Abstract
Many parasite populations are difficult to sample because they are non-uniformly distributed between several host species and are often not easily collected from the living host, limiting sample size and possibly distorting the representation of the population. For the parasite Schistosoma mansoni, we investigated the use of the aggregated eggs found in the stool of infected individuals as a simple and representative sample. Previously, we demonstrated that microsatellite allele frequencies can be accurately estimated from pooled DNA of cloned S. mansoni adults, and we show here that genotyping parasite populations from reproductively isolated laboratory strains can be used to identify these specific populations based on characteristic patterns of allele frequencies, as observed by polyacrylamide gel electrophoresis and automated sequencer analysis of fluorescently labeled PCR products. In addition, microsatellites used to genotype aggregates of eggs collected from stools of infected individuals produced results consistent with the geographic distribution of the samples. Direct analysis of total stool eggs can be an important approach to questions of population genetics for this parasite by increasing the sample size to thousands per individual and reducing bias.
Parasite population structure may influence transmission dynamics, host preference, and virulence, and all of these factors could significantly influence control strategies and drug design. Uncontrolled, schistosomiasis is one of the most common yet one of the most serious threats to health in the developing world (King and Dangerfield-Cha, 2008). It contributes to anemia and linear growth deficits, while the most serious outcome of infection is portal hypertension that results from the host’s response to eggs trapped in the liver. The causative organism, Schistosoma mansoni, is a multicellular parasite with a complex developmental life cycle distributed between 2 hosts. Adult male and female worms produce eggs in the mesenteric veins draining the intestines of mammalian hosts. Approximately half of these eggs pass out of the body in the stool, hatching as miracidia when they contact fresh water. The miracidia infect snails and undergo asexual reproduction, resulting in the release of cercariae that penetrate the skin of humans who come in contact with contaminated water. Because of the way they are distributed, a community of schistosomes is therefore a collection of discrete populations within a population. The infection within an individual host (infrapopulation) is not clonal, but is not a complete population in the genetic sense, since the viable members of the next generation are exported. This type of distribution is likely to obscure the true population structure of S. mansoni unless the infrapopulations are adequately sampled.
The development of two elements is critical for understanding schistosome population structure: a set of polymorphic neutral markers and an appropriate sampling strategy. Fortunately, microsatellite markers are common in schistosomes (Rodrigues et al., 2002), and their discovery has been aided by the S. mansoni Genome Project. Optimizing a sampling strategy has been more problematic. The exact numbers and distribution of snails is rarely if ever known, and their short life spans and low rates of active shedding also mean that the snails are usually only carrying the parasite population transmitted in the preceding few months by a limited segment of the human population—thus likely to underestimate the underlying variation across all hosts. Human activity is the major determinant for the distribution of the parasite. It is simpler and more accurate to sample all or nearly all of the human population of a community, since most individuals can be accounted for and their probability for infection can be accurately estimated. The most accessible and most representative parasite sample that can be obtained from humans is the eggs discharged in stool. One approach to sampling has been to produce cercariae or adult worms in the laboratory from eggs collected from stool. This is a time consuming process which samples relatively few organisms and introduces significant bias due to selection at the point of egg hatching, infection of laboratory snails, and infection of typical laboratory animals (LoVerde et al., 1985; Dalton et al., 1997; Sorensen et al., 2006). More recently, methods for assaying individual miracidia (obtained from eggs hatched in vitro) with multiple microsatellite markers have been described (Gower et al., 2007; Steinauer et al., 2008), but these studies are still limited in the number of individuals that may be tested within an infrapopulation and introduce the potential for bias toward eggs capable of hatching in vitro. By a recent estimate, a moderately infected individual with a typical parasite burden of 80-100 worm pairs would excrete 12000-15000 eggs daily (Wilson et al., 2006). Adequate sampling of enough eggs to represent the infrapopulation is unlikely to be achieved in laboratory passage of parasites or when the analysis proceeds egg by egg for hundreds of individuals.
The estimation of allele frequencies by microsatellite analysis in pooled DNA samples has been performed successfully in the past (see, e.g. Shaw et al., 1998; Collins et al., 2000; Schnack et al., 2004), and we previously demonstrated (Silva et al., 2006) that microsatellite allele frequencies can be estimated from the pooled DNA of S. mansoni clones. We show here how the same principles may be applied to naturally aggregated samples by using thousands of parasite eggs isolated from individual human infections to study the population structure of S. mansoni.
MATERIALS AND METHODS
S. mansoni laboratory strains were named according to the laboratory in which they were maintained and are represented by 300-500 adult worms from each. The laboratory strains were: CWRU (Case Western Reserve University, Cleveland, OH); BRI-1, BRI-2, BRI-3, and BRI-4 (Biomedical Research Institute, Gaithersburg, MD); CPGM (Centro de Pesquisa Gonçalo Moniz – Oswaldo Cruz Foundation, Salvador, Bahia, Brazil); IPR (Institute of Primate Research, Nairobi, Kenya); TBI (Theodor Bilhartz Institute, Cairo, Egypt); and York (University of York). The CWRU and BRI-1 strains were derived from the same founder population of Puerto Rican origin (Naval Medical Research Institute-NMRI strain) and have been separated for at least 30 years. BRI-4 may have also originated from the NMRI strain, while the BRI-2, BRI-3, and the York strains are of Puerto Rican origin but not from the NMRI strain. All strains were maintained in mice, except the IPR strain which represented a laboratory infection in baboons from wild caught snails. BRI-3 (Richards and Shade, 1987) and TBI are distinguished by having a more restricted snail host range. Schistosoma japonicum and Schistosoma hematobium laboratory strains were obtained from Biomedical Research Institute (NIAID Contract N01-A1-30026). Four hookworm samples (designated HK1-HK4) were obtained from eggs isolated from the stool of individual Kenyans with hookworm infections.
For the isolation of S. mansoni eggs, total morning stools were collected from children and adults as part of studies on schistosome morbidity in the communities of Itaquara and Salvador, Brazil, and Katheka, Kenya. In the Kenyan community and Salvador, previous chemotherapy was uncommon, while Itaquara had seen several campaigns with widespread treatment. Intensity of infection was quantified by the Kato-Katz method. A protocol based on published sieving methods (Dresden and Payne, 1981) and density gradients (Baltz et al., 1982; Dalton et al., 1997) for isolating eggs from the liver was used to isolate eggs from human feces. After homogenizing 200 g of feces in a large volume of cold 2% saline and a series of gravity sedimentations, the material was passed in succession through 420 μm mesh, 177 μm mesh, 107 μm mesh and 50 μm mesh sieves. The eggs (~100 μm × 70 μm) were washed off of the final 2 sieves. Some samples were further purified over a 10%:70% Percoll-saline step gradient spun at 400 × g for 10-20 min. After pelleting to the bottom of the tube, the eggs were washed with saline, and a final egg count was made.
DNA was isolated from samples by proteinase K digestion followed by phenol:chloroform extraction (Sambrook and Russell, 2001). A further purification step using QIAamp tissue kit spin columns (Qiagen, Inc., Valencia, CA) was performed on DNA extracted from stool egg samples to remove PCR inhibitors (Verweij et al., 2004).
Polymerase chain reaction (PCR) conditions and primer sequences for microsatellite loci SMMS 2 (S. mansoni microsatellite 2), SMMS 3, SMMS 13, SMMS 16, SMMS 17, SMMS 18, and SMMS 21 were as previously described (Silva et al., 2006). Primers for the human chemokine coreceptor gene CCR5 (Salkowitz et al., 2003) were used to test for human DNA contamination in DNA extracted from S. mansoni eggs. Primers previously used for randomly amplified polymorphic DNA (RAPD) analysis (Dias Neto et al., 1993) was used as controls for amplification. DNA samples from laboratory strains of S. mansoni and from eggs isolated from feces were genotyped by PCR amplification of the microsatellite loci and separated by polyacrylamide gel electrophoresis (PAGE) on 8% nondenaturing gels along with a 50 bp molecular weight marker (Fermentas, Glen Burnie, MD). Gels were stained with ethidium bromide and digital images were generated with a Doc-It gel documentation system and software (UVP, Upland, CA). To verify the identity of the presumed microsatellite amplicons from stool eggs, PCR products of some egg samples were purified from polyacrylamide gels using the QIAEX II kit (Qiagen, Inc., Valencia, CA) and sequenced.
We also evaluated capillary electrophoresis-based microsatellite analysis. The 11 Kenyan samples were amplified with fluorescently labeled forward primers SMMS 3, SMMS 16, SMMS 18, and SMMS 21 (using the dyes FAM, HEX, JOE, and ROX, respectively). The reaction products and the GS500LIZ molecular weight marker (Applied Biosystems) were separated on an Applied Biosystems 3730 genetic analyzer.
For PAGE separated PCR reaction products, DNA band intensities and migration were measured from digitized gel images using SigmaGel (Jandel Scientific) as described previously (Silva et al., 2006). Relative population allele frequencies were calculated by dividing the measured intensity of a particular band (representing an allele) by the summed intensities of all bands measured at that locus. For samples run on the AB 3730, output files were analyzed using Peak Scanner software v1.0 (Applied Biosystems), and relative allele frequencies were calculated using peak heights in a similar manner as band intensities from PAGE. Calculations were made using all observed alleles and repeated using only the alleles representing greater than 5% of the total allele intensities in each sample.
FST values were calculated with the program Arlequin 3.11 (Excoffier et al., 2005) using analysis of molecular variance (AMOVA), inputting the allele frequencies as relative proportions and using worm or final egg counts for sample sizes. The analysis was carried out with 16,000 permutations. Weighted average single locus estimators for FST values were calculated across loci (Reynolds et al., 1983) using the population variance values determined by Arlequin. Weighted averages have been shown to perform better than unweighted averages when combining FST values over multiple loci (Reynolds et al., 1983), although both were calculated for pairwise comparison of the laboratory strains. Effective population sizes (Ne) were calculated by a maximum likelihood method using the program MLNE (Wang and Whitlock, 2003). Other calculations were performed in Excel (Microsoft, Redmond, WA).
RESULTS
Microsatellite amplification of the S. mansoni laboratory strains
DNA extracted from S. mansoni populations maintained in several laboratories in the U.S. and abroad was amplified using the SMMS primer sets (Fig. 1, lanes 1-9). SMMS 2 primarily amplified two bands of approximately 228 and 261 bp, although the IPR strain exhibited several smaller bands more closely resembling the stepwise variation expected for microsatellites. The SMMS 3 amplification showed a greater degree of heterogeneity as well as greater differentiation between the strains than SMMS 2. A number of the laboratory strains appear to be monoallelic at several loci by PAGE, yet polymorphic for the IPR strain (Fig. 1, lane 7).
Figure 1.
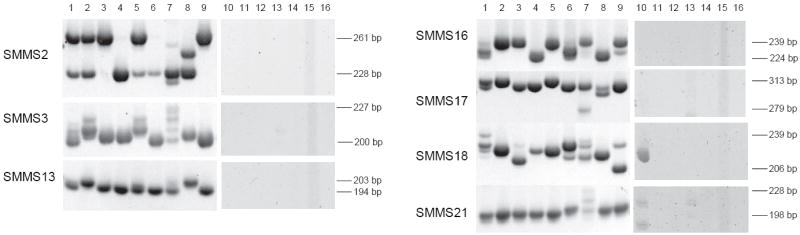
S. mansoni microsatellite specificity. Microsatellite amplification of laboratory strains of S. mansoni, and other organisms DNA. DNA templates are as follows: lanes 1-9, S. mansoni strains CWRU, BRI-1, BRI-2, BRI-3, BRI-4, CPGM, IPR, TBI, York; lane 10, human; lanes 11-14, hookworm isolates 1-4; lane 15, S. japonicum; lane 16, S. hematobium.
Band sizes and relative intensities were measured, and pairwise FST values were calculated between strains. The results indicate a high degree of differentiation between all laboratory strains (Table I); the weighted FST for all strains over all loci was 0.651 with the pairwise FST values ranging from 0.289 – 0.955.
Table I.
Average weighted and unweighted pairwise FST values* for S. mansoni laboratory strains.
| Case | BRI-1 | BRI-2 | BRI-3 | BRI-4 | CPGM | IPR | TBI | York | |
|---|---|---|---|---|---|---|---|---|---|
| Case | - | 0.366 | 0.420 | 0.553 | 0.267 | 0.387 | 0.303 | 0.449 | 0.348 |
| BRI-1 | 0.435 | - | 0.579 | 0.734 | 0.122 | 0.769 | 0.425 | 0.652 | 0.597 |
| BRI-2 | 0.498 | 0.789 | - | 0.545 | 0.443 | 0.433 | 0.369 | 0.718 | 0.135 |
| BRI-3 | 0.596 | 0.876 | 0.955 | - | 0.662 | 0.323 | 0.507 | 0.754 | 0.500 |
| BRI-4 | 0.340 | 0.289 | 0.741 | 0.843 | - | 0.696 | 0.359 | 0.648 | 0.464 |
| CPGM | 0.429 | 0.785 | 0.765 | 0.758 | 0.745 | - | 0.361 | 0.660 | 0.361 |
| IPR | 0.327 | 0.537 | 0.523 | 0.642 | 0.486 | 0.425 | - | 0.465 | 0.382 |
| TBI | 0.510 | 0.745 | 0.827 | 0.844 | 0.736 | 0.718 | 0.520 | - | 0.671 |
| York | 0.396 | 0.764 | 0.524 | 0.874 | 0.681 | 0.659 | 0.472 | 0.771 | - |
Unweighted values are presented above the diagonal
Specificity of the SMMS primers
The specificity of the primers for the 7 SMMS loci against other DNA templates likely to be encountered in stool samples (human, hookworm, S. japonicum, and S. hematobium) were used in PCR reactions with the 7 SMMS primer sets (Fig. 1, lanes 10-16). All these samples could be amplified by RAPD primers (data not shown) indicating the DNA was of sufficient quality and concentration for PCR. In Figure 1, the hookworm sample in lane 13 shows very weak bands for primers SMMS 3 and 21. Their intensity was much lower than that found in the S. mansoni PCR reactions and generally too low to register above background levels. Amplification did not occur with DNA template from S. japonicum and S. hematobium, demonstrating the specificity of the primers for S. mansoni. Given that the more closely related Schistosoma species did not amplify with the S. mansoni-specific primers, it is likely that the individual from whom the hookworm eggs were isolated also harbored a low-level infection of S. mansoni not detected by the stool examination.
Amplified bands for the SMMS 18 and 21 primer sets in the expected size range of the microsatellite amplicons were noted for human DNA. However, PCR using primers specific for a 312 bp fragment of the human CCR5 gene did not amplify DNA extracted from S. mansoni eggs (data not shown), indicating that the levels of human DNA are low or that methods used to isolate the eggs from fecal material are sufficient to minimize human DNA contamination.
Microsatellite amplification and analysis of Kenyan and Brazilian S. mansoni populations
DNA was extracted from S. mansoni eggs collected directly from stool samples of infected persons in Brazil and Kenya. Egg counts in stool ranged from 60 eggs per gram (epg) to greater than 3000 epg. The results of PCR amplification of these samples using the SMMS primer sets and PAGE analysis of the products is shown in Figure 2. The total eggs isolated from 2 individuals (Kenyan sample 1 and Brazilian sample 4) amplified poorly with most primer sets. For SMMS2 the samples from individual Brazilian infections amplified 2 alleles (228 and 261 bp) almost exclusively. This was consistent with the pattern of amplification with this marker for all of the laboratory strains originating from the Western Hemisphere. By contrast, populations from individual infections in Kenya amplified these 2 along with several additional alleles also observed in IPR, the only laboratory strain of Kenyan origin. Common allele sizes between the Kenyan and Brazilian populations were seen at all loci. Only one allele was observed by PAGE for loci SMMS 13 and 21 in the Brazilian samples, and in all but one of the Brazilian samples for locus 17. The observed number and size of alleles amplified and the FST, calculated using relative allele frequencies for each locus and egg counts of the samples, are indicated by geographic population in Table II. Division of the Brazilian population into its two component locales yielded FST values of 0.031 in Itaquara and 0.066 in Salvador. The effective population size likelihood point estimate for the Kenyan population (Ne = 840) was approximately twice that of the Brazilians sampled (Ne = 410).
Figure 2.
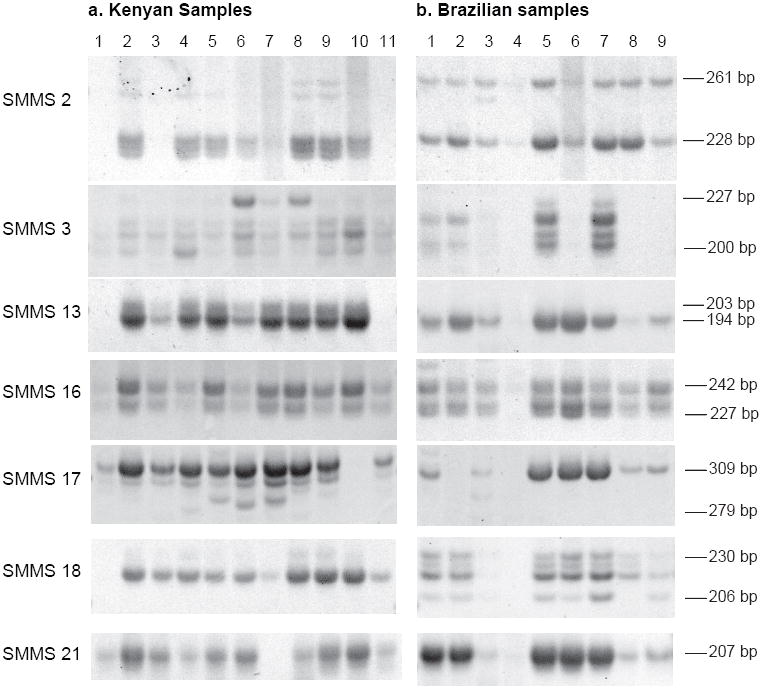
Microsatellite amplification of DNA extracted from S. mansoni total stool eggs from individual human infections analyzed on non-denaturing polyacrylamide gels. a. Kenyan samples 1-11. b. Brazilian samples 1-9.
Table II.
Microsatellite results summary for field egg populations.
| Kenya |
Brazil |
Combined |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Samples Analyzed | Alleles Observed | Band Sizes* | FST | Samples Analyzed | Alleles Observed | Band Sizes* | FST | Common Alleles | FST | ||
| SMMS 2 | 7 | 5 | 220 | 224 | 0.004 | 8 | 3 | 0.126 | 3 | 0.317 | ||
| 228 | 251 | 228 | 251 | |||||||||
| 261 | 261 | |||||||||||
| SMMS 3 | 11 | 4 | 0.043 | 4 | 6 | 200 | 203 | 0.043 | 3 | 0.153 | ||
| 200 | 209 | 209 | 215 | |||||||||
| 215 | 227 | 218 | 224 | |||||||||
| SMMS 13 | 9 | 2 | 194 | 203 | 0.006 | 8 | 1 | 194 | N.D.† | 1 | 0.627 | |
| SMMS 16 | 11 | 2 | 0.004 | 8 | 5 | 227 | 230 | 0.044 | 2 | 0.217 | ||
| 230 | 242 | 239 | 242 | |||||||||
| 251 | ||||||||||||
| SMMS 17 | 10 | 6 | 279 | 282 | 0.030 | 7 | 2 | 0.349 | 2 | 0.369 | ||
| 285 | 292 | 285 | 309 | |||||||||
| 298 | 309 | |||||||||||
| SMMS 18 | 10 | 4 | 218 | 224 | 0.096 | 7 | 4 | 206 | 218 | 0.020 | 3 | 0.247 |
| 227 | 230 | 224 | 230 | |||||||||
| SMMS 21 | 10 | 2 | 204 | 207 | 0.049 | 9 | 1 | 207 | N.D.† | 1 | 0.434 | |
| Weighted FST | 0.029 | 0.057 | 0.240 | |||||||||
PCR product sizes as estimated by SigmaGel software
N.D., FST not determined at monoallelic loci
Bands corresponding to several putative alleles for each locus were purified from gels and sequenced. In each case, sequences flanking the microsatellite regions from the field samples were identical to those obtained from GenBank Release 164 (data not shown). The one exception was the 19 bp indel in SMMS 2 which was noted previously (Silva et al., 2006) in cloned parasites from the CWRU life-cycle and was also seen in the field samples.
Capillary electrophoresis of total stool egg sample microsatellites
PCR products of Kenyan samples at loci SMMS 3, 16, 18, and 21 generated with fluorescently labeled primers were also analyzed by capillary electrophoresis. This method was able to resolve a greater number of separate alleles at each locus compared to PAGE (Fig. 3 and Table III) and was also more sensitive, providing data at locus SMMS 18 from one sample that did not produce visible bands by PAGE. Loci which appeared to be monomorphic in some samples by PAGE (e.g., SMMS 18 and SMMS 21 in Fig. 3) were observed to be polymorphic by capillary genotyping. Additionally, sizes of the PCR products as determined by the two methods were not in complete agreement. Despite this, the FST values calculated using peak heights as a measure of allele frequency (Table III) do not differ greatly from those calculated using PAGE band intensities. Calculation of FST values disregarding minor alleles (i.e., those contributing <5% of the summed total of peak heights in any one sample) also corresponded well with values generated using all measurable peaks. Analysis of serial dilutions of sample DNA showed that allele frequencies were maintained (as indicated by FST values ≤ 0.01 between dilution pairs) to dilutions of 1:25 and in some cases 1:125 (data not shown).
Figure 3.
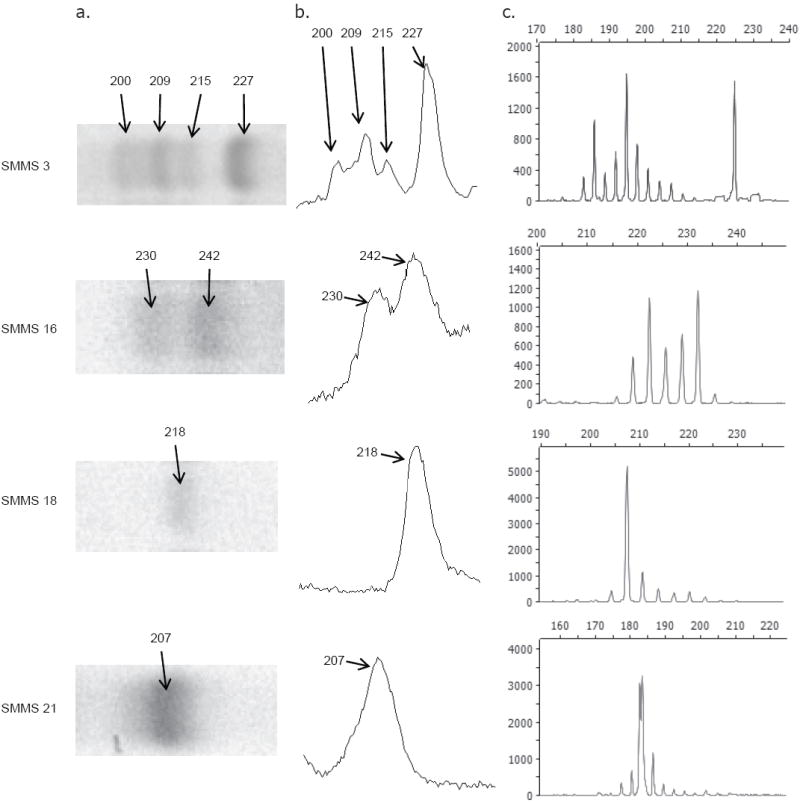
Microsatellite analysis of stool egg samples by capillary electrophoresis. a. Inverted digital image of EtBr stained polyacrylamide gel. b. Band intensity peaks as interpreted by gel analysis software. Allele size in bp as calculated by SigmaGel software in a and b. c. Capillary electrophoresis analysis of fluorescently labeled products. Vertical scale, fluorescence intensity; horizontal scale, size in bp.
Table III.
Microsatellite results summary for Kenya samples as determined by automated capillary electrophoresis
| All peaks |
Peaks ≥5% of total |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Samples Analyzed | Alleles Observed | Peak Sizes | FST | Alleles Observed | Peak Sizes | FST | ||||
| SMMS 3 | 11 | 12 | 183 | 186 | 189 | 0.013 | 9 | 183 | 186 | 189 | 0.013 |
| 192 | 195 | 198 | 192 | 195 | 198 | ||||||
| 201 | 204 | 206 | 201 | 204 | 206 | ||||||
| 209 | 212 | 215 | |||||||||
| SMMS 16 | 11 | 9 | 211 | 214 | 217 | 0.006 | 7 | 211 | 214 | 217 | 0.006 |
| 220 | 223 | 226 | 220 | 223 | 226 | ||||||
| 229 | 233 | 236 | 229 | ||||||||
| SMMS 18 | 11 | 12 | 194 | 197 | 201 | 0.090 | 6 | 0.084 | |||
| 204 | 208 | 211 | 204 | 208 | 211 | ||||||
| 214 | 217 | 220 | 214 | 217 | 220 | ||||||
| 224 | 230 | 240 | |||||||||
| SMMS 21 | 10 | 10 | 178 | 181 | 184 | 0.043 | 6 | 0.043 | |||
| 187 | 190 | 193 | 178 | 181 | 184 | ||||||
| 196 | 198 | 201 | 187 | 190 | 193 | ||||||
| 204 | |||||||||||
| Weighted FST | 0.028 | 0.027 | |||||||||
DISCUSSION
In this work, we demonstrate approaches for the investigation of S. mansoni population structure by microsatellite analysis of DNA from all eggs isolated from a single individual in a single sample as an enhanced sampling technique. This is likely to better represent the true infrapopulation composition and allow for an accurate determination of allele frequencies and subsequent calculation of the FST. Simple calculations suggest the potential magnitude of the sample size. Mean stool weight may range from 200-300 grams, so for a moderate infection of 100 eggs per gram there will be 20,000-30,000 eggs/stool. Recovering only 10% of these eggs will still yield a 2,000-3,000 egg sample from each infected individual.
We find that DNA from the total aggregate of eggs recovered from individual infections amplified as well as adult worm DNA to produce clear, interpretable patterns with microsatellite markers. Tri-and tetramer microsatellite repeats produce very little if any stuttering in PCR, a very important characteristic for analysis of aggregated samples, and this also appears to be the case with egg DNA from stool. Also critical to the use of microsatellites for analysis of eggs collected from stool is target specificity and scorability which this material demonstrates well. Scorability is improved, however, when samples are genotyped using an automated sequencer compared to PAGE. We also observed a discrepancy in PCR product sizes between capillary separations and PAGE (Tables II & III, Fig. 3). This is most likely due to the anomalous migration of double-stranded DNA observed in native polyacrylamide gels (Stellwagen, 1983). In comparing the 2 techniques, however, the major alleles can still be identified and measured with both, and the FST values for the Kenyan samples calculated from values based on both methods did not differ significantly. Thus, good approximations can be made in resource-limited laboratories without access to automated sequencers. Similarly, excluding minor alleles from analysis (Table III) demonstrates the robustness of this method, in that samples amplifying less efficiently (thereby underrepresenting the minor alleles) may still yield results that do not vary substantially from analysis with all measurable alleles, especially when extended across the thousands of eggs available in these samples.
Using natural pools or aggregates of worms from laboratory strains as an enhanced sampling technique we observed that each has a unique pattern of allele frequencies. While the egg samples produced expected FST results relative to geographic location and population history, the results of the laboratory strains was more difficult to reconcile. Schistosoma mansoni laboratory strains are limited populations of worms maintained as life cycles between Biomphalaria glabrata snails and a mammalian host, usually mice. Given their reproductive isolation, the founder effect, potential for bottlenecks, and selection in different laboratory hosts, it is not surprising that significant divergence has occurred for strains derived from a common source, obscuring the original relationships among strains.
Although we effectively sampled many thousands of eggs, we examined only a small number of human infections, so it would be premature to draw conclusions about schistosome population structure in the locations sampled, although general statements may be made from the infrapopulation results. It is likely that Brazilian populations arrived from West Africa and as such might be considered a founder population. Indeed, mitochondrial DNA data were used to demonstrate how New World S. mansoni populations were closely related to West African isolates, and the low number of New World haplotypes was hypothesized to be due to a founder effect (Morgan et al., 2005). However, if we suppose that allelic diversity is similar in East and West Africa, decreased allelic diversity was not observed in our Brazilian samples, since there were a total of 25 alleles in the Kenyan vs. 21 in the Brazilian field isolates over the 7 loci, and 15 alleles were common to both. The difference between our findings and those of Morgan et al. may be due to the nature of the mutational mechanisms in microsatellites and differences in selection pressure between the 2 types of markers (Jarne and Lagoda, 1996). The distribution of alleles for certain markers did demonstrate strong regional patterns. SMMS 2, for example showed primarily 2 alleles representing the indel of 19 bp in American laboratory strains as well as the Brazilian field population, while the Kenyan samples showed primarily the microsatellite variation (Figs. 1 and 2).
As with all approaches there are weaknesses and appropriate uses. Analysis of pooled or aggregated samples may not be ideal for evolutionary studies (Sorensen et al. 2007). Neither heterozygosity nor Hardy-Weinberg proportions can be measured for aggregated samples. Allelic diversity on the other hand is perhaps better measured because of the greater number of individuals sampled. FST, the most descriptive measurement of genetic differentiation, can be calculated from such data, and through it measures of gene flow, such as Nm (Wang and Whitlock, 2003) and genetic isolation by distance (Slatkin, 1993). The number of eggs appearing in the stool on any given day is highly variable and there may be variation in the relative contribution of each worm pair to the day’s output. This is likely to be minor over thousands of eggs, and stool could be collected over several days. Most importantly, the eggs in aggregate represent the generation being transmitted into the future. Some eggs remain trapped in tissues, but any worm pair whose progeny have this as an important characteristic are not likely to contribute significantly to continuation of the species. The range of potential diversity in the population is likely to be blunted by inclusion of worm clones or multiple siblings, but this is a faithful representation of the actual diversity or lack of it in the population. Sampling the full complement of S. mansoni eggs from the stool of infected humans, as in the present study, provides the most direct assessment of the infective worm population and produces a large sample size with reduced bias.
Acknowledgments
We wish to thank Fred Lewis at BRI and Alan Wilson at York University for samples and information on strains. This work was supported by NIH grant AI41680 and by CNPq grants 304713/2002-3 and 479839/2007-7.
LITERATURE CITED
- Baltz T, Lacassie I, Tribouley-Duret J, Tribouley J. Density-gradient separation of Schistosoma mansoni eggs. Journal of Parasitology. 1982;68:963–965. [PubMed] [Google Scholar]
- Collins HE, Li H, Inda SE, Anderson J, Laiho K, Tuomilehto J, Seldin MF. A simple and accurate method for determination of microsatellite total allele content differences between DNA pools. Human Genetics. 2000;106:218–226. doi: 10.1007/s004390051031. [DOI] [PubMed] [Google Scholar]
- Dalton JP, Day SR, Drew AC, Brindley PJ. A method for the isolation of schistosome eggs and miracidia free of contaminating host tissues. Parasitology. 1997;115:29–35. doi: 10.1017/s0031182097001091. [DOI] [PubMed] [Google Scholar]
- Dias Neto E, de Souza CP, Rollinson D, Katz N, Pena SD, Simpson AJ. The random amplification of polymorphic DNA allows the identification of strains and species of schistosome. Molecular and Biochemical Parasitology. 1993;57:83–88. doi: 10.1016/0166-6851(93)90246-t. [DOI] [PubMed] [Google Scholar]
- Dresden MH, Payne DC. A sieving method for the collection of schistosome eggs from mouse intestines. Journal of Parasitology. 1981;67:450–452. [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Gower C, Shrivastava J, Lamberton P, Rollinson D, Webster B, Emery A, Kabatereine N, Webster J. Development and application of an ethically and epidemiologically advantageous assay for the multi-locus microsatellite analysis of Schistosoma mansoni. Parasitology. 2007;134:523–536. doi: 10.1017/S0031182006001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarne P, Lagoda P. Microsatellites, from molecules to populations and back. Trends in Ecology & Evolution. 1996;11:424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- King CH, Dangerfield-Cha M. The unacknowledged impact of chronic schistosomiasis. Chronic Illness. 2008;4:65–79. doi: 10.1177/1742395307084407. [DOI] [PubMed] [Google Scholar]
- LoVerde PT, DeWald J, Minchella DJ, Bosshardt SC, Damian RT. Evidence for host-induced selection in Schistosoma mansoni. Journal of Parasitology. 1985;71:297–301. [PubMed] [Google Scholar]
- Morgan JAT, Dejong RJ, Adeoye GO, Ansa EDO, Barbosa CS, Bremond P, Cesari IM, Charbonnel N, Correa LR, Coulibaly G. Origin and diversification of the human parasite Schistosoma mansoni. Molecular Ecology. 2005;14:3889–3902. doi: 10.1111/j.1365-294X.2005.02709.x. [DOI] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS, Shade PC. The genetic variation of compatibility in Biomphalaria glabrata and Schistosoma mansoni. Journal of Parasitology. 1987;73:1146–1151. [PubMed] [Google Scholar]
- Rodrigues NB, Loverde PT, Romanha AJ, Oliveira G. Characterization of new Schistosoma mansoni microsatellite loci in sequences obtained from public DNA databases and microsatellite enriched genomic libraries. Memórias do Instituto Oswaldo Cruz. 2002;97(S1):71–75. doi: 10.1590/s0074-02762002000900015. [DOI] [PubMed] [Google Scholar]
- Salkowitz JR, Bruse SE, Meyerson H, Valdez H, Mosier DE, Harding CV, Zimmerman PA, Lederman MM. CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro. Clinical Immunology. 2003;108:234–240. doi: 10.1016/s1521-6616(03)00147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A laboratory manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schnack HG, Bakker SC, van’t Slot R, Groot BM, Sinke RJ, Kahn RS, Pearson PL. Accurate determination of microsatellite allele frequencies in pooled DNA samples. Eur J Hum Genet. 2004;12:925–934. doi: 10.1038/sj.ejhg.5201234. [DOI] [PubMed] [Google Scholar]
- Shaw S, Carrasquillo M, Kashuk C, Puffenberger E, Chakravarti A. Allele frequency distribution in pooled DNA samples: Applications to mapping complex disease genes. Genome Research. 1998;8:111–123. doi: 10.1101/gr.8.2.111. [DOI] [PubMed] [Google Scholar]
- Silva LK, Liu S, Blanton RE. Microsatellite analysis of pooled Schistosoma mansoni DNA: an approach for studies of parasite populations. Parasitology. 2006;132:331–338. doi: 10.1017/S0031182005009066. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Sorensen RE, Rodrigues NB, Oliveira G, Romanha AJ, Minchella DJ. Genetic filtering and optimal sampling of Schistosoma mansoni populations. Parasitology. 2006;133:443–451. doi: 10.1017/S0031182006000552. [DOI] [PubMed] [Google Scholar]
- Steinauer ML, Agola LE, Mwangi IN, Mkoji GM, Loker ES. Molecular epidemiology of Schistosoma mansoni: a robust, high-throughput method to assess multiple microsatellite markers from individual miracidia. Infection, Genetics, and Evolution. 2008;8:68–73. doi: 10.1016/j.meegid.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen NC. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983;22:6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. Journal of Clinical Microbiology. 2004;42:1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Whitlock MC. Estimating effective population size and migration rates from genetic samples over space and time. Genetics. 2003;163:429–446. doi: 10.1093/genetics/163.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RA, Van Dam GJ, Kariuki TM, Farah IO, Deelder AM, Coulson PS. The detection limits for estimates of infection intensity in schistosomiasis mansoni established by a study in non-human primates. International Journal for Parasitology. 2006;36:1241–1244. doi: 10.1016/j.ijpara.2006.07.002. [DOI] [PubMed] [Google Scholar]


