Abstract
Previous studies have revealed a major difference in the phylogenetic structure, extent of genetic diversity, and selection pressure between the surface glycoproteins and internal gene segments of avian influenza viruses (AIV) sampled from wild birds. However, what evolutionary processes are responsible for these strikingly different evolutionary patterns is unclear. To address this issue, we estimated the rate of evolutionary change and time of origin of each segment of AIV sampled globally. Strikingly, the internal segments of the sampled AIV strains possess common ancestors that existed less than 200 years ago. Similarly recent times of origin were observed for each of the individual subtypes within the HA, NA, and NS gene segments. Such a shallow history of genetic diversity suggests an evolutionary model in which the genetic structure of AIV is shaped by a combination of occasional selective sweeps in the HA and NA (and possibly NS) segments, coupled with transient genetic linkage to the internal gene segments.
Keywords: Avian influenza virus, Phylogeny, Hitchhiking, Reassortment, Natural selection
Introduction
Although influenza viruses are likely to have circulated in avian populations for millennia, a detailed description of these infectious agents has only been achieved recently. The first recorded outbreak of highly pathogenic avian influenza (HPAI) was in Italy in 1878 (Perroncito 1878), and then referred to as “fowl plague.” The causative agent of these disease outbreaks was not identified as influenza virus until 1955 (Schäfer 1955). The intensive surveillance of avian influenza viruses (AIVs) in wild birds did not begin in earnest until the 1970s (Easterday et al. 1968; Slepuskin et al. 1972; Slemons et al. 1974; Romváry et al. 1976; Fukumi et al. 1977), and since then a consensus has been reached that some species of wild aquatic birds are the natural reservoir of influenza A viruses (Webster et al. 1992). However, even given the current interest in AIVs, particularly their potential threat to human health, their long-term epidemiological dynamics and population history remain uncertain, especially at the global scale.
Recent research based on a combination of extensive surveillance and large-scale genome sequence analyses has provided important new insights into the patterns and processes of AIV evolution. First, a major geographic segregation is observed between viruses associated with bird species that utilize the Eurasia/Australia and North America flyways, respectively (Donis et al. 1989; Ito et al. 1991; Spackman et al. 2005), although some intercontinental genetic exchanges are observed on occasion (Dugan et al. 2008; Krauss et al. 2007; Widjaja et al. 2004). This geographical subdivision is reflected in a major phylogenetic split in trees representing five “internal” segments (PB2, PB1, PA, NP, and M) and in most, if not all, individual subtypes of phylogenies inferred for the more diverse HA, NA, NS segments (Dugan et al. 2008). Second, and more intriguing, the different segments of AIV exhibit two markedly different evolutionary patterns. For the two surface glycoprotein and major viral antigens, the hemagglutinin (HA) and neuraminidase (NA), there are multiple antigenically and phylogenetically distinct “subtypes” (16 and 9, respectively). A similar pattern is seen in the nonstructural segment (NS), in which the NS1 protein plays a role in evading host innate immunity (Baigent and McCauley 2003; Zhirnov and Klenk 2007; Li et al. 2006), and which is characterized by the highly divergent A and B alleles (Ludwig et al. 1991). The subtypes/alleles of these three segments differ substantially at the genetic level (e.g., the average inter-subtype amino acid pairwise identities are 45.5% for HA and 43.6% for NA; Dugan et al. 2008), and exhibit a relatively high ratio of nonsynonymous to synonymous substitutions per site for AIV (ratio dN/dS, range 0.14 to 0.16; Obenauer et al. 2006). In contrast, far shallower genetic diversity is observed within the five internal segments (with average pairwise identities ranging from 95 to 99%) and dN/dS values are considerably lower (0.03 to 0.04; Obenauer et al. 2006) than those seen in HA, NA, and NS, indicative of strong selective constraints (Dugan et al. 2008). That genetically similar internal gene segments can be associated with a diverse array of highly divergent HA and NA subtypes illustrates the high frequency of reassortment in AIV, such that the internal gene segments can be thought of as comprising a single gene pool (Dugan et al. 2008).
Less clear, however, are the evolutionary processes that are responsible for these very different evolutionary patterns. Given that all eight segments share broadly similar rates of nucleotide substitution, and particularly at synonymous sites (Bahl et al. 2009; Chen and Holmes 2006; see “Results and discussion” section), the dramatic differences in tree length suggest that the two groups of segments differ greatly in their divergence times, manifest in estimates of the Time to the Most Recent Common Ancestor (TMRCA). Indeed, as the origin of current circulating HA subtypes has been estimated to be within the last 3000 years using a variety of methods (Suzuki and Nei 2002; Chen and Holmes 2006), the TMRCAs for each of the five internal segments would then be remarkably recent. It is therefore important to determine what processes can explain such recent evolution, and also why the HA, NA, and NS segments have such different times to common ancestry compared to the five internal segments, particularly given the geographic segregation of the Eurasia/Australia and North America lineages. Answering these questions may help determine what factors control the transmission and establishment of viral lineages in new geographical localities.
In order to explain the discrepant evolutionary patterns of the two groups of gene segments, we estimated the time of common ancestry of each segment of AIV. From these data, we suggest that a complex combination of natural selection, genetic hitchhiking, and reassortment is responsible for the differing evolutionary patterns among the two groups of gene segments.
Materials and Methods
Sequence Data
Data sets of complete sequences from all eight genome segments of AIVs of all subtypes were downloaded from the NCBI influenza virus resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html). To avoid additional mutations caused by extensive passage, sequences obtained before 1970 were excluded. Also, to minimize biases in the estimation of substitution rates, HPAI H5N1 sequences were removed from the analysis, as it has previously been shown that these viruses evolve more rapidly than the Low Pathogenic Avian Influenza (LPAI) viruses (Chen and Holmes 2006; Vijaykrishna et al. 2008). As previous studies suggest that AIVs tend to form short-term local clusters without a significant species effect (Chen and Holmes 2009), identical sequences and those sampled from the same sample year and place were removed for computational tractability. In several cases where the sample size was still too large for a viable analysis, a number of other sequences were deleted randomly. This pruning process resulted in final data sets of following size: PB2 = 169 sequences; PB1 = 182; PA = 169; HA = 310; NP = 170; NA = 270; M = 180; NS = 180. For each segment, sequence alignments were created using MUSCLE (Edgar 2004) and adjusted manually using Se-Al (http://tree.bio.ed.ac.uk/software/seal/) according to the amino acid sequence. In the case of the HA and NA, some regions of the inter-subtype sequence alignment were extremely divergent such that they could not be aligned with certainty, in particular the HA signal peptide, the cleavage site insertions in HPAI H7, and the variable small stalk deletions in NA. To avoid any phylogenetic error due to mis-alignment, these small regions of ambiguity were deleted. This resulted in the following sequence alignments used for evolutionary analysis: PB2 = 2277 nt; PB1 = 2271 nt; PA = 2148 nt; HA = 1683 nt; NP = 1494 nt; NA = 1257 nt; MP = 979 nt; NS = 835 nt. All sequence alignments are available from the authors on request.
Evolutionary Analysis
We estimated the TMRCA for each segment data set using a Bayesian MCMC approach available in the BEAST program (v.1.4.7) (http://evolve.zoo.ox.ac.uk/Beast/; Drummond and Rambaut 2007). To be as robust as possible in our estimates of TMRCA, three sets of evolutionary models were used in each case, with the best-fit model determined using Bayes Factors on log likelihoods or inspection of the coefficient of variation (CoV) to asses the clock-like nature of evolutionary change (with CoV values > 0 indicating non-clock-like evolution). These three models were (i) the GTR + Γ4 substitution model under a strict molecular clock, (ii) the GTR + Γ4 substitution model under a relaxed (unrelated lognormal) molecular clock, and (iii) a model with different substitution rates for each of the three codon positions and a GTR substitution matrix incorporating a relaxed (unrelated lognormal) molecular clock. In all cases, we employed the Bayesian skyline population coalescent prior as this obviously best describes the fluctuating population dynamics characteristic of influenza virus (Rambaut et al. 2008). In each case, MCMC chains were run for sufficient time (ranging from 100 to 200 million generations) to achieve convergence (assessed using the TRACER program; http://www.evolve.zoo.ox.ac.uk), with uncertainty in parameter estimates reflected in the 95% highest probability density (HPD). The Maximum Clade Credibility (MCC) tree across all plausible trees was then computed from the BEAST trees using the TreeAnnotator program, with the first 10% trees removed as burn-in. To assess the reliability of our estimates of both the substitution rate and TMRCA, and to determine the extent of temporal structure in the AIV sequence data, we performed a regression analysis of tree root-to-tip genetic distance against sampling date using the program Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen/; Drummond et al. 2003) based on maximum likelihood (ML) trees for each segment.
Finally, we estimated ML trees for each segment using all AIV sequences available in GenBank (~2000 for each segment) and the PAUP* (version 4b10-MacOsX) package (Swofford 2003). Because of the very large number of sequences in each segment, NNI branch-swapping was utilized. In each case, the best-fit nucleotide substitution model was determined by MODELTEST (Posada and Crandall 1998) (parameter values available from the authors on request).
Results and Discussion
Age of Genetic Diversity
We present the most comprehensive analysis of the timescale of avian influenza virus evolution conducted to date. Inspection of the CoV revealed that strict clock-like evolution was rejected for each segment (Table 1). We therefore based all our results on a relaxed (uncorrelated lognormal) molecular clock. Likewise, the evolution of all segments was best described by a substitution model that did not partition the sequence alignment by codon position (results available from the authors on request). Similar to previous studies (Bahl et al. 2009; Chen and Holmes 2006), the LPAI viruses (mostly sampled from wild birds) exhibit relatively rapid rates of evolutionary change, ranging from 1.55 × 10−3 to 2.38 × 10−3 nucleotide substitutions per site, per year (subs/site/year). The evolutionary rates for each segment, as well as their 95% HPD values, are shown in Table 1.
Table 1.
Rates of nucleotide substitution and TMRCA of each segment of AIV
| Segment | CoV | Substitution rate (subs/site/year) × 10−3 | TMRCA (calendar year) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Median | 95% HPD lower | 95% HPD upper | Mean | Median | 95% HPD lower |
95% HPD upper |
||
| PB2 | 0.46 (0.40–0.51) | 2.10 | 2.10 | 1.90 | 2.29 | 1918 | 1917 | 1935 | 1900 |
| PB1 | 0.45 (0.38–0.52) | 2.10 | 2.10 | 1.90 | 2.31 | 1889 | 1892 | 1927 | 1848 |
| PA | 0.50 (0.43–0.57) | 2.02 | 2.02 | 1.83 | 2.23 | 1911 | 1913 | 1935 | 1885 |
| HA | 0.38 (0.33–0.42) | 2.23 | 2.23 | 1.98 | 2.49 | 897 | 908 | 1169 | 598 |
| NP | 0.46 (0.39–0.53) | 2.06 | 2.05 | 1.81 | 2.34 | 1880 | 1881 | 1914 | 1839 |
| NA | 0.35 (0.29–0.40) | 2.38 | 2.38 | 2.05 | 2.72 | 737 | 753 | 1053 | 396 |
| M | 0.53 (0.43–0.63) | 1.55 | 1.52 | 1.33 | 1.77 | 1907 | 1910 | 1934 | 1870 |
| NS | 0.72 (0.61–0.84) | 1.76 | 1.76 | 1.28 | 2.23 | 1450 | 1493 | 1748 | 1057 |
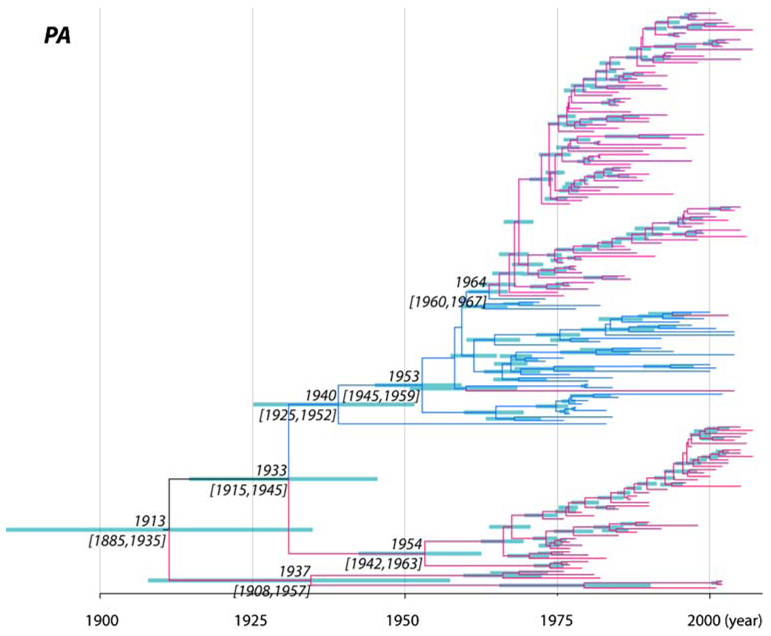
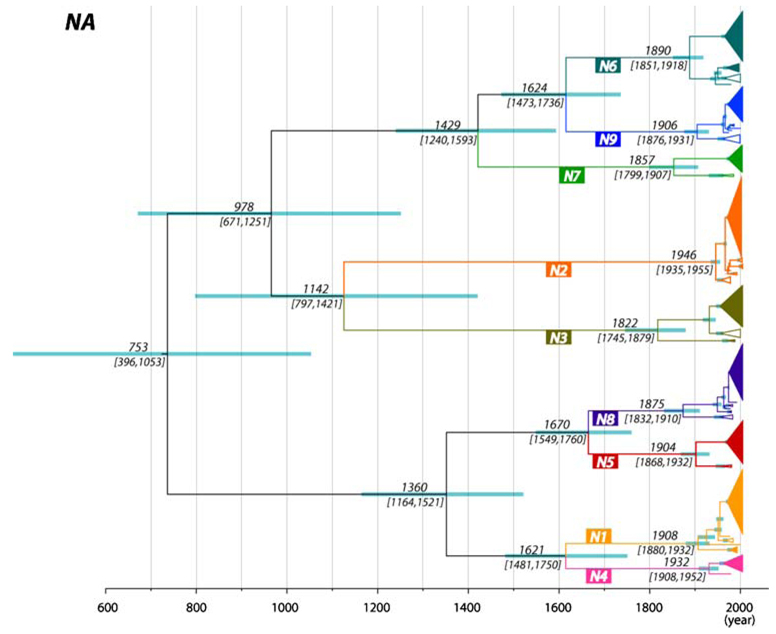
Another notable result, and one that also supports previous analyses, was that the root ages (i.e., TMRCAs) of the eight segment trees show two contrasting patterns. First, for the PB2/PB1/PA/NP/M segments, the mean TMRCAs of the sampled AIVs was only approximately 100 to 130 years before the present (range of 95% HPD values = 75–170 years; the example of the PA segment is shown in Fig. 1), placing their ancestry at the end of the nineteenth and start of the twentieth centuries (Table 1). Indeed, it is striking that the maximum age of any internal segment—that is, the oldest 95% HPD value—was only 170 years. In contrast, the mean TMRCAs for the HA/NA/NS segments were much older, namely 1097 (95% HPD = 840–1411 years), 1256 (95% HPD = 956– 1613 years), and 516 (95% HPD = 261–952 years) years before the present, respectively. Importantly, the individual subtypes (or alleles) of the HA/NA/NS segments have times of origin similar to those of the five internal segments, with the median TMRCAs ranging from the calendar years 1838 to 1976 (HA), 1822 to 1946 (NA), and 1907 to 1934 (NS) (the example of the NA is shown in Fig. 2; all other segments are shown in Supplementary Figs. 1–6). Very similar estimates for the substitution rate and TMRCA were observed using simple root-to-tip regression. Results for the PA segment are shown here (Fig. 3), whereas others are available as Supplementary Figs. 7–13. These regression analyses therefore confirm both that there is sufficient temporal structure within the AIV data to estimate rates and dates, and that our estimates of substitution rate are not merely a function of the particular coalescent model used.
Fig. 1.
MCC tree of the PA segment of AIV. Branches shaded pinkrepresent those from the Americas, whereas blue shaded samples represent those from Eurasia and Australia. The median node ages and their 95% HPD values are shown on the major nodes, with 95% HPD values shown as node bars
Fig. 2.
MCC tree of the NA segment of AIV. Subtypes are represented by different colors, with geographic clusters collapsed and depicted as triangles when possible. The filled triangles represent North America strains, whereas hollow triangles represent Eurasia/Australian strains. The median node ages and their 95% HPD values are shown on the major nodes, with 95% HPD values shown as node bars
Fig. 3.
Regression of root-to-tip distances against sampling date for the PA segment of AIV. The inferred rate of nucleotide substitution (slope), time to common ancestry (intercept), and correlation coefficient (r2) are also shown
Finally, it is intriguing that all segments other than H14 and H15 of the HA/NA/NS subtypes depict the geographic subdivision between the Eurasian/Australian and North American lineages, indicating that all intercontinental transmission events also occurred recently, and clearly after the separation of each individual subtype. Overall, all these analyses are indicative of a rapid epidemic turnover (Chen and Holmes 2006).
A Model of AIV Evolution
How can we explain these very different times of origin of the two groups of segments? One possibility is that it is the particular genome constellation of the five internal segments that was selected for, which then experienced an essentially global selective sweep approximately 100 years ago. Indeed, it is striking that the TMRCAs for the five internal segments are so similar, with strongly overlapping 95% HPD values. However, it is difficult to imagine how a particular combination of internal segments could be so selectively favored as to out-compete all other circulating variants, and in a wide array of avian species from diverse geographical locations, particularly given the low dN/dS values that usually characterize these segments (Obenauer et al. 2006; Suzuki 2006; Chen and Holmes 2006). It is therefore unlikely that the fixation of the five internal segments was due to a single event.
Perhaps the most telling insight into the process of influenza virus evolution is the general similarity in times of origin between the five internal segments and the individual subtypes of HA/NA/NS. This suggests that the fixation of the internal segments may be linked to the emergence and establishment of some, if not all, of the HA/NA/NS subtypes. In particular, the rapid fixation of the five internal segments may be caused by a process of hitchhiking to advantageous (and presumably antigenic) mutations in the HA and/or NA glycoproteins. Such a model has previously been proposed for the fixation of mutations in the M2 protein of human H3N2 influenza A virus that confer resistance to adamantane drugs (Simonsen et al. 2007).
We therefore propose that the genetic structure of AIV, and particularly the differing times of origin of the segments, is the result of a combination of occasional selective sweeps in the HA and NA, with transient genetic linkage to the internal gene segments. This process is represented schematically in Fig. 4. Accordingly, when a mutation in a specific HA and/or NA is selectively favored such that it spreads rapidly through the population, the frequency of the internal segments in that particular strain will also increase in frequency due to hitchhiking (i.e., that there is some degree of physical linkage). As the outbreak progresses, these advantageous strains will regularly co-infect animals carrying influenza viruses of different genome constellations, a process that seems to be a commonplace in nature (Dugan et al. 2008). Because reassortment is also likely to occur in these cases, individual segments will then become gradually unlinked from each other, eventually reaching the point at which different alleles of internal segments effectively act as a single gene pool. Finally, rising levels of herd immunity to the homologous subtype will allow new HA/NA subtypes to invade the population. However, because the internal segments will have spread to a large number of genetic backgrounds through reassortment, they will not experience such severe drops in frequency as the HA/NA segments. In sum, the genetic structure of AIV is dependent on the relative rates of natural selection, genetic linkage, reassortment, and population subdivision, although obtaining precise estimates for these rates in natural populations will be difficult.
Fig. 4.
Schematic representation of the population genetic structure and evolution of the internal gene segments of AIV, largely determined by a process of genetic hitchhiking to selectively advantageous variants in the HA and NA proteins, followed by frequent reassortment. Different colors represent different HA (or NA) alleles, whereas alleles of internal segments are shown as different shapes
The NS segment occupies an intermediate position under this evolutionary model: this segment is more diverse than the five other internal gene segments, being structured into the highly divergent A and B allelic lineages, but less so than the both HA and NA proteins. Even though the dN/dS value for the NS segment (0.147 for NS1) is similar to that of HA (0.130) and NA (0.206) (Chen and Holmes 2006), no positively selected sites have been identified in NS to date (Suzuki 2006). Further, although both the A and B alleles can be found in most HA/NA subtypes (H1–12 and N1–9) in nature, and can co-circulate in the same population (Dugan et al. 2008; Zohari et al. 2008), it is not known whether the two-allele pattern is necessary for the perpetuation of AIVs in wild birds. Therefore, it is possible that the division into the NS alleles is also the result of a series of hitchhiking events to emerging HA and/or NA subtypes, in the same manner as the other internal segments, rather than direct positive selection acting on this gene. Clearly, more work is needed to understand the nature of the selection pressures acting on NS.
The Global Phylogeography of AIV
A second important question relating to the evolutionary history of AIV is the nature of the phylogeographic processes that have resulted in largely distinct Eurasian/Australian and North American lineages. To state this question in another way, why do not we observe older viral strains in each continent? The key observation here is that although relatively infrequent, intercontinental transmission has clearly occurred multiple times. For example, two distinct North American N6 clades are observed (Fig. 2), one of which was seemingly introduced only recently into the North American shorebird population from Eurasia/Australia (95% HPD = 1963–1975). A similar pattern can be seen in some other HA/NA subtypes and internal segments (Supplementary Figs. 1–6). As a particular case in point, there are three major lineages of PA segments in North America, each of which originated in a different time period; lineage I between 1885 and 1935; lineage II between 1915 and 1945; and lineage III between 1960 and 1967 when it was introduced to North America from the Eurasian/Australian clade (Fig. 1). In this context, it is also worth noting that our expansive ML trees of the internal segments revealed some small clades transmitted either from Eurasia to America or vice versa (Supplementary Figs. 14–18).
The intercontinental transmission of AIV is clearly an ongoing process, although inherent sampling biases make it difficult to estimate the precise rates of viral gene flow. Such continual importation of viruses into different geographic regions facilitates a selective competition among resident and invasive lineages and insures that viral evolution on all continents occurs on roughly the same timescale (Bahl et al. 2009). This process is enhanced by the continual gene flow that appears to occur within individual continents, such as North America, and manifest as a strong temporal structure within AIV phylogenies (Chen and Holmes 2009). However, that multiple small clades of viruses have experienced trans-continental transmission suggests that not every viral lineage can become established in the new territory following importation. Indeed, it is likely that the successful establishment of new lineage in a new continent involves the introduction of HA/NA subtypes to which there is no prior immunity in the population (Bahl et al. 2009).
In conclusion, our analysis suggests that the evolutionary history of AIVs involves the continual birth and death of viral lineages. In this process, diversifying selection likely shaped by cross immunity on the immune-related proteins (HA, NA, and perhaps NS1) plays important role in generating both genetic and antigenic diversities. In contrast, the population genetic structure of the internal segments appears to be the result of a complex combination of serial hitchhiking on the competing viral strains with differing antigenicity, coupled with rapid gene flow on both the continental and global scales.
Supplementary Material
Acknowledgment
This study was funded in part by NIH grant GM080533. We thank two reviewers for useful comments.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00239-009-9312-8) contains supplementary material, which is available to authorized users.
Contributor Information
Rubing Chen, Center for Infectious Disease Dynamics, Department of Biology, The Pennsylvania State University, Mueller Laboratory, University Park, PA 16802, USA.
Edward C. Holmes, Center for Infectious Disease Dynamics, Department of Biology, The Pennsylvania State University, Mueller Laboratory, University Park, PA 16802, USA, ech15@psu.edu Fogarty International Center, National Institutes of Health, Bethesda, MD 20892, USA.
References
- Bahl J, Vijaykrishna D, Holmes EC, Smith GJC, Guan Y. Gene flow and competitive exclusion of avian influenza A in the natural reservoir hosts. Virology. 2009 doi: 10.1016/j.virol.2009.05.002. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. Bioessays. 2003;25:657–671. doi: 10.1002/bies.10303. [DOI] [PubMed] [Google Scholar]
- Chen R, Holmes EC. Avian influenza virus exhibits rapid evolutionary dynamics. Mol Biol Evol. 2006;23:2336–2341. doi: 10.1093/molbev/msl102. [DOI] [PubMed] [Google Scholar]
- Chen R, Holmes EC. Frequent inter-species transmission and geographic subdivision in avian influenza viruses from wild birds. Virology. 2009;383:156–161. doi: 10.1016/j.virol.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis RO, Bean WJ, Kawaoka Y, Webster RG. Distinct lineages of influenza virus H4 hemagglutinin genes in different regions of the world. Virology. 1989;169:408–417. doi: 10.1016/0042-6822(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Pybus OG, Rambaut A. Inference of viral evolutionary rates from molecular sequences. Adv Parasitol. 2003;54:331–358. doi: 10.1016/s0065-308x(03)54008-8. [DOI] [PubMed] [Google Scholar]
- Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterday BC, Trainer DO, Turnova B, Pereira HG. Evidence of infection with influenza viruses in migratory waterfowl. Nature. 1968;219:523–524. doi: 10.1038/219523a0. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumi H, Nerome K, Nakayama M, Ishida M. Serological and virological investigations of orthomyxovirus in birds in South-East Asian area. Dev Biol Stand. 1977;39:460–475. [PubMed] [Google Scholar]
- Ito T, Gorman OT, Kawaoka Y, Bean WJ, Webster RG. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S, Obert CA, Franks J, Walker D, Jones K, Seiler P, Niles L, Pryor SP, Obenauer JC, Naeve CW, Widjaja L, Webby RJ, Webster RG. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 2007;3:e167. doi: 10.1371/journal.ppat.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Min JY, Krug RM, Sen GC. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Ludwig S, Schultz U, Mandler J, Fitch WM, Scholtissek C. Phylogenetic relationship of the nonstructural (NS) genes of influenza A viruses. Virology. 1991;183:566–577. doi: 10.1016/0042-6822(91)90985-k. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- Perroncito E. Epizoozia tifoide nei gallinacei. Annali Accad Agri Torino. 1878;21:87–126. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romváry J, Mészáros J, Tanyi J, Rózsa J, Fábián L. Influenza infectedness of captured and shot wild birds on north-eastern and south-eastern parts of Hungary. Acta Vet Acad Sci Hung. 1976;26:363–368. [PubMed] [Google Scholar]
- Schäfer W. Vergleichender sero-immunologische Untersuchungen über die Viren der Influenza und klassischen Geflügelpest. Z Naturf. 1955;10b:81–91. [Google Scholar]
- Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol. 2007;24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- Slemons RD, Johnson DC, Osborn JS, Hayes F. Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Dis. 1974;18:119–124. [PubMed] [Google Scholar]
- Slepuskin AN, Pysina TV, Gonsovsky FK, Sazonov AA, Isacenko VA, Sokolova NN, Polivanov VM, Lvov DK, Zakstel’skaja LJ. Haemagglutination-inhibiting activity to type A influenza viruses in the sera of wild birds from the far east of the USSR. Bull World Health Organ. 1972;47:527–530. [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Stallknecht DE, Slemons RD, Winker K, Suarez DL, Scott M, Swayne DE. Phylogenetic analyses of type A influenza genes in natural reservoir species in North America reveals genetic variation. Virus Res. 2005;114:89–100. doi: 10.1016/j.virusres.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. Natural selection on influenza virus genome. Mol Biol Evol. 2006;23:1902–1911. doi: 10.1093/molbev/msl050. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nei M. Origin and evolution of influenza virus hemagglutinin genes. Mol Biol Evol. 2002;19:501–509. doi: 10.1093/oxfordjournals.molbev.a004105. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Version 4. Sunderland, MA: Sinauer Associates; 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) [Google Scholar]
- Vijaykrishna D, Bahl J, Riley S, Duan L, Zhang JX, Chen H, Peiris JS, Smith GJ, Guan Y. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 2008;4:e1000161. doi: 10.1371/journal.ppat.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja L, Krauss SL, Webby RJ, Xie T, Webster RG. Matrix gene of influenza a viruses isolated from wild aquatic birds: ecology and emergence of influenza A viruses. J Virol. 2004;78:8771–8779. doi: 10.1128/JVI.78.16.8771-8779.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhirnov OP, Klenk HD. Control of apoptosis in influenza virusinfected cells by up-regulation of Akt and p53 signaling. Apoptosis. 2007;12:1419–1432. doi: 10.1007/s10495-007-0071-y. [DOI] [PubMed] [Google Scholar]
- Zohari S, Gyarmati P, Ejdersund A, Berglöf U, Thorén P, Ehrenberg M, Czifra G, Belák S, Waldenström J, Olsen B, Berg M. Phylogenetic analysis of the non-structural (NS) gene of influenza A viruses isolated from mallards in Northern Europe in 2005. Virol J. 2008;5:147. doi: 10.1186/1743-422X-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.