Abstract
Systemic lupus erythematosus (SLE) is a systemic autoimmune disorder characterized by the presence of auto-antibodies to nuclear antigens, immune complex deposition, and subsequent tissue destruction. Early studies in twins suggested that SLE has, at least in part, a genetic basis, and a role for class II alleles in the major histocompatibility complex has been known for over 30 years. Through both linkage studies and candidate gene studies, numerous additional genetic risk factors have been identified. The recent publication of two SNP-based genome-wide association studies (GWAS) has resulted in the confirmation of a number of previously identified genetic risk loci and has identified new previously unappreciated loci conferring risk for development of SLE. A role for gene copy number variation (CNV) in SLE has also been appreciated through studies of the complement component 4 (C4) loci and more recent work in the IgG Fc receptor loci. The availability of large SNP-based GWAS datasets will undoubtedly lead to the genome-wide analysis and identification of copy number variants related to genetic susceptibility for development of SLE. We review current studies of CNV in SLE susceptibility that include reports of association between SLE and CNV in C4, IgG Fc receptors, TLR7, and CCL3L1.
Introduction
Systemic lupus erythematosus (SLE) is a complex genetic disease characterized by an autoimmune response to nuclear antigens, immune complex deposition, and subsequent tissue destruction. SLE affects approximately 1 in 2,000 people with a female-to-male ratio of 9:1 (Hochberg, 1997a). The American College of Rheumatology has established eleven diagnostic criteria for SLE (four of which must be present for participation in a clinical trial) (Tan et al., 1982; Hochberg, 1997b). These criteria include presence of a malar rash, discoid rash, photosensitivity, oral ulcers, arthritis, serositis (pleuritis, pericarditis), renal disorder (persistent proteinuria, cellular casts), neurologic disorder (seizures, psychoses), hematologic disorder (hemolytic anemia, leukopenia, lymphopenia, thrombocytopenia), immunologic disorder (anti-DNA, anti-Smith, anti-phospholipid antibodies present), and abnormal antinuclear antibody titer. Consequently, the SLE phenotype is extremely heterogeneous with variable phenotypes even among related individuals.
Although the etiology of SLE is still unclear, both genetic and environmental factors are involved. For example medications, ultraviolet radiation exposure, and infections have all been associated with SLE (Sarzi-Puttini et al., 2005). Candidate gene searches for SLE susceptibility have led to multiple findings in various ethnic groups. Strong and consistent association has been observed with class II alleles of the major histocompatibility complex (MHC). In addition, consistent association of non-MHC variants, some of which are functionally relevant, have been reported (Croker and Kimberly, 2005; Gregersen and Behrens, 2006; Brown et al., 2007). Recent genome-wide association studies (GWAS) have confirmed some previously associated loci and revealed new genetic regions related to the disease. A GWAS, conducted by SLEGEN (International Consortium for SLE Genetics), reported association with multiple sites of the MHC and with polymorphisms of interferon regulatory factor 5 (IRF5), integrin alpha M (ITGAM), KIAA1542 (PHRF1), the PX domain containing serine/threonine kinase (PXK), an individual marker on chromosome 1q25, an IgG Fc receptor (FCGR2A), protein tyrosine phosphatase non-receptor type 22 (PTPN22), and signal transducer and activator of transcription 4 (STAT4) (Harley et al., 2008). Another GWAS, conducted by Genentech also reported strong associations in the MHC cluster and the ITGAM-ITGAX region in addition to B lymphoid tyrosine kinase (BLK) (Hom et al., 2008).
One topic currently under intense investigation is the role of copy number variations (CNVs) in the genetic contribution to complex diseases. Copy number polymorphisms arise when an entire gene or gene segment has been duplicated or when a gene is absent in some individuals (Eichler et al., 2007). Extra copies of genes may contribute to overexpression of proteins, and deletion of a gene may lead to a deficiency and functional consequences. Entire gene duplications allow new genes with new functions to evolve while keeping a functional backup copy of the original, ancestral gene (Ohno, 1970).
Copy number variation in SLE
Although the broad significance of CNV in disease has only recently been explored in detail, examples of CNVs associated with SLE have been known for more than a decade. Originally, these CNVs were detected as deficiencies of certain proteins, but recent work has shown that the genes coding for these proteins can have a copy number (CN) >2 (duplication), in addition to a CN <2 (deletion or deficiency) (Table 1).
Table 1.
Known CNVs affecting SLE in humans or similar autoimmune phenotypes in rodent models. CN: copy number.
| Protein | Gene(s) | Species | Chromosome | CNV | Significance |
|---|---|---|---|---|---|
| Complement component 4 | C4A, C4B | Human | 6p21 | 0 to >4 | Deficiency: high risk for SLE; low CN: risk factor for SLE; high CN: protective against SLE |
| Toll-like receptor 7 | Tlr7 | Mouse | XqF5 | >2 | High CN: increased susceptibility to autoimmune phenotype |
| Fcγ receptor 3B | FCGR3B | Human | 1q23 | 0 to >3 | High CN: protective against SLE; low CN: risk factor (SLE and glomerulonephritis) |
| Fcγ receptor 3 | Fcgr3 | Rat | 13q24 | 0 to >3 | Low CN: risk factor for crescentic glomerulonephritis |
CNV has also been associated with autoimmune phenotypes in rodent models that are similar to and used as models of human SLE. Association of the genes that vary in CN with the autoimmune phenotype in murine models is not always recapitulated in human SLE. However, these genes may offer clues to common mechanisms in human SLE and rodent autoimmune phenotypes.
Complement component 4 (C4)
The complement system consists of more than 30 plasma and cell-surface proteins and has many roles in the innate immune system, including general defense against infection, clearance of immune complexes and apoptotic debris, and interfacing with the adaptive immune system (Walport, 2001a). Complement's role in clearance of immune complexes and apoptotic debris is thought to be important in the development of or protection against SLE. Genetic deficiency of complement proteins has been known to predispose to development of SLE (Walport, 2001b; Cook and Botto, 2006). For example, homozygous deletions of the early complement components C1, C2, and C3 are strongly associated with development of an SLE-like phenotype (Walport, 2001b). Furthermore, polymorphisms of several genes for complement proteins associated with SLE have already been reported (Walport, 2006).
Aside from homozygous deletions, copy number variation in the C4 locus, a key component in the classical pathway of the complement system, has been extensively studied in the context of SLE. C4 is involved in clearing of immune complexes, apoptotic cells, and infectious agents (Walport, 2001a). Most people have two C4 genes that code for two variants of C4 (C4A and C4B), both of which are found in the class III region of the MHC on human chromosome 6 (Yang et al., 1999). Deficiency of C4 has been reported in SLE as early as the 1970s (Hauptmann et al., 1974; Schaller et al., 1977; Clark and Klebanoff, 1978). The C4 genes have been shown to vary in CN via duplications and deletions of multi-gene cassettes (including RP1, the C4 genes, a CYP21 pseudogene, and TNX) referred to as RCCX modules (Yang et al., 1999). A recent paper analyzing CNVs of RCCX modules demonstrated that low CN predisposed toward SLE while high CN protected against SLE in a population of European Americans (Yang et al., 2007). It is thought that increased CN of the C4 genes protects against SLE by increasing the clearance of immune complexes and apoptotic cell debris, while decreased CN of the C4 genes is a risk factor for SLE due to decreases in the same processes.
Fcγ Receptor 3B (FCGR3B) and other Fcγ receptors
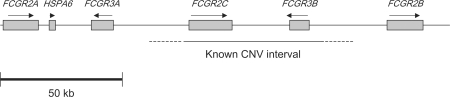
Fcγ receptors (FCGRs) are receptors that bind the Fc domain of IgG antibodies, serving as a link between the adaptive and innate immune response. FCGRs regulate immune responses via tyrosine phosphorylation of their activating or inhibitory cytoplasmic domains or of associated accessory or signaling chains (Nimmerjahn and Ravetch, 2007). Genetic variants of Fc receptor genes, especially those in the classical low affinity Fc receptor cluster on human chromosome 1q23 (Fig. 1), are known to have a role in both susceptibility to and severity of SLE (Croker and Kimberly, 2005; Alarcon et al., 2006; Brown et al., 2007).
Fig. 1.
The human 1q23 classical FCGR gene cluster. Shaded boxes indicate the relative locations and sizes of genes in the cluster, and arrows indicate the direction of transcription. This figure was based on output from the UCSC genome browser (Kent et al., 2002).
FCGR3B codes for the neutrophil-specific, low-affinity FCGR3 that is glycosylphosphatidyl-inositol (GPI) linked to the membrane (Nimmerjahn and Ravetch, 2007). Although it is GPI linked, unlike the other FCGRs which are transmembrane proteins, FCGRB3 is a functional regulator of neutrophil activation (Huizinga et al., 1990a; Hundt and Schmidt, 1992; Edberg and Kimberly, 1994; Kocher et al., 1997). Functional alleles of FCGR3B alter the binding of IgG that subsequently results in different quantitative levels of neutrophil activation in donors homozygous for neutrophil antigen 1 (NA1) versus NA2 (Croker and Kimberly, 2005; Brown et al., 2007). Evidence of increased copy number of FCGR3B was originally suggested through the detection of three allele patterns of the receptor in three individuals (NA1, NA2, and SH, now known as HNA-1a, HNA-1b, and HNA-1c, respectively) (Koene et al., 1998). NA-null individuals have been known for many years, and clinically it can result in isoimmune neutropenia in children born to NA-null mothers (Schnell et al., 1989). The NA1 and NA2 alloantigens have been localized to FcγRIIIb on human neutrophils (Stroncek, 2002; Bux, 2008). Molecular and genetic characterization of individuals homozygous for FCGR3B deficiency was first reported in 1990 in a healthy individual and in a patient with SLE (Clark et al., 1990; Huizinga et al., 1990b). A high copy number of FCGR3B has more recently been reported to be protective against glomerulonephritis and lupus nephritis in humans, while low CN was a risk factor (Aitman et al., 2006). FCGR3B CNV has also been found to be similarly associated with SLE (but not organ specific lupus) and other autoimmune disorders (Fanciulli et al., 2007). The frequencies of the three copy number variant FCGR3B allelesare different across ethnic groups (Kissel et al., 2000), and these differences may contribute to ethnic and geographical differences in SLE (Lau et al., 2006).
While there is no rodent homologue of FCGR3B, rodent Fcgr3 may be considered to be a functional homologue of the human gene. Fcgr3 is expressed in neutrophils and participates in neutrophil activation (Coxon et al., 2001), and CNV of Fcgr3 has been reported to be associated with risk for glomerulonephritis in rats (Aitman et al., 2006).
CNV of FCGR2C has been reported to be linked to CNV of FCGR3B (de Haas et al., 1995) and to predispose to idiopathic thrombocytopenic purpura (Breunis et al., 2008). However, CNV of the other FCGR genes in SLE remains unexplored. Further characterization of the FCGR gene cluster on human chromosome 1q23 and their relationship to SLE promises to be a fruitful area for future studies.
The Yaa locus and toll-like receptor 7 (TLR7)
The BSXB strain of mice spontaneously develops an autoimmune phenotype similar to human SLE, which includes production of anti-nuclear antibodies. Unlike human SLE, the autoimmune phenotype was enhanced in males rather than females. This observation led to the discovery of a Y-linked autoimmune accelerator locus, referred to as Yaa, which increases the severity of the autoimmune phenotype in this mouse model (Murphy and Roths, 1979; Izui et al., 1995).
While the identity of the Yaa gene is not known, CNVs of Yaa were recently reported and duplications of Yaa were shown to further increase susceptibility to autoimmunity in mice (Pisitkun et al., 2006). The authors attempted to identify the causative gene by quantifying mRNA and genomic DNA within the Yaa locus. Of the genes with amplified mRNA and genomic DNA, Pisitkun et al. (2006) proposed Tlr7 as the causative gene because of its biological properties including the binding of RNA. B-cell hyperactivity has been previously reported in Yaa mice. The authors also demonstrated that Tlr7 interacts with Bruton's tyrosine kinase (Btk) and that Btk signaling is necessary for the autoimmune response to nucleolar antigens in Yaa mice. Tlr7 is part of a family of the toll-like receptors that recognize nucleic acids and nucleic acid-like structures (Takeda et al., 2003). It is possible that Tlr7 has some reactivity to nuclear material from apoptotic debris and that increased CN of Tlr7 would cause increased autoimmune response to nuclear material.
CNVs of the human TLR7 gene have recently been examined in SLE, but no association between TLR7 CN and the SLE phenotype was observed (Kelley et al., 2007). It is possible that Tlr7 is not the autoimmunity-predisposing gene in the Yaa locus. The Tlr8 gene is contiguous to Tlr7 and is therefore within the same interval identified by Pisitkun et al. (2006). It is also possible that Tlr7 is the causative gene in the Yaa locus and that this is an example of a disparity between the genetics of human SLE and murine models of autoimmunity (Mestas and Hughes, 2004; Vyse, 2008).
Other CNV loci in SLE
It is likely that CNVs in other loci will be important contributors to both susceptibility to and severity of SLE and other autoimmune diseases. Indeed, recent studies in Crohn's disease, an autoimmune inflammatory disease of the gastrointestinal tract, have demonstrated a role for CNV in the β-defensin gene cluster on human chromosome 8 (Fellermann et al., 2006). The defensins are endogenous antibiotic and cytotoxic peptides that form a chemical barrier at the epithelial surface, and their relative deficiency may lead to bacterial adherence to the mucosa, slow invasion, and secondary mucosal inflammation. The chromosomal region containing the defensin genes is characterized by segmental duplication, and there is extensive CNV in the defensin genes. Patients with Crohn's disease are characterized as having lower copy numbers of the defensin genes suggesting a functional role for these genes in the development of the disease (Fellermann et al., 2006). In SLE, a recent report has suggested that CNV in the chemokine (C-C motif) ligand 3-like 1 (CCL3L1) locus is associated with risk of disease development (Mamtani et al., 2008). In summary, there are already tantalizing clues that CNVs will be important in understanding the genetic basis of SLE and other autoimmune phenotypes.
Current genome-wide association studies and CNVs
Although impressive progress has been made in defining candidate CNVs associated with SLE, there are still many open questions and technical challenges. At present, given the availability of two independent GWAS in SLE using single nucleotide polymorphisms (SNPs), the use of the GWAS data for analysis of CNV is most promising. CNVs can be detected through either direct scoring or through identification of SNP markers known to be in linkage disequilibrium with CNVs. While there are certainly many methodological issues to be overcome in doing such analyses, these data would permit the first genome-wide assessment of CNVs in SLE. Early successes in using SNP-based arrays for assessment of CNVs in complex genetic phenotypes include recent reports in amyotrophic lateral sclerosis (ALS) (Blauw et al., 2008; Cronin et al., 2008), schizophrenia (Stefansson et al., 2008), ischemic stroke (Matarin et al., 2008), and autism (Marshall et al., 2008). While the currently available GWAS in SLE have not yet reported CNV data, it is likely that such analyses will be completed soon and will yield additional insights into the role of genetic variants in the development and prognosis of SLE.
Significance of CNVs in basic and clinical SLE studies
There are a number of different mechanisms by which CNVs could be causative variants in disease susceptibility and severity (Beckmann et al., 2007). CNVs might have functional consequences through direct structural disrupting/alteration of gene(s). Certainly, CNV would be expected to alter gene dosage, and this might amplify the impact of SNP variants that may or may not occur in regions of CNV. For example, it is well established that CCL3L1 gene dose (i.e., CNV) and CCR5 genotype combinations have a substantial impact on cellular immunity, HIV-AIDS pathogenesis, and responsiveness to antiretroviral therapy in patients with HIV-AIDS (Dolan et al., 2007; Ahuja et al., 2008). In SLE, similar interactions between CCL3L1 CNV and CCR5 genotypes have been suggested (Mamtani et al., 2008). It is also interesting to note that CNVs in CCL3L1 may also be associated with other autoimmune diseases such as rheumatoid arthritis (McKinney et al., 2008).
Another area of potential interest for the role of CNVs is drug response. As suggested by the example of CCL3L1 and antiretroviral treatment in HIV-AIDS (Gonzalez et al., 2005; Ahuja et al., 2008), it is likely that there are CNVs that will influence treatment of SLE and other autoimmune diseases. One provocative example could be the use of intravenous immunoglobulins (IVIGs) as a treatment for immune deficiency, autoimmune, and inflammatory disorders (Negi et al., 2007). Aside from the expense, a significant problem in its clinical use is the variability in its therapeutic efficacy in different individuals. Mechanisms of IVIG's immunoregulatory effects are complicated and still remain unclear, although recent studies suggest a significant role for IgG Fc receptors. Both FCGR2B and FCGR3A have been proposed as key factors in the mechanism of action of IVIG's anti-inflammatory properties (Samuelsson et al., 2001; Siragam et al., 2006; Park-Min et al., 2007). In this context, recently identified CNVs in FCGR3A and FCGR2C (Breunis et al., 2008) and potential CNVs in other FCGR genes may result in different levels of FCGR protein expression in different individuals, thus enhancing or reducing regulatory functions of IVIG.
Conclusion
Demonstration of the importance of CNVs in SLE will undoubtedly yield new insights into the pathogenic mechanisms in SLE and other complex autoimmune diseases. Although remarkable progress has been made in both initial GWASs and target gene studies in SLE, there are still significant gaps in our understanding of the genetic basis of the disease. Further, more dense screens in multiple ethnic groups will be a necessary follow-up to the initial GWAS successes in SLE. It is still not known if there will be common CNV variants that associate with disease and/or whether there will be less common/rare variants that are found in affected individuals as observed in the recent genome-wide CNV analysis in ALS (Blauw et al., 2008). Current and future CNV candidate regions/genes will require methodological development for independent replication studies. Finally, integrating our knowledge about SNPs and CNVs will be required to gain a complete understanding of the genetic basis of SLE and related autoimmune diseases.
Acknowledgements
We thank Dr. Robert P. Kimberly for his continued support.
Footnotes
This work was supported by grants from the National Institutes of Health RO1-AR33062, RO1-AR42476, PO1-AR49084, U54-RR02577, and MO1-RR00032.
References
- Ahuja SK, Kulkarni H, Catano G, Agan BK, Camargo JF, et al. CCL3L1-CCR5 genotype influences durability of immune recovery during antiretroviral therapy of HIV-1-infected individuals. Nat Med. 2008;14:413–420. doi: 10.1038/nm1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Alarcon GS, McGwin G, Jr, Petri M, Ramsey-Goldman R, Fessler BJ, et al. Time to renal disease and end-stage renal disease in PROFILE: a multiethnic lupus cohort. PLoS Med. 2006;3:1949–1956. doi: 10.1371/journal.pmed.0030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- Blauw HM, Veldink JH, van Es MA, van Vught PW, Saris CG, et al. Copy-number variation in sporadic amyotrophic lateral sclerosis: a genome-wide screen. Lancet Neurol. 2008;7:319–326. doi: 10.1016/S1474-4422(08)70048-6. [DOI] [PubMed] [Google Scholar]
- Breunis WB, van Mirre E, Bruin M, Geissler J, de Boer M, et al. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. 2008;111:1029–1038. doi: 10.1182/blood-2007-03-079913. [DOI] [PubMed] [Google Scholar]
- Brown EE, Edberg JC, Kimberly RP. Fc receptor genes and the systemic lupus erythematosus diathesis. Autoimmunity. 2007;40:567–581. doi: 10.1080/08916930701763710. [DOI] [PubMed] [Google Scholar]
- Bux J. Human neutrophil alloantigens. Vox Sang. 2008;94:277–285. doi: 10.1111/j.1423-0410.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- Clark MR, Liu L, Carkson SB, Ory PA, Golstein IM. An abnormality of the gene that encodes neutrophil Fc receptor III in a patient with systemic lupus erythematosus. J Clin Invest. 1990;86:341–346. doi: 10.1172/JCI114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RA, Klebanoff SJ. Role of the classical and alternative complement pathways in chemotaxis and opsonization: studies of human serum deficient in C4. J Immunol. 1978;120:1102–1108. [PubMed] [Google Scholar]
- Cook HT, Botto M. Mechanisms of disease: the complement system and the pathogenesis of systemic lupus erythematosus. Nat Clin Pract Rheum. 2006;2:330–337. doi: 10.1038/ncprheum0191. [DOI] [PubMed] [Google Scholar]
- Coxon A, Cullere X, Knight S, Sethi S, Wakelin MW, et al. FcγRIII mediates neutrophil recruitment to immune complexes: a mechanism for neutrophil accumulation in immune-mediated inflammation. Immunity. 2001;14:693–704. doi: 10.1016/s1074-7613(01)00150-9. [DOI] [PubMed] [Google Scholar]
- Croker JA, Kimberly RP. Genetics of susceptibility and severity in systemic lupus erythematosus. Curr Opin Rheumatol. 2005;17:529–537. doi: 10.1097/01.bor.0000169360.15701.27. [DOI] [PubMed] [Google Scholar]
- Cronin S, Berger S, Ding J, Schymick JC, Washecka N, et al. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet. 2008;17:769–774. doi: 10.1093/hmg/ddm361. [DOI] [PubMed] [Google Scholar]
- de Haas M, Kleijer M, van Zwieten R, Roos D, von dem Borne AE. Neutrophil FcγRIIIb deficiency, nature and clinical consequences: a study of 21 individuals from 14 families. Blood. 1995;86:2403–2412. [PubMed] [Google Scholar]
- Dolan MJ, Kulkarni H, Camargo JF, He W, Smith A, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. [DOI] [PubMed] [Google Scholar]
- Edberg JC, Kimberly RP. Modulation of Fcγ and complement receptor function by the glycosyl-phosphatidylinositol-anchored form of FcγRIII. J Immunol. 1994;152:5826–5835. [PubMed] [Google Scholar]
- Eichler EE, Nickerson Da, Altshuler D, Bowcock AM, Brooks LD, et al. Completing the map of human genetics variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Gregersen PK, Behrens TW. Genetics of autoimmune diseases – disorders of immune homeostasis. Nat Rev Genet. 2006;7:917–928. doi: 10.1038/nrg1944. [DOI] [PubMed] [Google Scholar]
- Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptmann G, Grosshans E, Heid E. Lupus erythematosus syndrome and complete deficiency of the fourth component of complement. Boll Ist Sieroter Milan. 1974;53:228. [PubMed] [Google Scholar]
- Hochberg MC. The epidemiology of systemic lupus erythematosus. In: Wallace DJ, Hahn BH, editors. Dubois' Lupus Erythematosus. Baltimore: Williams and Wilkins; 1997a. pp. 49–65. [Google Scholar]
- Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997b:40–1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- Huizinga TW, Dolman KM, van der Linden NJ, Kleijer M, von dem Borne AE, et al. Phosphatidylinositol-linked FcrIII mediates exocytosis of neutrophil granule proteins but does not mediate initiation of the respiratory burst. J Immunol. 1990a;144:1432–1437. [PubMed] [Google Scholar]
- Huizinga TW, Kuijpers RW, Kleijer M, Schulpen TW, Cuypers HT, et al. Maternal genomic neutrophil FcRIII deficiency leading to neonatal isoimmune neutropenia. Blood. 1990b;76:1927–1932. [PubMed] [Google Scholar]
- Hundt M, Schmidt RE. The glycosylphosphatidylinositol-linked Fc receptor III represents the dominant receptor structure for immune complex activation of neutrophils. Eur J Immunol. 1992;22:811–816. doi: 10.1002/eji.1830220327. [DOI] [PubMed] [Google Scholar]
- Izui S, Iwamoto M, Fossati L, Merino R, Takahashi S, Ibnou-Zekri N. The Yaa gene model of systemic lupus erythematosus. Immunol Rev. 1995;144:137–156. doi: 10.1111/j.1600-065x.1995.tb00068.x. [DOI] [PubMed] [Google Scholar]
- Kelley J, Johnson MR, Alarcon GS, Kimberly RP, Edberg JC. Variation in the relative copy number of the TLR7 gene in patients with systemic lupus erythematosus and healthy control subjects. Arthritis Rheum. 2007;56:3375–3378. doi: 10.1002/art.22916. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. The human genome browswer at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel K, Hofmann C, Gittinger FS, Daniels G, Bux J. HNA-1a, HNA-1b and HNA-1c (NA1, NA2, SH) frequencies in African and American blacks and Chinese. Tissue Antigens. 2000;56:143–148. doi: 10.1034/j.1399-0039.2000.560205.x. [DOI] [PubMed] [Google Scholar]
- Kocher M, Siegel ME, Edberg JC, Kimberly RP. Cross-linking of Fc gamma receptor IIa and Fc gamma receptor IIIb induces different proadhesive phenotypes on human neutrophils. J Immunol. 1997;159:3940–3948. [PubMed] [Google Scholar]
- Koene HR, Kleijer M, Roos D, de Haas M, Von dem Borne AE. FcγRIIIB gene duplication: evidence for presence and expression of three distinct FcγRIIIB genes in NA(1+,2+)Sh(+) individuals. Blood. 1998;91:673–679. [PubMed] [Google Scholar]
- Lau CS, Yin G, Mok MY. Ethnic and geographical differences in systemic lupus erythematosus: an overview. Lupus. 2006;15:715–719. doi: 10.1177/0961203306072311. [DOI] [PubMed] [Google Scholar]
- Mamtani M, Rovin B, Brey R, Camargo JF, Kulkarni H, et al. CCL3L1 gene-containing segmental duplications and polymorphisms in CCR5 affect risk of systemic lupus erythematosus. Ann Rheum Dis. 2008;67:1076–1083. doi: 10.1136/ard.2007.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarin M, Simon-Sanchez J, Fung HC, Scholz S, Gibbs R, et al. Structural genomic variation in ischemic stroke. Neurogenetics. 2008;9:101–108. doi: 10.1007/s10048-008-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Merriman ME, Chapman PT, Gow PJ, Harrison AA, et al. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67:409–413. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Murphy ED, Roths JB. A Y chromosome associated factor in strain BXSB producing accelerated autoimmunity and lymphoproliferation. Arthritis Rheum. 1979;22:1188–1194. doi: 10.1002/art.1780221105. [DOI] [PubMed] [Google Scholar]
- Negi VS, Elluru S, Siberil S, Graff-Dubois S, Mouthon L, et al. Intravenous immunoglobulin: an update on the clinical use and mechanisms of action. J Clin Immunol. 2007;27:233–245. doi: 10.1007/s10875-007-9088-9. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Fc-receptors as regulators of immunity. Adv Immunol. 2007;96:179–204. doi: 10.1016/S0065-2776(07)96005-8. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- Park-Min KH, Serbina NV, Yang W, Ma X, Krystal G, et al. FcgRIII-dependent inhibition of interferon-gamma responses mediates suppressive effects of intravenous immune globulin. Immunity. 2007;26:67–78. doi: 10.1016/j.immuni.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Pisitkun P, Dean JA, Difilippantonio MJ, Tarasenko T, Satterthwait AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- Samuelsson A, Towers TL, Ravetch JV. Anti-inflammatory activity of IVIG mediated through the inhibitory Fc receptor. Science. 2001;291:484–486. doi: 10.1126/science.291.5503.484. [DOI] [PubMed] [Google Scholar]
- Sarzi-Puttini P, Atzeni F, Iaccarino L, Doria A. Environment and systemic lupus erythematosus: an overview. Autoimmunity. 2005;38:465–472. doi: 10.1080/08916930500285394. [DOI] [PubMed] [Google Scholar]
- Schaller JG, Gilliland BG, Ochs HD, Leddy JP, Agodoa LC, Rosenfeld SI. Severe systemic lupus erythematosus with nephritis in a boy with deficiency of the fourth component of complement. Arthritis Rheum. 1977;20:1519–1525. doi: 10.1002/art.1780200812. [DOI] [PubMed] [Google Scholar]
- Schnell M, Halligan G, Herman J. A new granulocyte antibody directed at a high-frequency antigen causing neonatal alloimmune neutropenia. Transfusion. 1989;29:46S. [Google Scholar]
- Siragam V, Crow AR, Brinc D, Song S, Freedman J, Lazarus AH. Intravenous immunoglobulin ameliorates ITP via activating Fc gamma receptors on dendritic cells. Nat Med. 2006;12:688–692. doi: 10.1038/nm1416. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroncek D. Neutrophil alloantigens. Transfus Med Rev. 2002;16:67–75. doi: 10.1053/tmrv.2002.29406. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Akria S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- Vyse TJ. Understanding lupus: fishing genes out of mice and men. Immunity. 2008;28:8–10. doi: 10.1016/j.immuni.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Walport MJ. Complement, first of two parts. N Engl J Med. 2001a;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- Walport MJ. Complement, second of two parts. N Engl J Med. 2001b;344:1140–1144. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- Walport MJ. Complement and systemic lupus erythematosus. Arthritis Res. 2006;4:S279–S293. doi: 10.1186/ar586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–1054. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Mendoza AR, Welch TR, Zipf WB, Yu CY. Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module) J Biol Chem. 1999;274:12147–12156. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]