Abstract
Ionotropic glutamate receptors are major excitatory receptors in the central nervous system and also have far reaching influence in other areas of the body. Their modular nature has allowed for the isolation of the ligand binding domain and subsequent structural studies using a variety of spectroscopic techniques. This review will discuss the role of specific ligand:protein interactions in mediating activation in the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) subtype of glutamate receptors as established by various spectroscopic investigations of the GluR2 and GluR4 subunits of this receptor. Specifically, this review will provide an introduction of the insight gained from X-ray crystallography and nuclear magnetic resonance (NMR) investigations and then go on to focus on studies utilizing vibrational spectroscopy and fluorescence resonance energy transfer (FRET) to study the behavior of the isolated ligand binding domain in solution and discuss the importance of specific ligand:protein interactions in the mechanism of receptor activation.
Keywords: vibrational spectroscopy, FRET, AMPA receptors, agonist activation, ligand binding
Introduction
Ionotropic glutamate receptors are ligand-gated ion channels activated by the binding of the neurotransmitter glutamate. They are responsible for the majority of excitatory synaptic signaling in the mammalian central nervous system (1–7). This class of receptors has also been shown to be involved in many diverse neuropathologies, such as ischemia, Parkinson’s disease, and amyotrophic lateral sclerosis (3,8–12) as well as in other varying roles throughout the body (13–17). Their wide ranging effects in the body make this class of receptors an important focus of current research in an effort to elucidate their mechanism of activation.
Ionotropic glutamate receptors are homo- or heteromeric tetramers that assemble in a dimer-of-dimers fashion. They are classified based on their affinity for three different types of agonists: α-amino-3-hydroxy-5-methyl-4-isoxaxolepropionic acid (AMPA), N-methyl-D-aspartate (NMDA), and kainate. AMPA receptors are composed of a combination of subunits GluR1-GluR4, NMDA of NR1-NR3, and kainate of GluR5-7 and KA1-2. Agonist binding to the extracellular ligand binding domain results in conformational changes that allow for the formation of a cation specific transmembrane channel and flow of ions across the cell membrane (4–7,18).
Ionotropic glutamate receptors exhibit a modular topology and consist of an N-terminal domain that has a variety of functions including subunit assembly and receptor trafficking; a bilobed extracellular ligand binding domain known as S1S2; three transmembrane segments and a pore loop segment; and an intracellular C-terminus that is involved in modulatory functions and interactions with other proteins in the cytosol (4–7,18). This modular nature of the receptor has allowed for investigations of the isolated ligand binding domain. The large scale expression of this domain has paved the way for detailed structural investigations, using techniques such as X-ray crystallography (19–28), nuclear magnetic resonance (NMR) (18,29–31), vibrational spectroscopy (32–37), and fluorescence resonance energy transfer (FRET) (38). This review will focus primarily on spectroscopic investigations of AMPA receptors and insights gained through these investigations with a brief overview of the insights from the X-ray and NMR structures obtained for this subtype.
Agonists mediate activation through a cleft closure conformational change
Electrophysiological studies have shown varying degrees of activation depending on the particular agonist activating the channel. Activation of wild type homomeric GluR2 AMPA receptors by saturating concentrations of full agonists glutamate and AMPA results in large peak currents (38), while peak currents due to activation of these channels by saturating concentrations of partial agonists kainate (38) and the 5-substitued willardiines (21) are smaller. Single channel studies of wild type GluR2 homomeric receptors utilizing full agonist glutamate and partial agonists (S)-5-bromowillardiine and (S)-5-iodowillardiine have indicated that both full and partial agonists activate the same set of conductance states but differ in the frequency of occupying of these states. Glutamate will preferentially engage the higher conductance states but also activate the lower conductance states while the partial agonists will preferentially engage the lower conductance states but also occupy the high conductance states (21). Extensive studies have been done utilizing the isolated ligand binding domain of GluR2 to investigate the structural changes in this portion of the receptor associated with these differing extents of agonism.
The crystal structures of the isolated ligand binding domain of the GluR2 subunit showed a bilobed ligand binding cleft, where ligands bind between the two lobes (Figure 1). In addition, they also showed a greater amount of closure of the ligand binding cleft with full agonists, such as glutamate, AMPA, and quisqualate, a lesser amount of cleft closure with partial agonists, such as kainate and the 5-substituted willardiines, and minimal or no cleft closure in the apo or antagonist bound states (19,21,23,26). These structures in all but one case (24) showed that the degree of cleft closure of the ligand binding domain correlated to the extent of agonism by a given ligand (19,21,23,26). These results suggested a multistate induced fit or KNF (39) model of allosteric activation for AMPA receptors (19,21,24), with the cleft closure conformational change being the primary mediator between the ligand binding domain and the channel segments.
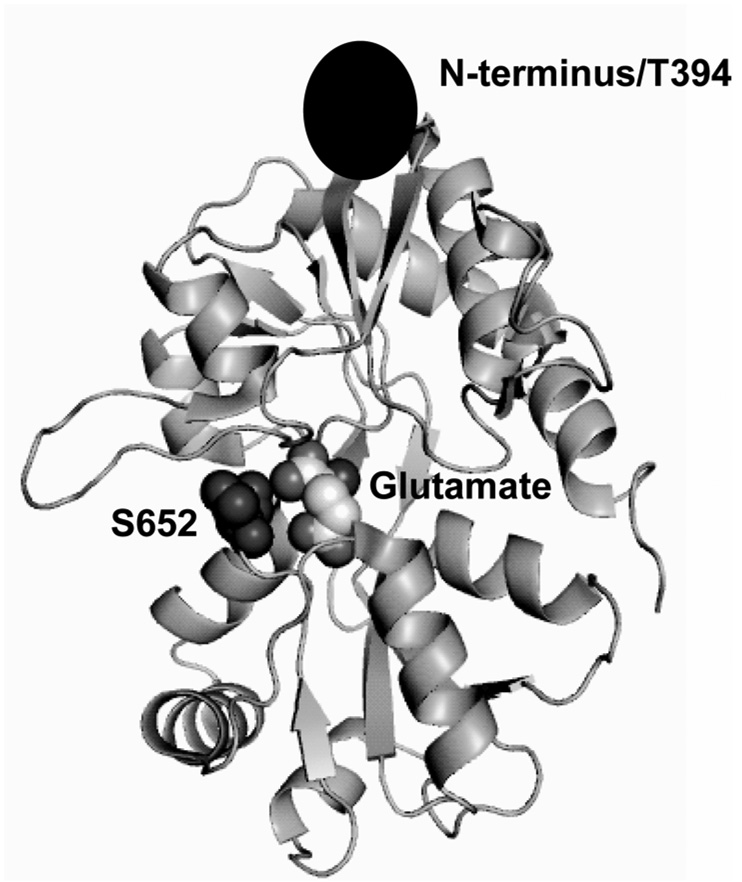
Figure 1.
Crystal structure of GluR2-S1S2 protein with glutamate bound (19). Sites that were tagged with fluorescent labels for FRET measurements are highlighted.
The information and insight provided by these crystal structures is no doubt a significant step in the understanding of the mechanism of activation of these receptors. However, the crystal structures represent the protein in a static state and cannot provide dynamic information about the protein due to the binding of these agonists. Complementary spectroscopic investigations have provided insight into these motions and also suggest that there may be other mechanisms in addition to the cleft closure conformational change that control the subtleties of the extent of activation by a given ligand.
Role of backbone dynamics in agonist mediated receptor activation
NMR studies looking at the GluR2 ligand binding domain in complex with glutamate have focused on characterizing the backbone dynamics of this domain. McFeeters and Oswald in their study describe the binding pocket for the GluR2-S1S2 protein as containing two components: one that interacts with the α-carboxylate and α-amine groups of the ligand, located in domain 1, and one that interacts with the γ-carboxylate of glutamate, located in domain 2 (29). McFeeters and Oswald found that the residues interacting with the α-carboxylate and α-amine groups of the ligand had little or no dynamic motion, while those residues interacting with the γ-carboxylate of glutamate had motion on the µs-ms time scale.
The µs-ms time scale dynamics involved a β-sheet core in domain 2, and since the channel segments are directly connected to β-sheet segments that are a part of this core, fine changes in the dynamics in this region could be a mechanism by which the agonist controls the subtleties of the extent of receptor activation. Additionally, the two linkers that connect domains 1 and 2 to each other are also connected to this β-sheet core of domain 2, indicating a role for these components in the domain closure conformational change (18,29). While the dynamics and conformational changes have been studied by X-ray and NMR methods, vibrational spectroscopy has been used to zoom in and investigate the role of specific ligand:protein interactions in controlling the dynamics and conformational changes and ultimately the function of the receptor.
Role of specific ligand:protein interactions
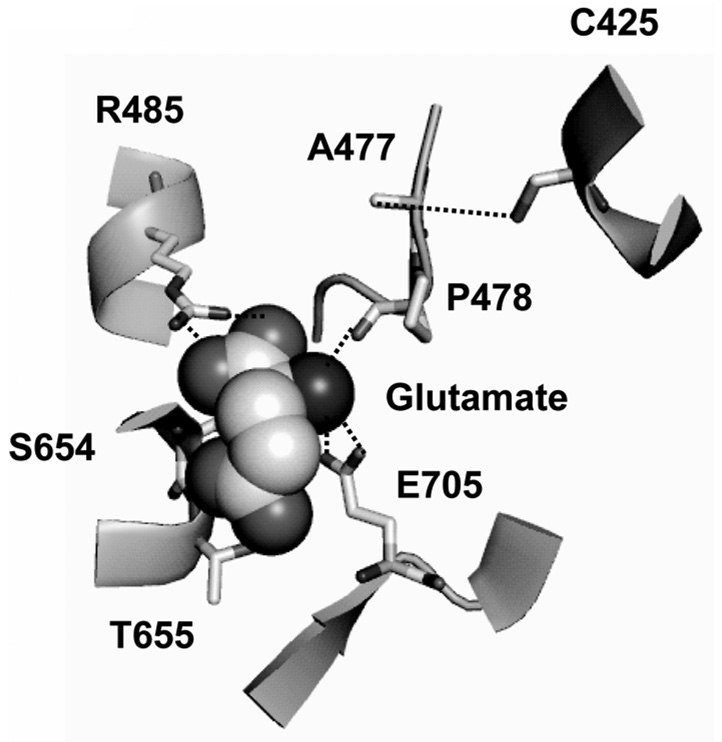
The vibrational signatures of specific moieties are very sensitive to the atomic-level chemical environment of the protein, and hence small changes in the environment of the ligand or in the protein as a consequence of ligand binding can be probed in more detail using vibrational spectroscopy. This technique is particularly suited for investigating glutamate receptors since the carboxylate groups of glutamate and the other ligands have signature vibrational modes in the region of 1400–1800 cm−1, which allow for characterization of the bound ligand. Additionally, due to the proximity of the Cys 425 residue to the α-amine group of the ligands, the S-H stretching frequency of this residue can be monitored to provide information about the environment of this group (Figure 2) (33,34,36,37). Specifically, the frequency of the asymmetric carboxylate stretching mode has been established to have a linear dependence on the enthalpy of association at the carboxylate moiety, with a 3 cm−1 downshift in frequency corresponding to a 1 kcal/mol decrease in the enthalpy of association (36,40). Using this correlation, and by comparing the frequencies of the α-carboxylate moiety for glutamate in the protein versus in solution (Table 1), it was established that the α-carboxylate moiety of glutamate exhibited a 1.3 kcal/mol decrease in energy bound to the protein relative to in buffer, consistent with the electrostatically favorable environment due to the presence of a positively charged R485 within hydrogen bonding distance of this group as observed in the crystal structure (Figure 2). The γ-carboxylate of the glutamate, on the other hand, exhibited an increase in the frequency of the asymmetric carboxylate stretching mode which corresponds to a 3.7 kcal/mol increase in energy. These results suggested that while the crystal structure showed favorable hydrogen bonding interactions with residues S654 and T655, the overall energetics at the γ-carboxylate were unfavorable. The E705D mutant interestingly did not exhibit this unfavorable interaction and showed a downshift in the asymmetric carboxylate stretching mode of the γ-carboxylate, thus suggesting that the E705 residue was contributing to the unfavorable interaction, and that while the crystal structures showed the E705 residue 7 Å away from the γ-carboxylate, this carboxylate was probably in closer proximity to the E705 residue in solution (Figure 2) (33).
Figure 2.
The environment of the ligand glutamate in GluR2-S1S2 (19). The side chain residues interacting with the ligand are shown.
Table 1.
Shift in the frequency of the asymmetric α-carboxylate vibration for the agonist bound to the GluR2-S1S2 wild type, L650T, and Y450F proteins relative to the frequency in buffer and of the Cys 425 S-H stretching vibration in the agonist-bound state relative to the frequency of the vibration in the apo state (37). Reproduced with permission from Biochemistry 2007; 46: 1343–1349. Copyright 2007 American Chemical Society.
| Protein-Agonist | α-carboxylate shift (cm−1) |
SH shift (cm−1) |
|---|---|---|
| WT-AMPA | 9 | 18 |
| Y450F-AMPA | 9 | 20 |
| L650T-AMPA | 10 | 0 |
| WT-Glutamate | 4 | 18 |
| Y450F-Glutamate | 5 | 0 |
| L650T-Glutamate | 4 | 20 |
| WT-Kainate | 18 | 0 |
| Y450F-Kainate | 14 | 11 |
| L650T-Kainate | 13 | 16 |
The vibrational spectroscopic investigations were also used to compare the environment of the α-carboxylate groups of AMPA and kainate with the ligand binding domain. As seen with glutamate, there was a decrease in the frequencies of the asymmetric α-carboxylate vibrations for the bound ligands, suggesting favorable interactions at this moiety for all three ligands, and the strength of the interactions followed the order kainite>AMPA>glutamate (Table 1). The S-H stretching vibration of Cys 425, which due to its proximity acts as a good probe of the interactions at the α-amine group of the ligand, showed no shift between the apo and kainate-bound forms, indicating no significant perturbation in the environment of this residue due to kainate binding, while a large downshift was seen with the full agonists AMPA and glutamate bound, indicating a large perturbation and a stronger hydrogen bond at the S-H group of Cys 425. These results suggested that the interactions at the α-amine group follow the order AMPA=glutamate>kainate (Table 1). (33).
Tug of war
The vibrational studies with full agonists glutamate and AMPA and partial agonist kainate suggest that a tug of war occurs between the α-carboxylate and α-amine groups of the agonists. Full agonists glutamate and AMPA have stronger interactions at their α-amine groups and weaker interactions at their α-carboxylate groups, while partial agonist kainate has a stronger interaction at its α-carboxylate group and a weaker interaction at its α-amine group (33,37). To extend these correlations, mutant receptors that exhibited a wide range of activations for these three agonists were studied (37). The changes in the ligand:protein interactions as studied by vibrational spectroscopy were correlated to the extent of cleft closure in these mutant receptors as determined using fluorescence resonance energy transfer (FRET) and ultimately to the functional consequences.
Fluorescence resonance energy transfer (FRET) to measure cleft closure
Two strategies were utilized to measure the extent of cleft closure in the GluR2-S1S2 protein (Figure 1). In the first strategy, residue S652 (near the ligand binding cleft but not involved in the ligand binding process) was mutated to a cysteine and labeled with diethylenetriaminepentaacetic acid chelate of Tb (DTPA-Tb) as the donor while a histidine tag was put at the N-terminus and labeled with a Cy3 derivative of nitrilotriaacetic acid chelate of Ni as the acceptor. In the second strategy, residues S652 and T394 (at the N-terminus) were mutated to cysteines and labeled with a 1:1 ratio of either DTPA-Tb or triethylenetetraaminehexaacetic acid chelate of Tb (TTHA-Tb) as the donor and fluorescein as the acceptor. The inherent cysteines (425 and 436) were not mutated because they are inaccessible to the fluorophores. The donor was then excited at 337 nm and emission lifetimes were collected at either 488 nm (to measure donor only lifetimes), 515 nm (fluorescein) or 575 nm (Cy3 Ni chelate) for the protein in the apo and glutamate-, AMPA-, and kainate-bound states (38).
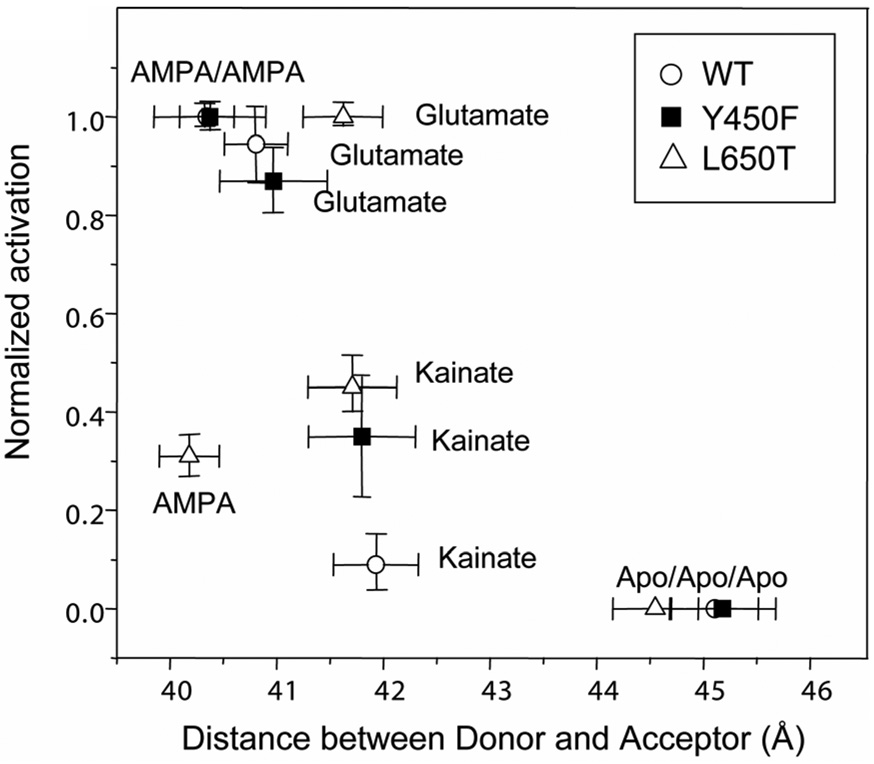
The FRET distances as determined based on the sensitized emission of the acceptor fluorophore in the presence of the donor fluorophore followed the order apo>kainate>glutamate≈AMPA. The order was consistent with the crystal structures; however, the changes as determined by FRET between the various ligated states were smaller than those observed in the crystal structures (Figure 3). Since the FRET investigations probe the average distances occupied by the fluorophores, it can be concluded that in solution on average the cleft closure changes are not as large as those observed in the crystal structures (38).
Figure 3.
Distance between N-terminus/residue 394 and residue S652 as determined by FRET for the GluR2-S1S2 wild type and L650T and Y450F mutants plotted as a function of extent of activation for the apo and AMPA-, glutamate-, and kainate-bound states (37). Reproduced with permission from Biochemistry 2007; 46: 1343–1349. Copyright 2007 American Chemical Society.
Extending these FRET investigations to the L650T and Y450F mutations that exhibit a wide range of activations by the three ligands kainate, glutamate and AMPA, it was found that the Y450F mutation followed the same trends as the wild type (37), where cleft closure correlated to the degree of activation, but that in the L650T mutant that was not the case (Figure 3). In the L650T mutant, the AMPA-bound form was more closed than the glutamate-bound form, while glutamate exhibited a greater amount of activation than AMPA (38). The L650T mutant hence provided an excellent system to study if the correlations between the interactions at the α-carboxylate and α-amine groups of the ligands as determined by the vibrational spectroscopic investigations of the wild type protein tracked the changes in the extent of cleft closure or the changes in activation.
Vibrational spectroscopic investigations in both of these mutants showed a correlation in all but one case between the strength of the interactions at the α-amine group of the agonist and degree of activation, with stronger interactions at the α-amine group resulting in a large activation of the channel (Table 1). Specifically, in the L650T mutant the strength of the interactions at the α-amine group of the agonist did not track the extent of cleft closure but could be correlated to the extent of activation. These results suggest that the interaction of the α-amine group of the agonist could be another pathway by which the ligand controls channel activation (37). It would be interesting to determine the correlations between the strength of the interactions at the α-amine group and the dynamics of the protein specifically by studying the L650T mutation. This would provide insight into whether the two mechanisms are coupled.
Kinetic steps in ligand binding: time-resolved FTIR spectroscopy
One of the additional advantages of using vibrational spectroscopy is that it can be applied in a time-resolved manner to investigate the steps of the ligand binding process as the protein proceeds from the unligated to the ligated state (36). Because the static end state structures of the GluR2 S1S2 protein have been characterized (33), the changes in the vibrational modes through time can be monitored to give structural information about the protein as it progresses through the various stages of ligand binding (36). Stopped flow fluorescence measurements with the GluR4 excised ligand binding domain have indicated a two step ligand binding process: a “ligand docking” step followed by a “protein locking” conformational change through the use of a series of mutants on both lobes of the ligand binding cleft (41). However, these stopped flow fluorescence measurements are limited by temporal constraints and structural ambiguity. Time-resolved FTIR investigations have the submillisecond time resolution required to study the ligand binding process in glutamate receptors and also the ability to provide structural information about the intermediates formed during the process. By using an inert caged precursor of glutamate, the ligand can be photolytically released from the cage with a laser pulse to initate the reaction, and measurements can be taken thereafter during the ligand binding process (32,36).
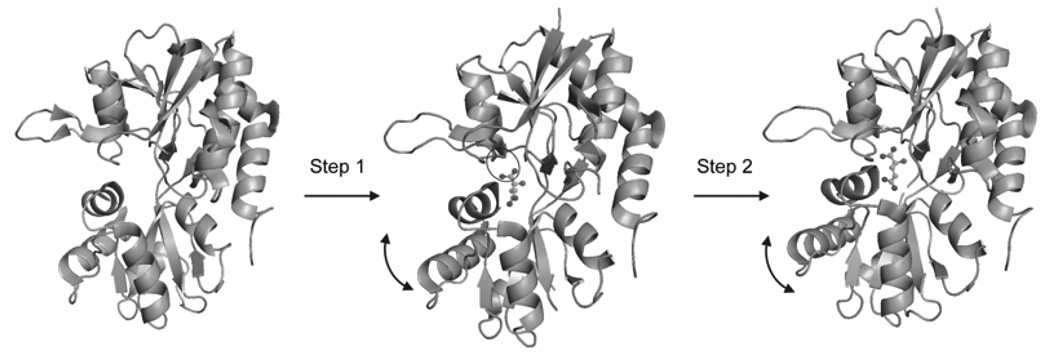
The time-resolved FTIR difference spectra for the E705D mutant (32) showed a two-step binding process, in agreement with the fluorescence measurements. However, unlike the fluorescence measurements, the FTIR spectra are able to determine the structural changes associated with the ligand binding steps (Figure 4). In the first step, interactions between the α-carboxylate of glutamate and the protein are established. In the second step, interactions at the γ-carboxylate of glutamate are formed with rearrangements in the protein backbone (32). The time-resolved spectra were able to not only confirm the two step process seen with the fluorescence measurements but were also able to structurally characterize the process to give insight into the interactions formed between glutamate and the S1S2 domain during the binding process.
Figure 4.
Schematic representation of the structural changes associated with the two-phase ligand binding process for the GluR2-S1S2 domain binding to glutamate (32). Reproduced with permission from Nat. Chem. Biol. 2005;1: 329–332. Copyright 2005 Nature Publishing Group.
Isolated ligand binding domain ⬄ full receptor?
All of the studies discussed so far in this review have been done using the isolated ligand binding domain S1S2 from the full receptor. The accuracy of the conclusions made by these studies is dependent on the inference that the S1S2 protein is a good model of this domain in the full receptor. The assumption that the behavior of the isolated S1S2 domain is equivalent to that of the full receptor has been based on studies showing similar ligand binding affinities for both (19,42,43) and similar electronic environments for the antagonist 6-cyano-7-nitro-2,3-dihydroxyquinoxaline (CNQX) (44). FRET investigations on the GluR4 subunit expressed in HEK cells have also been performed to determine if the amount of cleft closure in the full receptor is equal to that in the soluble ligand binding domain (45).
In these investigations, the N-terminus was deleted from the full length GluR4 subunit, which has been shown not to affect functioning of the receptor (46), and the inherent cysteines at positions 426 and 529 (equivalent to 425 and 528 in GluR2) were mutated to serines. GFP (acceptor) was then fused to the N-terminus of the receptor and residues 444 and 446 (443 and 445 in GluR2) in domain 1 and 653 and 686 (652 and 685 in GluR2) in domain 2 were individually mutated to cysteines and labeled with the malemide derivative of Tb chelate as the donor fluorophore. The emission lifetimes in the absence and presence of donor in the apo and glutamate- and kainate-bound states were then measured to determine the distances between the selected residues and the N-terminus.
When the domain 1 sites were tagged with terbium, no changes in distance were observed between the apo, glutamate-, and kainate-bound states of the receptor. However, when the domain 2 sites were tagged with terbium, the distances measured between the donor and acceptor followed the order of apo>kainate>glutamate (45). This is in agreement with the crystal structures (19), with the cleft being most open in the apo state, partially closed in the kainate-bound state, and fully closed in the glutamate-bound state. In addition, no changes in distance were observed for the glutamate-bound structure with and without the desensitization blocker cyclothiazide, supporting the hypothesis that receptor activation (channel opening) is mediated primarily by cleft closure while desensitization is propagated by interactions at the dimer interface (45). These results provided the first conclusive proof that the changes in the isolated ligand binding domain can be correlated to changes in the full receptor and strengthens the hypothesis that the isolated ligand binding domain is a good model for the binding domain in the full receptor.
Conclusions
The large scale expression of the isolated ligand binding domain has paved the way for detailed structural and spectroscopic investigations, which have teased apart the contributions of specific ligand:protein interactions in mediating conformational changes and dynamics in the ligand binding domain and ultimately activation of AMPA receptors. While the ligand binding domain has been studied extensively, the changes in the transmembrane and intracellular segments associated with the activation and desensitization are still largely unknown, and investigations on these segments or the full receptor are essential to gain a complete understanding of the allosteric mechanism in this group of proteins.
Acknowledgments
Acknowledgement of research grants: V.J. was supported by grants from the National Science Foundation (MCB-04444352) and the National Institutes of Health (R01GM073102 and R21NS051378). K.A.M. was funded by National Institutes of Health Molecular Biophysics Training Grant T32 GM008280.
References
- 1.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharm. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 2.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 3.Parsons CG, Danysz W, Zieglgansberger W. Excitatory amino acid neurotransmission. Handb Exp Pharmacol. 2005:249–303. doi: 10.1007/3-540-28082-0_10. [DOI] [PubMed] [Google Scholar]
- 4.Madden DR. The structure and function of glutamate receptor ion channels. Nat Rev Neurosci. 2002;3:91–101. doi: 10.1038/nrn725. [DOI] [PubMed] [Google Scholar]
- 5.Oswald RE. Ionotropic glutamate receptor recognition and activation. Adv Protein Chem. 2004;68:313–349. doi: 10.1016/S0065-3233(04)68009-0. [DOI] [PubMed] [Google Scholar]
- 6.Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- 7.Palmer CL, Cotton L, Henley JM. The molecular pharmacology and cell biology of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Pharmacol Rev. 2005;57:253–277. doi: 10.1124/pr.57.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cull-Candy S, Kelly L, Farrant M. Regulation of Ca2+-permeable AMPA receptors: synaptic plasticity and beyond. Curr Opin Neurobiol. 2006;16:288–297. doi: 10.1016/j.conb.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Van Den Bosch L, Van Damme P, Bogaert E, Robberecht W. The role of excitotoxicity in the pathogenesis of amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:1068–1082. doi: 10.1016/j.bbadis.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Gardoni F, Di Luca M. New targets for pharmacological intervention in the glutamatergic synapse. Eur J Pharmacol. 2006;545:2–10. doi: 10.1016/j.ejphar.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 11.Palmada M, Centelles JJ. Excitatory amino acid neurotransmission. Pathways for metabolism, storage and reuptake of glutamate in brain. Front Biosci. 1998;3:d701–d718. doi: 10.2741/a314. [DOI] [PubMed] [Google Scholar]
- 12.Kwak S, Weiss JH. Calcium-permeable AMPA channels in neurodegenerative disease and ischemia. Curr Opin Neurobiol. 2006;16:281–287. doi: 10.1016/j.conb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Hinoi E, Takarada T, Ueshima T, Tsuchihashi Y, Yoneda Y. Glutamate signaling in peripheral tissues. Eur J Biochem. 2004;271:1–13. doi: 10.1046/j.1432-1033.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 14.Boldyrev AA, Carpenter DO, Johnson P. Emerging evidence for a similar role of glutamate receptors in the nervous and immune systems. J Neurochem. 2005;95:913–918. doi: 10.1111/j.1471-4159.2005.03456.x. [DOI] [PubMed] [Google Scholar]
- 15.Dingledine R, Conn PJ. Peripheral glutamate receptors: molecular biology and role in taste sensation. J Nutr. 2000;130:1039S–1042S. doi: 10.1093/jn/130.4.1039S. [DOI] [PubMed] [Google Scholar]
- 16.Chenu C. Glutamatergic innervation in bone. Microsc Res Tech. 2002;58:70–76. doi: 10.1002/jemt.10120. [DOI] [PubMed] [Google Scholar]
- 17.Salt TE. Glutamate receptor functions in sensory relay in the thalamus. Philos Trans R Soc Lond B Biol Sci. 2002;357:1759–1766. doi: 10.1098/rstb.2002.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFeeters RL, Oswald RE. Emerging structural explanations of ionotropic glutamate receptor function. Faseb J. 2004;18:428–438. doi: 10.1096/fj.03-0873rev. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong NA, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 21.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 23.Jin R, Horning M, Mayer ML, Gouaux E. Mechanism of activation and selectivity in a ligand-gated ion channel: structural and functional studies of GluR2 and quisqualate. Biochemistry. 2002;41:15635–15643. doi: 10.1021/bi020583k. [DOI] [PubMed] [Google Scholar]
- 24.Armstrong N, Mayer M, Gouaux E. Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic adjustment of agonist-induced conformational changes. Proc Natl Acad Sci U S A. 2003;100:5736–5741. doi: 10.1073/pnas.1037393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. Embo J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin R, Gouaux E. Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal. Biochemistry. 2003;42:5201–5213. doi: 10.1021/bi020632t. [DOI] [PubMed] [Google Scholar]
- 27.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 29.McFeeters RL, Oswald RE. Structural mobility of the extracellular ligand-binding core of an ionotropic glutamate receptor. Analysis of NMR relaxation dynamics. Biochemistry. 2002;41:10472–10481. doi: 10.1021/bi026010p. [DOI] [PubMed] [Google Scholar]
- 30.McFeeters RL, Swapna GV, Montelione GT, Oswald RE. Semi-automated backbone resonance assignments of the extracellular ligand-binding domain of an ionotropic glutamate receptor. J Biomol NMR. 2002;22:297–298. doi: 10.1023/a:1014954931635. [DOI] [PubMed] [Google Scholar]
- 31.Valentine ER, Palmer AG., 3rd Microsecond-to-millisecond conformational dynamics demarcate the GluR2 glutamate receptor bound to agonists glutamate, quisqualate, and AMPA. Biochemistry. 2005;44:3410–3417. doi: 10.1021/bi047984f. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Q, Du M, Ramanoudjame G, Jayaraman V. Evolution of glutamate interactions during binding to a glutamate receptor. Nat Chem Biol. 2005;1:329–332. doi: 10.1038/nchembio738. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Q, Jayaraman V. Chemistry and conformation of the ligand-binding domain of GluR2 subtype of glutamate receptors. J Biol Chem. 2004;279:26346–26350. doi: 10.1074/jbc.M403111200. [DOI] [PubMed] [Google Scholar]
- 34.Jayaraman V, Keesey R, Madden DR. Ligand--protein interactions in the glutamate receptor. Biochemistry. 2000;39:8693–8697. doi: 10.1021/bi000892f. [DOI] [PubMed] [Google Scholar]
- 35.Madden DR, Thiran S, Zimmermann H, Romm J, Jayaraman V. Stereochemistry of quinoxaline antagonist binding to a glutamate receptor investigated by Fourier transform infrared spectroscopy. J. Biol. Chem. 2001;276:37821–37826. doi: 10.1074/jbc.M106171200. [DOI] [PubMed] [Google Scholar]
- 36.Jayaraman V. Spectroscopic and kinetic methods for ligand-protein interactions of glutamate receptor. Methods Enzymol. 2004;380:170–187. doi: 10.1016/S0076-6879(04)80008-9. [DOI] [PubMed] [Google Scholar]
- 37.Mankiewicz KA, Rambhadran A, Du M, Ramanoudjame G, Jayaraman V. Role of the Chemical Interactions of the Agonist in Controlling alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid Receptor Activation. Biochemistry. 2007;46:1343–1349. doi: 10.1021/bi062270l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramanoudjame G, Du M, Mankiewicz KA, Jayaraman V. Allosteric mechanism in AMPA receptors: A FRET-based investigation of conformational changes. Proc Natl Acad Sci U S A. 2006;103:10473–10478. doi: 10.1073/pnas.0603225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Q, Thiran S, Yernool D, Gouaux E, Jayaraman V. A vibrational spectroscopic investigation of interactions of agonists with GluR0, a prokaryotic glutamate receptor. Biochemistry. 2002;41:1602–1608. doi: 10.1021/bi015729e. [DOI] [PubMed] [Google Scholar]
- 41.Abele R, Keinänen K, Madden DR. Agonist-induced isomerization in a glutamate receptor ligand-binding domain: a kinetic and mutagenetic analysis. J. Biol. Chem. 2000;275:21355–21363. doi: 10.1074/jbc.M909883199. [DOI] [PubMed] [Google Scholar]
- 42.Arvola M, Keinänen K. Characterization of the ligand-binding domains of glutamate receptor (GluR)-B and GluR-D subunits expressed in Escherichia coli as periplasmic proteins. J. Biol. Chem. 1996;271:15527–15532. doi: 10.1074/jbc.271.26.15527. [DOI] [PubMed] [Google Scholar]
- 43.Kuusinen A, Arvola M, Keinänen K. Molecular dissection of the agonist binding site of an AMPA receptor. EMBO J. 1995;14:6327–6332. doi: 10.1002/j.1460-2075.1995.tb00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deming D, Cheng Q, Jayaraman V. Is the isolated ligand binding domain a good model of the domain in the native receptor? J Biol Chem. 2003;278:17589–17592. doi: 10.1074/jbc.C300105200. [DOI] [PubMed] [Google Scholar]
- 45.Du M, Reid SA, Jayaraman V. Conformational changes in the ligand-binding domain of a functional ionotropic glutamate receptor. J Biol Chem. 2005;280:8633–8636. doi: 10.1074/jbc.C400590200. [DOI] [PubMed] [Google Scholar]
- 46.Pasternack A, Coleman SK, Jouppila A, Mottershead DG, Lindfors M, Pasternack M, Keinanen K. Alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor channels lacking the N-terminal domain. J Biol Chem. 2002;277:49662–49667. doi: 10.1074/jbc.M208349200. [DOI] [PubMed] [Google Scholar]