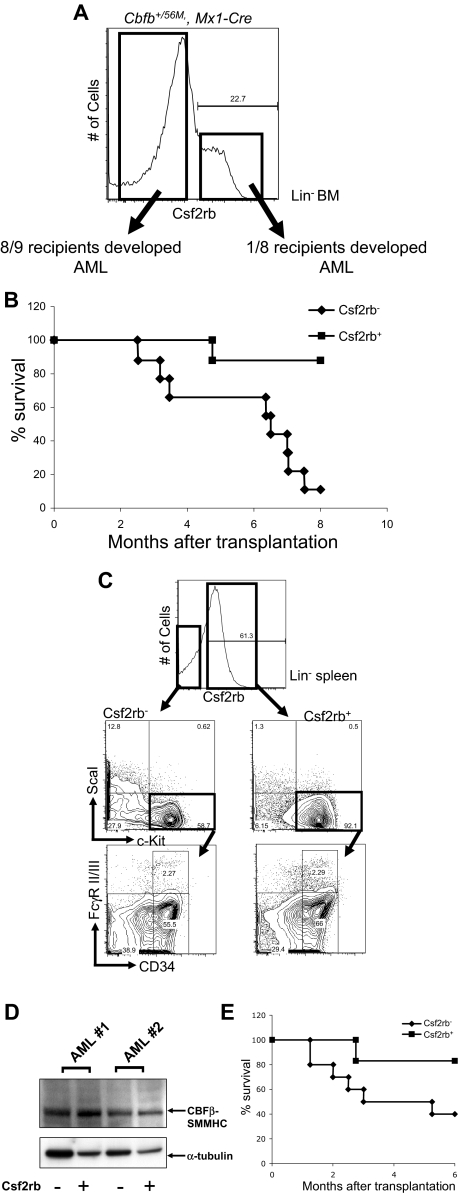
Therefore, we asked whether this population was also enriched for cells with the potential to acquire leukemia-initiating activity, referred to as preleukemic progenitors. We tested this hypothesis in our conditional Cbfb-MYH11 knockin mouse model. We treated Cbfb+/56M, Mx1-Cre+ mice with ENU, and induced expression of Cbfb-MYH11 by pI:pC injection.24 Fourteen days after induction of Cbfb-MYH11 and well before leukemic transformation, the lin− bone marrow cells were isolated and sorted based on Csf2rb expression. By PCR, we tested the excision efficiency of the conditional Cbfb-56M allele (supplemental Figure 7) in the sorted Csf2rb− and Csf2rb+ cells. We found that the ratio of the excised Cbfb-56M allele and the Cbfb allele was close to 1 (Csf2rb−: 1.15 ± 0.25; Csf2rb+: 1.12 ± 0.07), indicating approximately 100% excision efficiency in both populations. Either 25 000 Csf2rb− or 25 000 Csf2rb+ cells were then transplanted into congenic recipient mice, which were monitored for leukemia development. Strikingly, 8 of the 9 mice that received a transplant of Csf2rb− cells developed leukemia within 8 months after transplantation (Figure 6A-B); in contrast, only 1 of the 8 mice that received a transplant of Csf2rb+ cells developed leukemia during the same time period.
Figure 6.
Csf2rb−/lin− BM cells in mice expressing Cbfb-MYH11 are enriched for preleukemic progenitors and leukemia-initiating cells. (A) FACS staining of lineage-depleted bone marrow cells from ENU-treated, preleukemic Cbfb+/56M, Mx1-Cre mice 14 days after induction of Cbfb-MYH11. Cells were sorted for Csf2rb expression as indicated by the boxes, and transplanted into sublethally irradiated mice via retro-orbital injection. (B) Kaplan-Meier survival curves of mice that received a transplant of preleukemic Csf2rb− or Csf2rb+ cells. (C) FACS staining for the indicated differentiation markers in the Csf2rb− and Csf2rb+ leukemic spleen cells from a representative mouse that developed AML after transplantation of preleukemic Csf2rb− cells. (D) Western blot of CBFβ-SMMHC expression in the Csf2rb− and Csf2rb+ leukemic cells of 2 different recipient animals. (E) Kaplan-Meier survival curves of secondary transplant recipient mice that underwent transplantation as described in panel A with leukemic Csf2rb− and Csf2rb+ cells.
The leukemia that developed from the transplanted Csf2rb− cells was indistinguishable from those that developed in ENU-treated Cbfb+/56M, Mx1-Cre+ mice, in that the majority of the leukemic spleen cells expressed Csf2rb (Figure 6C). Interestingly, the Csf2rb− and the Csf2rb+ leukemic cells showed similar populations of abnormal myeloid progenitors, as defined by CD34 and FcgR II/III in the LKS− compartment, which are similar to the abnormal myeloid progenitors described previously.23 By Western blot (Figure 6D), both the Csf2rb− and Csf2rb+ cells showed abundant levels of CBFβ-SMMHC, with the Csf2rb+ cells expressing slightly more of the fusion protein.
For secondary transplantation, 500 000 Csf2rb− or 500 000 Csf2rb+ leukemic spleen cells were injected into each congenic recipient. Only 1 of 6 mice receiving the Csf2rb+ cells developed AML, whereas 6 of 10 mice that received a transplant of Csf2rb− cells developed AML at 6 months after transplantation (Figure 6E). This result indicates that the leukemia-initiating cells, like the preleukemic progenitor cells, are enriched in the Csf2rb− population.
Discussion
It has been proposed that the Cbfb-MYH11 fusion gene acts primarily by inhibiting normal Cbfb/Runx1 activity.10–13 This model is supported by the finding that mice expressing Cbfb-MYH11 have a definitive hematopoietic phenotype indistinguishable from both Cbfb−/− and Runx1−/− embryos.22 However, in this report, we demonstrate that Cbfb+/MYH1 embryos have defects in primitive hematopoiesis, a phenotype not initially described in either the Cbfb−/− or the Runx1−/− embryos.15–20 The Cbfb/Runx1 repression–independent defects during primitive hematopoiesis include more sustained proliferation and increased apoptosis, in addition to a severe delay in differentiation characterized by continued expression of c-Kit and 3 additional early progenitor markers: Csf2rb, Il1rl1, and Gata2. Because the differentiation delay was detected at an earlier embryonic stage than the proliferation and apoptosis defects, we speculate that it is the primary defect. As withdrawal from the cell cycle is a common feature of differentiation, it is likely that the sustained high levels of proliferation of the primitive blood cells in the Cbfb+/MYH11 embryos reflect the differentiation blockage in those cells.
We showed that Csf2rb, Il1rl1, and Gata2 were also expressed in Cbfb-MYH11+ mouse and human leukemic samples, suggesting that the gene expression changes induced by the Cbfb/Runx1 repression–independent mechanism also take place in leukemogenesis by Cbfb-MYH11. Therefore, these 3 genes may play important roles in leukemia development, especially in the differentiation blockage of leukemia cells. However, as we and others have shown (Figure 2, supplemental Figure 5),41,42 all 3 of these genes are expressed in early hematopoietic progenitors. In addition, these genes are not part of the unique inv(16) gene expression signature identified by Valk et al,28 implying that they are also expressed in non–CBFB-MYH11 AMLs. Therefore, it is equally likely that expression of these genes is the result and not the cause of the Cbfb-MYH11–mediated block in differentiation. Further experimentation will be required to address whether expression of these genes is a direct effect of Cbfb-MYH11 expression, and their role in leukemogenesis.
Currently, we can only speculate as to the mechanism of CBFB-MYH11's CBFB/RUNX repression–independent activity. One possible explanation is that the protein product of the CBFB-MYH11 fusion gene, CBFβ-SMMHC, dimerizes with RUNX1, but rather than repressing its activity, causes an alteration in its activity. The CBFβ-SMMHC/RUNX1 complex could effect altered gene expression by interaction with a novel set of target genes. As has been shown for the related fusion protein AML1-ETO,43 the CBFβ-SMMHC/RUNX1 heterodimer may preferentially associate with different promoter sequences than the CBFβ/RUNX1 heterodimer. It is also possible that the CBFβ-SMMHC/RUNX1 dimer forms a complex with other DNA-binding transcription factors and, through this interaction, is recruited to promoters of novel target genes, which do not require DNA binding by RUNX1. Such DNA-binding–independent activities by RUNX1 have been proposed previously,44,45 which may explain the mechanism of certain RUNX1 mutations in M0 AML samples.46 CBFβ-SMMHC/RUNX1 may also alter RUNX1 activity by recruiting a novel set of cofactors to the promoters of genes normally regulated by CBFβ/RUNX1. It has been shown that, depending on the cofactors recruited, RUNX1 can act as either an activator or a repressor.47 Perhaps the presence of the SMMHC tail influences cofactor binding so that the CBFβ-SMMHC/RUNX1 complex acts as a strong activator of transcription. This model is consistent with our observation that the majority of the gene expression changes observed in Cbfb+/MYH11 embryos were increases in expression. Alternatively, the CBFβ-SMMHC/RUNX1 complex could be recruiting transcriptional repressors that result in more potent repression of target genes than loss of Runx1 activity alone. The loss of expression of key target genes may in turn result in the delayed differentiation and continued expression of genes associated with early progenitors. However, by any of these models, CBFβ-SMMHC is altering RUNX1 gene regulation through a gain-of-function activity, rather than just inhibiting normal RUNX1 function as proposed in earlier models.
It has been shown previously that, before oncogenic transformation, Cbfb-MYH11 causes an accumulation of LKS− cells, termed AMPs.23 The AMPs have similar leukemic potential to the hematopoietic progenitors and stem cells in the LKS+ compartment. We observed a similar accumulation of LKS− cells (Figure 4), which was short lasting (supplemental Figure 6), whereas the previously described AMPs persisted much longer.23 The reason for this difference is not clear, but could be related to the age at which mice were treated with pI:pC. In the current study, we treated older mice (5-8 weeks of age), whereas the previous study used younger mice (3 weeks old), which may lead to different efficiencies of Cre-mediated excision or in different cell compartments. Alternatively, this difference may be due to differences in HSC properties between young and old mice. It has been shown that there is a sudden decrease in HSC self-renewal between 3 and 4 weeks after birth in mice.48 Therefore, it is likely that Cre excision in younger mice would result in a greater pool of HSCs expressing Cbfb-MYH11 and, therefore, a more prolonged differentiation defect.
In this report, we showed that Cbfb-MYH11 expression also leads to an accumulation of abnormal Csf2rb-expressing cells. The Csf2rb+ cells do not directly correspond to the AMPs, as this population contained both LKS+ and LKS− cells. In addition, we found that the Csf2rb+ cells had very low progenitor activity compared with the Csf2rb− cells. Similar to the AMP population, the induced Csf2rb+ cells in preleukemic mice were short lived; they disappeared from the bone marrow and peripheral blood 6 weeks after Cbfb-MYH11 expression. However, after leukemic transformation, most leukemia cells express Csf2rb.
More importantly, we showed that Csf2rb expression could be used to isolate preleukemic progenitors and leukemia-initiating cells from those that are not capable of initiating leukemia. Specifically, the Csf2rb− cells from the lin− fraction of Cbfb+/56M; Mx1-Cre bone marrow, which contain both LKS+ and LKS− cells, were enriched for both preleukemic progenitors, as well as leukemia-initiating cells.
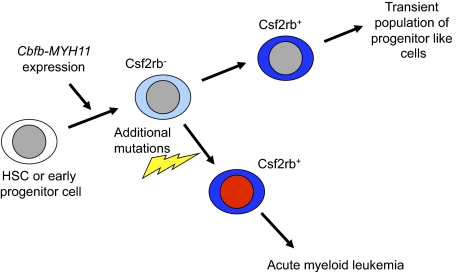
Based on these data, we propose the following model for Cbfb-MYH11–induced leukemogenesis (Figure 7). We propose that expression of Cbfb-MYH11 in HSCs and progenitors induces an abnormal differentiation process in a subset of these cells that culminates in a population of Csf2rb-expressing cells. In the absence of additional mutations, this Csf2rb+ population has low self-renewal capacity and does not have long-term consequences. However, a population of Cbfb-MYH11–expressing, Csf2rb− cells is retained in the bone marrow, likely at a very low frequency. If these Csf2rb− cells, which we have shown include the preleukemic progenitor cells, acquire additional mutations, they follow a similar abnormal differentiation process to produce Csf2rb+ cells (Figures 3B and 6C). However, this population is no longer transient, but expands indefinitely and represents the bulk of leukemia cells.
Figure 7.
Model of Cbfb-MYH11–induced aberrant differentiation and leukemia initiation. We propose that expression of Cbfb-MYH11 in HSCs or early progenitors causes an abnormal and incomplete differentiation that culminates in a Csf2rb+ cell population. Our results indicate that by the time the Cbfb-MYH11–expressing cells have reached the Csf2rb+ stage, they are no longer capable of initiating leukemia.
In summary, we have shown in this study that the fusion gene Cbfb-MYH11 effects a block in differentiation via a novel, Cbfb/Runx1-independent mechanism. Among these abnormally differentiated cells, Csf2rb can be used to isolate cells with leukemia-initiating potential from those that do not. These results have important implications for the study of Cbfb-MYH11–induced leukemogenesis, and the characterization of preleukemic progenitors and leukemia-initiating cells.
Acknowledgments
The authors thank Abdel Elkahloun and the NHGRI microarray core facility for their assistance with microarray hybridization and reading, Chenwei Wang and the NHGRI bioinformatics core facility for their assistance with the microarray analysis, Stephen Wincovitch and the NHGRI Cytogenetics and Fluorescent Microscopy Core for their assistance in imaging cytospin slides, and Lucio Castilla for his insight and advice in the preparation of this paper.
This work was supported by the Intramural Research Program of National Human Genome Research Institute, National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.K.H. designed and performed experiments, analyzed the data, and wrote the paper; Y.K., S.A., M.K., and L.Z. performed experiments and analyzed the data; L.A. performed experiments and P.P.L. designed the experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: P. Paul Liu, 49 Convent Dr, 49/3A26, Bethesda, MD 20892; e-mail: pliu@nhgri.nih.gov.
References
- 1.Le Beau MM, Larson RA, Bitter MA, Vardiman JW, Golomb HM, Rowley JD. Association of an inversion of chromosome 16 with abnormal marrow eosinophils in acute myelomonocytic leukemia: a unique cytogenetic-clinicopathological association. N Engl J Med. 1983;309(11):630–636. doi: 10.1056/NEJM198309153091103. [DOI] [PubMed] [Google Scholar]
- 2.Liu PP, Hajra A, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood. 1995;85(9):2289–2302. [PubMed] [Google Scholar]
- 3.Liu P, Tarlé SA, Hajra A, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261(5124):1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 4.Castilla LH, Garrett L, Adya N, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23(2):144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 5.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 6.Ogawa E, Maruyama M, Kagoshima H, et al. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993;90(14):6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogawa E, Inuzuka M, Maruyama M, et al. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993;194(1):314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13(6):3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang YY, Shi J, Zhang L, et al. Energetic and functional contribution of residues in the core binding factor beta (CBFbeta) subunit to heterodimerization with CBFalpha. J Biol Chem. 2000;275(50):39579–39588. doi: 10.1074/jbc.M007350200. [DOI] [PubMed] [Google Scholar]
- 10.Lukasik SM, Zhang L, Corpora T, et al. Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat Struct Biol. 2002;9(9):674–679. doi: 10.1038/nsb831. [DOI] [PubMed] [Google Scholar]
- 11.Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18(12):7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutterbach B, Hou Y, Durst KL, Hiebert SW. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc Natl Acad Sci U S A. 1999;96(22):12822–12827. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durst KL, Lutterbach B, Kummalue T, Friedman AD, Hiebert SW. The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol Cell Biol. 2003;23(2):607–619. doi: 10.1128/MCB.23.2.607-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9(2):129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84(2):321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93(8):3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Stacy T, Miller JD, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87(4):697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 18.Okada H, Watanabe T, Niki M, et al. AML1(−−) embryos do not express certain hematopoiesis-related gene transcripts including those of the PU.1 gene. Oncogene. 1998;17(18):2287–2293. doi: 10.1038/sj.onc.1202151. [DOI] [PubMed] [Google Scholar]
- 19.Niki M, Okada H, Takano H, et al. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc Natl Acad Sci U S A. 1997;94(11):5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki K, Yagi H, Bronson RT, et al. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci U S A. 1996;93(22):12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokomizo T, Hasegawa K, Ishitobi H, et al. Runx1 is involved in primitive erythropoiesis in the mouse. Blood. 2008;111(8):4075–4080. doi: 10.1182/blood-2007-05-091637. [DOI] [PubMed] [Google Scholar]
- 22.Castilla LH, Wijmenga C, Wang Q, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87(4):687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 23.Kuo YH, Landrette SF, Heilman SA, et al. Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9(1):57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Yagi T, Morimoto A, Eguchi M, et al. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102(5):1849–1856. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
- 27.Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350(16):1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 28.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350(16):1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 29.Gutiérrez NC, Lopez-Perez R, Hernandez JM, et al. Gene expression profile reveals deregulation of genes with relevant functions in the different subclasses of acute myeloid leukemia. Leukemia. 2005;19(3):402–409. doi: 10.1038/sj.leu.2403625. [DOI] [PubMed] [Google Scholar]
- 30.Verhaak RG, Wouters BJ, Erpelinck CA, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94(1):131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Center for Biotechnology Information. Gene Expression Omnibus (GEO). [Accessed March 2009]. http://www.ncbi.nlm.nih.gov/geo.
- 32.Hayashida K, Kitamura T, Gorman DM, Arai K, Yokota T, Miyajima A. Molecular cloning of a second subunit of the receptor for human granulocyte-macrophage colony-stimulating factor (GM-CSF): reconstitution of a high-affinity GM-CSF receptor. Proc Natl Acad Sci U S A. 1990;87(24):9655–9659. doi: 10.1073/pnas.87.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitamura T, Sato N, Arai K, Miyajima A. Expression cloning of the human IL-3 receptor cDNA reveals a shared beta subunit for the human IL-3 and GM-CSF receptors. Cell. 1991;66(6):1165–1174. doi: 10.1016/0092-8674(91)90039-2. [DOI] [PubMed] [Google Scholar]
- 34.Tavernier J, Devos R, Cornelis S, et al. A human high affinity interleukin-5 receptor (IL5R) is composed of an IL5-specific alpha chain and a beta chain shared with the receptor for GM-CSF. Cell. 1991;66(6):1175–1184. doi: 10.1016/0092-8674(91)90040-6. [DOI] [PubMed] [Google Scholar]
- 35.Jordan CT, Upchurch D, Szilvassy SJ, et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia. 2000;14(10):1777–1784. doi: 10.1038/sj.leu.2401903. [DOI] [PubMed] [Google Scholar]
- 36.Kumar S, Minnich MD, Young PR. ST2/T1 protein functionally binds to two secreted proteins from Balb/c 3T3 and human umbilical vein endothelial cells but does not bind interleukin 1. J Biol Chem. 1995;270(46):27905–27913. doi: 10.1074/jbc.270.46.27905. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz J, Owyang A, Oldham E, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23(5):479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 38.Lee ME, Temizer DH, Clifford JA, Quertermous T. Cloning of the GATA-binding protein that regulates endothelin-1 gene expression in endothelial cells. J Biol Chem. 1991;266(24):16188–16192. [PubMed] [Google Scholar]
- 39.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 40.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 41.Orlic D, Anderson S, Biesecker LG, Sorrentino BP, Bodine DM. Pluripotent hematopoietic stem cells contain high levels of mRNA for c-kit, GATA-2, p45 NF-E2, and c-myb and low levels or no mRNA for c-fms and the receptors for granulocyte colony-stimulating factor and interleukins 5 and 7. Proc Natl Acad Sci U S A. 1995;92(10):4601–4605. doi: 10.1073/pnas.92.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miranda-Saavedra D, De S, Trotter MW, Teichmann SA, Gottgens B. BloodExpress: a database of gene expression in mouse haematopoiesis. Nucleic Acids Res. 2009:D873–D879. doi: 10.1093/nar/gkn854. database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okumura AJ, Peterson LF, Okumura F, Boyapati A, Zhang DE. t(8;21)(q22;q22) Fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood. 2008;112(4):1392–1401. doi: 10.1182/blood-2007-11-124735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pabst T, Mueller BU, Harakawa N, et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001;7(4):444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]
- 45.Wheeler JC, VanderZwan C, Xu X, Swantek D, Tracey WD, Gergen JP. Distinct in vivo requirements for establishment versus maintenance of transcriptional repression. Nat Genet. 2002;32(1):206–210. doi: 10.1038/ng942. [DOI] [PubMed] [Google Scholar]
- 46.Cammenga J, Niebuhr B, Horn S, et al. RUNX1 DNA-binding mutants, associated with minimally differentiated acute myelogenous leukemia, disrupt myeloid differentiation. Cancer Res. 2007;67(2):537–545. doi: 10.1158/0008-5472.CAN-06-1903. [DOI] [PubMed] [Google Scholar]
- 47.Satoh Y, Matsumura I, Tanaka H, et al. AML1/RUNX1 works as a negative regulator of c-Mpl in hematopoietic stem cells. J Biol Chem. 2008;283(44):30045–30056. doi: 10.1074/jbc.M804768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowie MB, Kent DG, Dykstra B, et al. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci U S A. 2007;104(14):5878–5882. doi: 10.1073/pnas.0700460104. [DOI] [PMC free article] [PubMed] [Google Scholar]