Abstract
Recent transcriptomics efforts have revealed that numerous protein-coding messenger RNAs have natural antisense transcript partners, most of which seem to be noncoding RNAs. Here we identify a conserved noncoding antisense transcript for β-secretase-1 (BACE1), a crucial enzyme in Alzheimer’s disease pathophysiology. The BACE1-antisense transcript (BACE1-AS) regulates BACE1 mRNA and subsequently BACE1 protein expression in vitro and in vivo. It seems that the argument for concordant regulation can only be made in the experiments with the siRNA against BACE1-AS. This convention has been followed throughout the manuscript. Please check carefully.]. Upon exposure to various cell stressors including amyloid-β 1–42 (Aβ 1–42), expression of BACE1-AS becomes elevated, increasing BACE1 mRNA stability and generating additional Aβ 1–42 through a post-transcriptional feed-forward mechanism. BACE1-AS concentrations were elevated in subjects with Alzheimer’s disease as well as in amyloid precursor protein transgenic mice. These data show that BACE1 mRNA expression is under the control of a regulatory noncoding RNA that may drive Alzheimer’s disease–associated pathophysiology. In summary, we report that a long noncoding RNA is directly implicated in the increased abundance of Aβ 1–42 in Alzheimer’s disease.
Sequential cleavage of amyloid precursor protein (APP) by BACE1, the β-site cleaving enzyme essential for Aβ 1–42 and amyloid-β 1–40 (Aβ 1–40) biosynthesis1, and γ-secretase initiates the ‘amyloid cascade’ that is central to Alzheimer’s disease pathophysiology2,3. Oligomers of Aβ 1–42 produced by BACE1 influence key aspects of Alzheimer’s disease4-9. Studies have revealed elevated brain BACE1 concentrations in subjects with Alzheimer’s disease compared with normal controls10-15. However, controversy exists concerning the extent of BACE1 upregulation and whether this upregulation involves BACE1 mRNA or protein16-18.
Loss of BACE1 results in numerous behavioral and physiological deficits, including memory loss19, emotional deficits20, myelination defects in peripheral nerves21,22 and loss of synaptic plasticity20. Thus, the subtle but crucial boundaries between BACE1 physiology and pathology indicate that BACE1 expression must be tightly regulated, allowing the enzyme to perform its physiological functions while avoiding the serious consequences of over- or underexpression.
Here we report that BACE1-AS, a natural antisense transcript, plays a part in determining BACE1 expression. BACE1-AS rapidly and reversibly upregulates BACE1 levels in response to a variety of stresses, including Aβ 1–42 exposure. Furthermore, we show elevated BACE1-AS in several brain regions of individuals with Alzheimer’s disease. These data suggest that this previously unexamined noncoding RNA has a role in regulating BACE1 and in driving Alzheimer’s disease pathology.
RESULTS
Identification of BACE1 natural antisense transcript
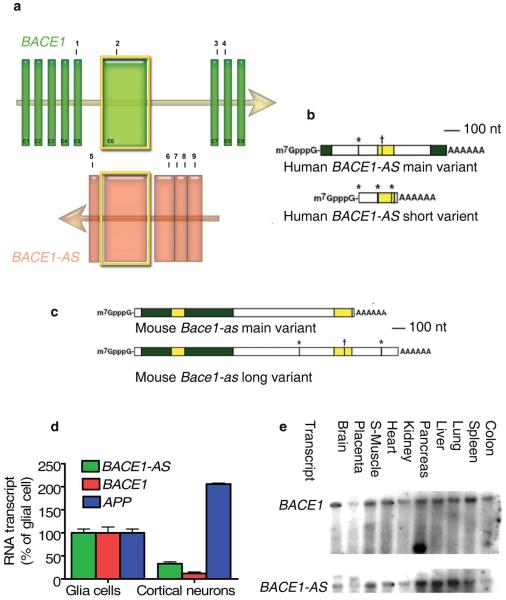
BACE1-AS was originally identified through the FANTOM large-scale transcriptomics efforts as one member of some 1,000 sense-antisense pairs conserved between human and mouse23. BACE1-AS is a conserved ~2-kb RNA transcribed from the positive strand of chromosome 11, on the opposite strand of the BACE1 locus (11q 23.3), including 104 nucleotides of full complementarity to exon 6 of human BACE1 mRNA (Fig. 1a). We performed rapid amplification of cDNA ends (RACE) for directional sequencing of 5′ and 3′ ends and identified two splice variants for human and mouse BACE1-AS that overlap the BACE1 sense transcript (Fig. 1b,c). We found a poly-A tail and cap structure in both human and mouse sequences, indicating that BACE1-AS is a fully processed transcript of RNA polymerase II. However, there is no apparent open reading frame. Sequencing data for human and mouse BACE1-AS are illustrated in Supplementary Data 1 and 2 online.
Figure 1.
Genomic organization and expression analysis of BACE1 and BACE1-AS. (a) Genomic sequences of BACE1 and BACE1-AS; arrows show the direction of transcription. BACE1 exons are depicted as vertical bars and marked E1–E9. Human BACE1-AS is transcribed from the same region in chromosome 11, but on the opposite strand. Yellow highlighted exons are the overlapping region (104 base pairs) of BACE1 and BACE1-AS, which are conserved across species. Sites numbered 1, 3 and 4 are BACE1 siRNAs target sites, and site 2 is the northern blot probe site. Sites 5, 6 and 7 are the target sites of the BACE1-AS siRNAs and site 8 is the RT-PCR probe target region, which are all in the non-overlapping part of the BACE1-AS transcript. Sites 5 and 9 represent the primers for 3′ and 5′ RACE, respectively. (b,c) RACE sequencing data revealed that BACE1-AS contains cap structure and a poly-A tail and that this transcript undergoes differential splicing in both human and mouse. The yellow highlighted segments are the overlap region to the BACE1 sense transcript and the green highlighted segments are additional nucleotides observed from our sequencing data. Point mismatches to the genomic sequence are indicated by stars (*) for A to G and crosses (†) for C to T changes. Nt, nucleotides. (d) Expression of BACE1-AS, BACE1 and APP mRNA in human cortical neurons (HCN1A) compared to glial cells (M059K). (e) Northern blot expression analysis of BACE1 (top) and BACE1-AS (bottom) in ten human tissues. S-muscle, skeletal muscle
Expression analysis of BACE1 and BACE1-AS
BACE1 mRNA expression levels were 25–75% greater than BACE1-AS transcript levels across all samples examined from various tissues and cell lines, in contrast to the relative concentrations in Alzheimer’s disease brain (described below). We also observed Bace1 and Bace1-AS transcripts in various regions of the mouse brain. (Supplementary Figs. 1 and 2 online).
In cultures of glia and cortical neurons of human origin, APP mRNA was about twice as abundant in neuronal cultures compared to glial cultures, whereas BACE1 and BACE1-AS transcripts were two to three times more abundant in the glial cells (Fig. 1d). Northern blot analysis with in vitro–transcribed strand-specific RNA probes confirmed that various human tissues express both BACE1 and BACE1-AS (Fig. 1e). This observation suggests that BACE1 and BACE1-AS expression may be regulated concordantly, as was recently shown for other sense-antisense pairs24,25.
BACE1-AS regulates BACE1 RNA and protein in vitro
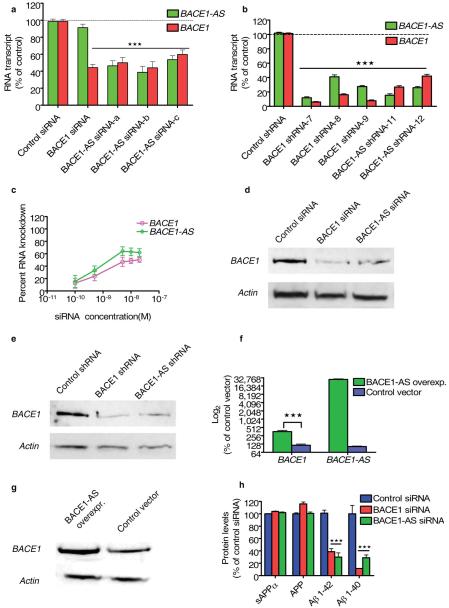
We next investigated whether the BACE1-AS transcript regulates expression of BACE1 mRNA. Unexpectedly, transfection of human SH-SY5Y cells with any one of three distinct small interfering RNA (siRNA) sequences targeting nonoverlapping regions of the BACE1-AS transcript resulted in a statistically significant knockdown of not only the targeted BACE1-AS transcript, but also BACE1 mRNA (Fig. 2a). There are broadly two types of regulation between sense and antisense transcripts. In concordant regulation, like in the case of BACE1-AS, the antisense transcripts change the level of the sense RNA, or corresponding protein levels, in a positive way. In contrast, in discordant regulation, the antisense transcripts have negative (opposing) effects on sense transcripts. We also observed this concordant pattern of regulation in human HEK293T and HEK-SW cells, in which siRNA-mediated knockdown of BACE1-AS resulted in a similar reduction in BACE1 mRNA (data not shown). In a control experiment in HEK-SW cells, the level of BACE2 did not change with BACE1-AS siRNA treatment, lending further support to the specificity of the observed BACE1 regulation by BACE1-AS (Supplementary Fig. 3 online). Furthermore, because three distinct siRNA molecules that exclusively target the BACE1-AS transcript resulted in concomitant reduction of sense BACE1 transcript, it is highly unlikely that the siRNAs acted to knock down BACE1 transcript through a nonspecific (or ‘off-target’) mechanism.
Figure 2.
BACE1-AS regulates BACE1 mRNA and protein expression in vitro. (a) Targeting BACE1-AS transcript with three different siRNAs caused decreases (P < 0.0001) in both BACE1 and BACE1-AS transcripts in neuroblastoma cells (SH-SY5Y). (b) Stable transfection of HEK293T cells with shRNA for BACE1, BACE1-AS and a control shRNA shows that knockdown of BACE1 for an extended period of time leads to reduction of BACE1-AS and knockdown of BACE1-AS also leads to reduction of BACE1 mRNA levels (P < 0.001). (c) HEK-SW cells were transfected with 100 pM, 500 pM, 5 nM, 10 nM or 20 nM BACE1-AS siRNA. BACE1-AS knockdown ranged from 10–60%, and BACE1 downregulation ranged between 10–50% with increasing concentrations of siRNA. (d,e) Western blot showing that knockdown of either BACE1 or BACE1-AS with siRNA or shRNA leads to reduction of the BACE1 protein. (f,g) Overexpression of BACE1-AS but not an empty control vector leads to increased BACE1 mRNA (P < 0.001) and protein concentrations. (h) HEK-SW cells were transfected with siRNA for BACE1, BACE1-AS or a control siRNA and analyzed for Aβ 1–40, Aβ 1–42, sAPPα and total APP concentrations by ELISA. Aβ 1–40 and Aβ 1–42 abundance was reduced (P < 0.0001) after transfection of siRNA targeting of either BACE1 or BACE1-AS. Total APP or sAPPα, an enzymatic product of α-secretase, were not changed.
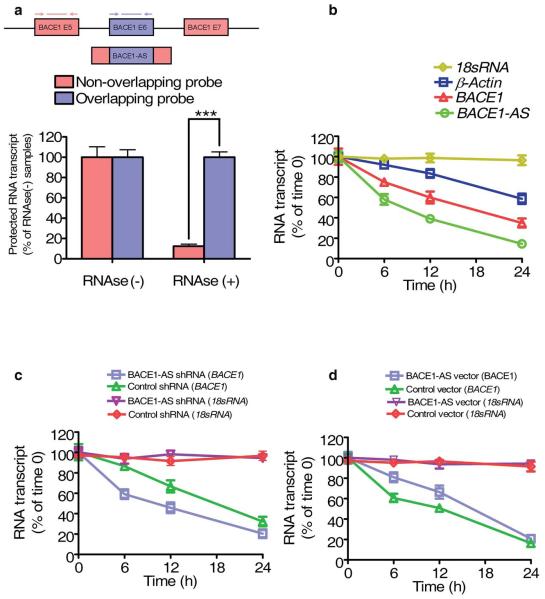
To investigate the effects of long-term siRNAs directed against BACE1-AS on BACE1 expression, we generated stable HEK293T cell lines expressing four distinct short hairpin RNAs (shRNAs) to BACE1 or BACE1-AS transcripts. Three of the BACE1 shRNAs and two of the BACE1-AS shRNAs were functional and induced sustained reduction of BACE1 and BACE1-AS, respectively (Fig. 2b). Cells expressing BACE1-AS shRNA showed reduced BACE1 levels, and vice versa (Fig. 2b).
To examine the dose-response relationship of BACE1 mRNA and BACE1-AS siRNA, we measured the reduction in BACE1 mRNA expression across a range of concentrations of BACE1-AS siRNA (100 pM–20 nM) in HEK-SW cells. The resulting data confirmed that siRNA targeting BACE1-AS reduces the expression of BACE1 mRNA in a concentration-dependent manner (Fig. 2c).
Next, we assessed BACE1 protein abundance after transfection of HEK293T cells with BACE1-AS siRNA or shRNA. Western blotting showed that siRNA against BACE1-AS as well as siRNA against BACE1, (but not control siRNAs), reduced the expression of BACE1 protein (Fig. 2d,e). Thus, BACE1-AS seems to control the expression of BACE1 at both the mRNA and the protein levels. Additionally, for accurate quantification of the effects of siRNA treatments on BACE1 protein expression, we established a method for protein quantification by enzyme complementation assay (ECA, Methods and Supplementary Fig. 4 online). We then measured the changes in BACE1 protein concentration after treatment with siRNAs and shRNAs and observed that siRNAs against either BACE1 or BACE1-AS reduce BACE1 protein abundance by 40–60% (Supplementary Fig. 5 online).
Moreover, overexpression of BACE1-AS led to a fourfold increase in BACE1 mRNA (Fig. 2f). When measured by western blotting, the overexpression of BACE1-AS resulted in increased BACE1 protein abundance (Fig. 2g). These observations further confirm the regulation of BACE1 expression by BACE1-AS, not only at the mRNA but also at the protein level.
Knockdown of BACE1-AS reduces Aβ 1–40 and Aβ 1–42 production
We measured by ELISA the amount of Aβ 1–40 and Aβ 1–42 after depletion of BACE1-AS in the HEK-SW cell line that contains mutated APP, swAPP751, so-called Swedish mutation26. We found a reduced concentration of both Aβ 1–40 and Aβ 1–42 in cells treated with BACE1-AS siRNA (Fig. 2h). To rule out possible nonspecific effects of siRNA treatment on APP concentration that may account for the observed decreases in Aβ 1–40 and Aβ 1–42, we assayed the amount of soluble APPα (sAPPα, soluble product of α-secretase in the supernatants of the HEK-SW cells) and total APP abundance by ELISA. Neither sAPPα nor total APP abundance was altered upon BACE1 siRNA or BACE1-AS siRNA treatment (Fig. 2h). These results suggest that BACE1-AS siRNA treatment results in reduced BACE1 protein function without affecting APP or α-secretase products.
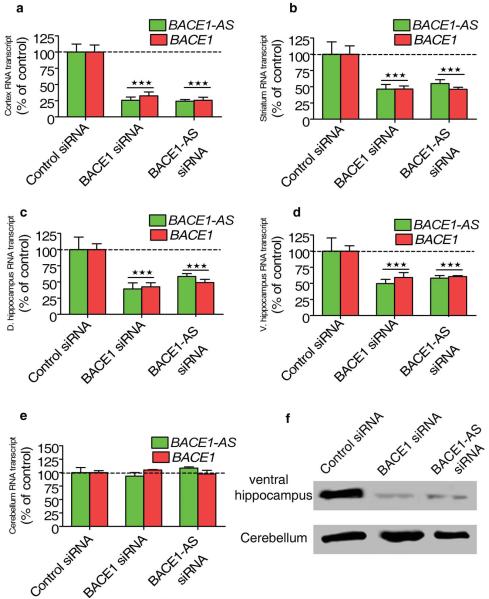
Knockdown of BACE1-AS reduces BACE1 levels in vivo
The above data support a role for BACE1-AS in regulating BACE1 function in vitro in human cells. Next, we assessed whether orthologous Bace1-AS also regulates Bace1 mRNA and protein in vivo in mouse brain. After 14 d of continuous siRNA infusion, Bace1 mRNA levels were reduced across forebrain regions located adjacent to the third ventricle in mice treated with either Bace1 siRNA or Bace1-AS siRNA, compared to levels unaltered by control siRNA (Fig. 3a–d). Bace1 and Bace1-AS transcripts were unaltered in the cerebellum, a structure that is spatially restricted from the third ventricle, of siRNA-treated mice, consistent with previous work that indicates limited penetration of pump-mediated infusion of siRNA into the brain27 (Fig. 3e). We also measured the amount of Bace1 protein in the ventral hippocampus and the cerebellum by western blotting after siRNA treatment. We found that both Bace1 siRNA and Bace1-AS siRNA treatment resulted in reduced Bace1 protein abundance (Fig. 3f). Our in vivo findings agree with the in vitro data described above and indicate that reduced Bace1-AS expression results in reduction of Bace1 mRNA and protein expression in vivo.
Figure 3.
Bace1-AS regulates Bace1 in vivo. (a–e) Synthetic unmodified siRNAs designed to target the nonoverlapping region of either Bace1 or Bace1-AS and a control siRNA were constantly infused into three groups of mice over a period of two weeks. The siRNAs directed against either Bace1 or Bace1-AS but not the control siRNA resulted in decrease in both Bace1 and Bace1-AS levels (P < 0.0001) in cortex (a), striatum (b), dorsal hippocampus (c) and ventral hippocampus (d). In the cerebellum (e) both transcripts were unchanged, as expected for a tissue that is not directly connected to the third ventricle of the brain. (f) Western blot showing decreases of Bace1 protein abundance in the ventral hippocampus but not in the cerebellum after in vivo treatment with siRNA against either Bace1 or Bace1-AS.
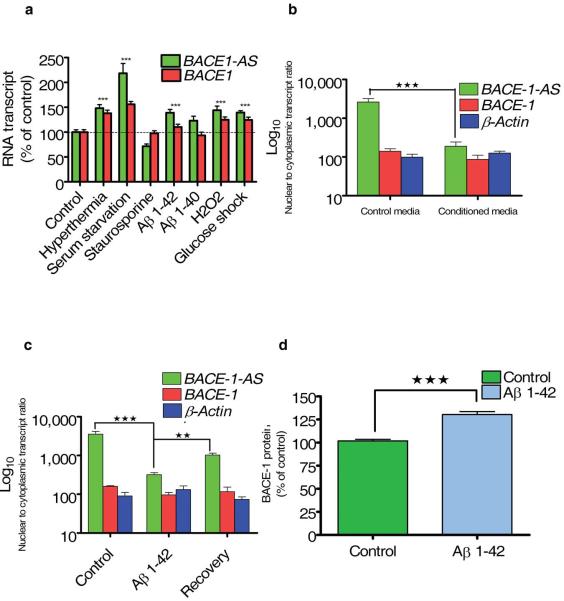
Cell stressors increase BACE1-AS and BACE1 protein
Different cell stressors have long been implicated in the pathogenesis of Alzheimer’s disease12,28. We exposed HEK-SW cells to hyperthermia, serum starvation, staurosporine, Aβ 1–42, Aβ 1–40, hydrogen peroxide (H2O2) or high glucose for 12 h. We found that exposure of the cells to high temperature, serum starvation, Aβ 1–42, H2O2 or high glucose resulted in a ~30–130% increase in BACE1-AS levels and a ~20–60% increase in BACE1 mRNA levels (Fig. 4a). Serum starvation generated the strongest response, whereas Aβ 1–40 and staurosporine exposure did not significantly alter BACE1-AS expression levels(Fig. 4a). These results suggest that many, but not all, cell stressors can contribute to the pathogenesis of Alzheimer’s disease by altering BACE1-AS expression and subsequently BACE1 enzyme activity.
Figure 4.
Effect of cell stressors on BACE1 and BACE1-AS. (a) HEK-SW cells were exposed to cell stressors. BACE1-AS and BACE1 transcripts were measured by RT-PCR and normalized to ACTB as an endogenous control. Hyperthermia, serum starvation, Aβ 1–42, hydrogen peroxide or glucose shock caused elevation of BACE1-AS and, to a lesser degree, elevation of BACE1 transcripts. Staurosporine or Aβ 1–40 did not increase either transcript level. (b) The 7PA2-CHO cells were previously shown to overproduce Aβ 1–42 dimers and oligomers. Conditioned media from these cells or control parental CHO cells were collected and added to SH-SY5Y cells for 2 h after removal of the regular media. Conditioned media from 7PA2-CHO cells, but not control media, caused the BACE1-AS transcript to relocate to the cytoplasm (P < 0.0001). (c) Exposure of SH-SY5Y cells to 1 μM Aβ 1–42 for 2 h caused an increase in cytoplasmic BACE1-AS (P < 0.001). The nuclear-cytoplasmic ratio was recovered upon removal of the peptides and maintenance of the cells in regular media for 1 h. (d) Exposure to 1 μM Aβ 1–42 peptide caused an elevation in BACE1 protein abundance (P < 0.001). Protein amounts were measured with a β-galactosidase ECA.
Accumulating evidence describes Aβ 1–42 itself as a potent cell stressor10,29-32. To test the hypothesis that Aβ 1–42 increases BACE1 expression by a BACE1-AS dependent mechanism, we exposed SH-SY5Y cells for 2 h to conditioned media from CHO-7PA2 cells, which overexpress APP and generate Aβ 1–42 dimers and oligomers33. Exposure of the SH-SY5Y cells to conditioned media from CHO-7PA2 cells, but not conditioned media from control parental CHO cells resulted in an increase in cytoplasmic concentrations of BACE1-AS transcript (Fig. 4b). We obtained similar results when incubating SH-SY5Y cells with synthetic Aβ 1–42 (1 μM for 2 h; Fig. 4c). Removal of the cell stressors normalized BACE1-AS expression patterns. Using an ECA, we found that synthetic Aβ 1–42 (1 μM for 12 h) elicited an increase in BACE1 protein abundance as well (Fig. 4d). Taken together, the above data indicate that cell stress increases BACE1-AS levels, which in turn increases BACE1 levels; this may result in an increase in APP processing and Aβ 1–42 production. Subsequently, increased Aβ 1–42 levels can further increase BACE1-AS expression, driving the APP processing cascade in a feed-forward manner.
BACE1-AS forms RNA duplex and increases stability of BACE1
We used an RNase protection assay (RPA) on RNA from SH-SY5Y cells to test the possibility of RNA duplex formation (Supplementary Methods). RT-PCR data showed that the overlapping part of both transcripts was protected from degradation, indicating that BACE1 and BACE1-AS indeed form a RNA duplex (Fig. 5a). We also validated the RPA data on a 10% Tris-borate-EDTA–urea gel using radiolabeled BACE1-AS probes (data not shown).
Figure 5.
BACE1-AS increases the stability of BACE1 mRNA. (a) RPA performed on RNA samples from SH-SY5Y cells. Depicted here are RT-PCR results from two sets of primers and probes covering overlapping and nonoverlapping regions of BACE1 mRNA. The overlapping region of BACE1 transcript is protected from degradation by RNase A+T, suggesting RNA duplex formation. (b) Stability of BACE1 and BACE1-AS transcripts over time was measured by RT-PCR relative to time 0 after blocking new RNA synthesis with α-amanitin (50 μM) in HEK293T cells. BACE1-AS showed a shorter half-life than BACE1 and ACTB. 18s RNA, which is a product of RNA polymerase I, was unchanged. (c) The stability of BACE1 mRNA was measured in stably transfected HEK293T cells expressing a BACE1-AS shRNA and a second cell line expressing a negative control shRNA. The stability of BACE1 mRNA was decreased in cells expressing BACE1-AS shRNA relative to the control cell line (P < 0.01). (d) The stability of BACE1 mRNA increased in HEK293T cells overexpressing BACE1-AS in comparison to a cell line transfected with an empty vector.
RNA duplex formation may act to alter the secondary or tertiary structure of BACE1 and thereby increase its stability. We assessed the stability of BACE1 and BACE1-AS transcripts by blocking new RNA synthesis with α-amanitin and measuring the loss of BACE1, BACE1-AS, β-actin (ACTB) and 18s RNA over a 24-h period. We found that BACE1-AS had a shorter half-life than BACE1 mRNA (8.5 h versus 17.5 h Fig. 5b). 18s ribosomal RNA, which is a product of RNA polymerase I, was not affected by α-amanitin treatment (Fig. 5b). In a cell line that constitutively expresses BACE1-AS shRNA and thereby has depleted BACE1-AS levels, we found decreased stability of BACE1 mRNA compared with cells transfected with a control shRNA (Fig. 5c). Conversely, cells that overexpress BACE1-AS showed increased stability of BACE1 (Fig. 5d). Collectively, our data demonstrate that BACE1-AS increases the stability of BACE1 mRNA.
BACE1-AS is elevated in subjects with Alzheimer’s disease
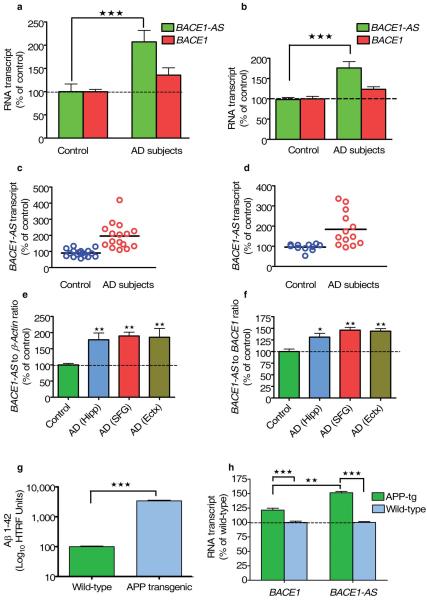
Elevated BACE1-AS concentrations may facilitate increased BACE1 activity and disease progression in the brains of human subjects with Alzheimer’s disease. To examine this question, we assessed BACE1-AS and BACE1 mRNA abundance in RNA samples prepared from parietal lobes and cerebellum from five postmortem brains of human subjects with Alzheimer’s disease and from five age- and sex-matched control brains. In the Alzheimer’s disease samples, the relative quantity of BACE1-AS transcript was increased by two to three times, along with a smaller increase in BACE1 transcript (Fig. 6a and Supplementary Fig. 6a online). In a separate group of 35 subjects with Alzheimer’s disease and 35 age- and sex-matched controls34, we examined RNA samples derived from cerebellum (25 Alzheimer’s disease samples and 21 control samples), hippocampus (13 Alzheimer’s disease samples and 11 control samples), entorhinal cortex (13 Alzheimer’s disease samples and 11 control samples) and superior frontal gyrus (16 Alzheimer’s disease samples and 17 control samples). The BACE1-AS transcript concentrations were elevated in subjects with Alzheimer’s disease by up to sixfold, with an average elevation of about twofold across all brain regions (Fig. 6b–d and Supplementary Fig. 6b,c). We detected a smaller (~30%) increase in BACE1 mRNA concentrations in these subjects compared to their matched controls (Fig. 6a,b) [AU: Figure callout or ‘data not shown’?]. Taken together, these results support our hypothesis that increases in BACE1-AS expression, probably related to cell stressors, drives upregulation of BACE1 mRNA and protein level, thereby facilitating Aβ 1–42 biosynthesis in human Alzheimer’s disease brain.
Figure 6.
BACE1-AS and BACE1 expression is elevated in the brain of individuals with Alzheimer’s disease. (a) The relative quantity of BACE1-AS transcript was elevated by two to three times in parietal cortex and cerebellum of five human subjects with Alzheimer’s disease (AD subjects) (P < 0.0001) as compared to matched control individuals (20 RNA samples). To a lesser degree, BACE1 mRNA was also increased, by approximately 30%. (b) The relative quantity of BACE1-AS transcript was elevated by almost twofold (P < 0.0001) and BACE1 mRNA elevated about 30% in four brain regions (cerebellum, superior frontal gyrus, entorhinal cortex and hippocampus) of 35 individuals with Alzheimer’s disease compared to the average of 35 control individuals (128 RNA samples in total). (c) Scatter plot of BACE1-AS transcript expression in superior frontal gyrus of 17 control subjects and 16 subjects with Alzheimer’s disease. Upregulation (P < 0.0001) of BACE1-AS was observed in subjects with Alzheimer’s disease. (d) Scatter plot of BACE1-AS transcript expression in hippocampus of 11 controls and 13 individuals with Alzheimer’s disease. Upregulation (P < 0.001) of BACE1-AS was observed in the subjects with Alzheimer’s disease. (e,f) BACE1-AS to ACTB ratio (e) and BACE1-AS to BACE1 ratio (f) in hippocampus (Hipp), superior frontal gyrus (SFG), entorhinal cortex (Ectx) of subjects with Alzheimer’s disease compared to control individuals. Both ratios are higher (P < 0.001) in subjects with Alzheimer’s disease. (g) The concentrations of human Aβ 1–42 peptide are elevated in the brains of APP-tg19599 mice (P < 0.0001). (h) Bace1-AS is elevated by 50% (P < 0.0001) in whole brains of APP-tg19599 (APP-tg) mice.
BACE1-AS may have utility as a new biomarker of Alzheimer’s disease35. To this end, we calculated the ratio of BACE1-AS relative to BACE1 and ACTB mRNA in different brain regions of control subjects and subjects with Alzheimer’s disease. We found that the BACE1-AS to ACTB ratio was increased in various brain regions in subjects with Alzheimer’s disease as compared to control individuals (Fig. 6e). A smaller increase in the BACE1-AS to BACE1 ratio was also observed in the brains of individuals with Alzheimer’s disease (Fig. 6f). These data demonstrate that the ratio between BACE1-AS and other RNA transcripts, including BACE1, could potentially be used as a biomarker of Alzheimer’s disease.
APP transgenic mice have increased levels of Bace1-AS
Tg19959 mice, considered a mouse model of Alzheimer’s disease, overexpress a doubly mutated human APP (APP-tg19959)36 and consequently have increased levels of Aβ 1–42 (ref. 37). Samples from whole brains excised from four six-week-old male mice had increased (~300-fold) levels of Aβ 1–42 compared with samples from matched wild-type controls, as measured by homogeneous time resolved fluorescence (HTRF) assay (Fig. 6g and Supplementary Methods). Expression of the Bace1-AS transcript was increased by about 45%, and Bace1 mRNA expression was increased by about 25% in the brains of the APP-tg19959 mice compared with wild-type control mice (Fig. 6h), similar to the measurements in the human samples.
DISCUSSION
The contrast between BACE1’s essential role in cognitive, emotional and synaptic functions19,20 and its pathophysiological dysregulation in Alzheimer’s disease38,39 highlights the regulatory complexity of this protein. Owing to the consequences of its dysregulation, BACE1 gene expression must normally maintain tight robust regulatory control.
In this study, we have characterized a conserved noncoding antisense transcript for BACE1, called BACE1-AS, which functions as a regulator of BACE1 gene expression. We present data showing that BACE1-AS is widely co-expressed with BACE1 in cell lines, tissues and Alzheimer’s disease–sensitive brain regions and that it regulates BACE1 expression in vitro and in vivo. We found that selective siRNA targeting of the nonoverlapping regions of the BACE1-AS resulted in reduction of BACE1 mRNA and protein abundance in vitro. Administration of siRNA that selectively targeted either Bace1 or Bace1-AS into mouse brains reduced the levels of both transcripts, indicating that this concordant regulation also occurs in vivo.
In addition, we have shown that alterations in BACE1-AS RNA concentrations can alter Aβ 1–40 and Aβ 1–42 production. Considering the narrow window between essential levels and excessive levels (as in Alzheimer’s disease) of BACE1 protein, we believe that neuronal cells must maintain precise physiological regulation of BACE1 expression by using both pre- and post-transcriptional regulatory mechanisms. The RNA transcript, BACE1-AS, seems to function as a regulatory component of this machinery.
Because BACE1-AS regulates BACE1 expression in vivo, we propose that the elevation of BACE1-AS, resulting from the actions of Alzheimer’s disease–related cell stressors, forms a basis for a deleterious feed-forward cycle of Alzheimer’s disease progression. Even small changes in BACE1 activity may lead to a long-lasting and chronic process of Aβ 1–42 accumulation in the Alzheimer’s disease brain38,40. Our current findings provide further evidence for a feed-forward mechanism of stress-dependent and activity-dependent41 Aβ 1–42 production. Recent studies have shown that amyloid plaques induce elevation of BACE1 protein expression in adjacent neurons by a post-transcriptional mechanism10. This finding is consistent with the present data in which Aβ 1–42 was shown to induce increased levels of BACE1-AS, thereby driving BACE1-mediated APP processing and further accumulation of Aβ 1–42. In support of the above interpretation, we found that two independent sets of human Alzheimer’s disease brain samples as well as an animal model of Alzheimer’s disease express elevated levels of both BACE1-AS transcript and, to a lesser degree, BACE1 sense transcript. In contrast to the downregulation of most transcripts reported to date in Alzheimer’s disease brains42,43, BACE1-AS upregulation may be the driving force behind Alzheimer’s disease–related BACE1 dysregulation15,44,45. Thus, our results implicate a noncoding RNA in the control of gene expression central to the delicate balance between healthy stress response and the pathophysiological β-amyloid cascade.
Aβ 1–42 induces synaptic depression by triggering endocytosis of glutamatergic N-methyl d-aspartate receptors from the post-synaptic membrane6. Synaptic activity–dependent production of Aβ 1–42 achieves this depressive effect of N-methyl d-aspartate receptor endocytosis by a series of common mechanisms that implicate Aβ 1–42 in the establishment of some forms of long-term depression46. Although they contribute to the depth and richness of mammalian memory, if not precisely controlled, these mechanisms may lead to chronic neuronal stress and the onset of Alzheimer’s disease. We have previously speculated that noncoding RNAs may be required for some forms of long-term depression47, and BACE1-AS could well be involved in such a function.
Treatment with BACE1-AS siRNA may achieve a preferential reduction of stress-induced increases in BACE1 expression without disturbing physiologically essential basal expression levels. Thus, we propose that BACE1-AS could potentially constitute a drug target candidate well suited to mediate the transition between the essential physiological functions of BACE1 and its pathological dysregulation in the chronically stressed setting of early Alzheimer’s disease48. Our in vivo experiments using infusion of unmodified synthetic siRNA over an extended period of time in experimental mice support the validity of an siRNA approach to decrease BACE1 expression, perhaps in humans as well. A recent technological breakthrough suggests that systemic administration of modified siRNA may cross the blood-brain barrier and thereby target RNA transcripts in the brain49. Alternatively, proteins involved in BACE1-AS localization or turnover could serve as potential targets for therapeutic interventions.
METHODS
Enzyme complementation assay
ECA is a technology developed by DiscoveRx that allows for the measurement of changes in protein abundance (Supplementary Fig. 2). We cloned the cDNA of BACE1 into a pCMV-ProLabel vector upstream of the ProLabel. We transfected vector into HEK293T cells to produce a fusion protein (BACE1 and the enzyme donor fragment of β-galactosidase) and made a stable cell line, which we called C3. In our experiments, we treated HEK293T cells with Aβ 1–42 peptides for 12 h and then added the lysis buffer (DiscoveRx), which includes the enzyme acceptor fragment of β-galactosidase. When the two fragments of the β-galactosidase combine in solution, the enzyme becomes active and hydrolyzes a substrate that produces a chemiluminescent signal. The strength of this signal is proportional to the protein being produced (in this case, BACE1). In a separate experiment, we transfected the stable cell line C3 overexpressing BACE1 with siRNA or shRNA against BACE1, BACE1-AS or control siRNA and measured protein expression 72 h later with this methodology. We plotted data as a percentage of control siRNA.
Human samples
The first set of human brain samples was prepared at the USC Alzheimer’s Disease Research Center. The USC Alzheimer’s Disease Research Center obtained informed consent from all subjects and the USC Institutional Review Board approved the use of the human tissue. RNA was extracted from parietal lobes and cerebellum of postmortem brains of five subjects with Alzheimer’s disease and five matched controls. The average age of subjects with Alzheimer’s disease was 85 years (range 75–92 years) and 91.8 years (90–95 years) for controls. The postmortem interval ranged from 3.75–10.1 h with a mean of 5.87 h. We treated RNA samples with DNase and purified them with RNeasy mini columns (QIAGEN). We prepared cDNA from 400 ng of RNA samples and used RT-PCR for relative quantification of different transcripts. The second set of human brain samples was prepared from rapid autopsy brain tissue that had been obtained from J. Rogers (Sun Health Research Institute); all enrolled subjects or legal representatives had signed a Sun Health Research Institutional Review Board–approved informed consent form allowing both clinical assessments during life and several options for brain and bodily organ donation after death. These cases included 35 autopsy-confirmed cases of Alzheimer’s disease with an average age of 81.8 years (range 64–92 years) and 35 controls with an average age of 72.3 years (range 53–91 years). The postmortem interval ranged from 1.25–5 h with a mean of 2.5 h. The average duration of disease in the subjects with Alzheimer’s disease was 9.2 years. Total RNA was isolated via CsCl purification from tissue dissected from specific regions of brain. Although not all regions were available from all cases, we examined a total of 128 RNA samples from superior frontal gyrus, entorhinal cortex, hippocampus and cerebellum for BACE1 and BACE1-AS expression by RT- PCR.
Mouse studies
We obtained approval for mouse studies from the Institutional Animal Care and Use Committee at The Scripps Research Institute.
We used 18 six-month-old male mice for in vivo siRNA infusion experiments. We prepared mice with chronic indwelling cannulae in the dorsal third ventricle implanted subcutaneously with osmotic minipumps that delivered continuous infusions (0.25 μl/h) of synthetic unmodified siRNA directed against Bace1, Bace1-AS or control siRNA (previously shown to have no effects on the expression of human and mouse genes) at a dose of 0.4 mg/d for 2 weeks. We connected tubing to the exit port of the osmotic minipump and tunneled it subcutaneously to the indwelling cannula, such that siRNAs were delivered directly into the brain.
Pump-mediated infusion of siRNA was previously shown to significantly and specifically knock down expression of targeted mRNAs in the brain, but with a limited tissue penetration27. Indeed, RNA knockdown upon ventricular infusion of siRNAs for 14 consecutive d was usually obtained in brain regions immediately adjacent to the ventricle, with diminishing effects of the siRNA as the distance from the ventricle increased.
We excised five tissues from each mouse for RNA quantitative measurement—the dorsal hippocampus, ventral hippocampus, cortex, dorsal striatum and cerebellum. RNA extraction is described in Supplementary Methods.
Tg19959 mice were produced by pronuclear microinjection of (FVB × 129S6F1) embryos with a cosmid insert containing human APP with two familial Alzheimer’s disease mutations (KM670/671NL and V717F) under the control of the hamster PrP promoter. We euthanized four Tg19959 mice and four control male littermates at 6 weeks old. We used brain tissues for RNA measurements and Aβ 1–42 detection by HTRF. In a separate experiment, we euthanized three wild-type male mice and excised their tissues for expression profiling of Bace1 and Bace1-AS by RT-PCR.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Rogers (Sun Health Research Institute) for autopsy brain tissue. We are grateful to M. Leissring (Mayo Clinic, Jacksonville) for helpful discussions and for kindly providing cell lines and APP-tg19959 mouse materials. We also thank D. Willoughby for his help in RNA purification from human Alzheimer’s disease samples. S. Brothers provided valuable help in manuscript preparation. M.A.Faghihi. is partly supported by a scholarship from the Ahwaz University of Medical Sciences, Ministry of Health I.R. Iran. This study has been supported in part by the US National Institutes of Health (AG 029290).
References
- 1.Goedert M, Spillantini MG. A century of Alzheimer’s disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 2.Faghihi MA, Mottagui-Tabar S, Wahlestedt C. Genetics of neurological disorders. Expert Rev. Mol. Diagn. 2004;4:317–332. doi: 10.1586/14737159.4.3.317. [DOI] [PubMed] [Google Scholar]
- 3.Monaco S, Zanusso G, Mazzucco S, Rizzuto N. Cerebral amyloidoses: molecular pathways and therapeutic challenges. Curr. Med. Chem. 2006;13:1903–1913. doi: 10.2174/092986706777585022. [DOI] [PubMed] [Google Scholar]
- 4.Zhu D, et al. Phospholipases A2 mediate amyloid-β peptide–induced mitochondrial dysfunction. J. Neurosci. 2006;26:11111–11119. doi: 10.1523/JNEUROSCI.3505-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esposito G, et al. CB1 receptor selective activation inhibits β-amyloid-induced iNOS protein expression in C6 cells and subsequently blunts tau protein hyperphosphorylation in co-cultured neurons. Neurosci. Lett. 2006;404:342–346. doi: 10.1016/j.neulet.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Snyder EM, et al. Regulation of NMDA receptor trafficking by amyloid-β. Nat. Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 7.Matsuyama S, Teraoka R, Mori H, Tomiyama T. Inverse correlation between amyloid precursor protein and synaptic plasticity in transgenic mice. Neuroreport. 2007;18:1083–1087. doi: 10.1097/WNR.0b013e3281e72b18. [DOI] [PubMed] [Google Scholar]
- 8.Abramov AY, Canevari L, Duchen MR. β-amyloid peptides induce mitochondrial dysfunction and oxidative stress in astrocytes and death of neurons through activation of NADPH oxidase. J. Neurosci. 2004;24:565–575. doi: 10.1523/JNEUROSCI.4042-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohyagi Y, et al. Intracellular Aβ42 activates p53 promoter: a pathway to neurodegeneration in Alzheimer’s disease. FASEB J. 2005;19:255–257. doi: 10.1096/fj.04-2637fje. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, et al. β-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer’s disease pathogenesis. J. Neurosci. 2007;27:3639–3649. doi: 10.1523/JNEUROSCI.4396-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun X, et al. Hypoxia facilitates Alzheimer’s disease pathogenesis by up-regulating BACE1 gene expression. Proc. Natl. Acad. Sci. USA. 2006;103:18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong Y, et al. Oxidative stress potentiates BACE1 gene expression and Aβ generation. J. Neural Transm. 2005;112:455–469. doi: 10.1007/s00702-004-0255-3. [DOI] [PubMed] [Google Scholar]
- 13.Li R, et al. Amyloid β peptide load is correlated with increased β-secretase activity in sporadic Alzheimer’s disease patients. Proc. Natl. Acad. Sci. USA. 2004;101:3632–3637. doi: 10.1073/pnas.0205689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holsinger RM, McLean CA, Collins SJ, Masters CL, Evin G. Increased β-secretase activity in cerebrospinal fluid of Alzheimer’s disease subjects. Ann. Neurol. 2004;55:898–899. doi: 10.1002/ana.20144. [DOI] [PubMed] [Google Scholar]
- 15.Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. β-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch. Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- 16.Johnston JA, et al. Expression and activity of β-site amyloid precursor protein cleaving enzyme in Alzheimer’s disease. Biochem. Soc. Trans. 2005;33:1096–1100. doi: 10.1042/BST20051096. [DOI] [PubMed] [Google Scholar]
- 17.Ohno M, et al. BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer’s disease. Neuron. 2004;41:27–33. doi: 10.1016/s0896-6273(03)00810-9. [DOI] [PubMed] [Google Scholar]
- 18.Tesco G, et al. Depletion of GGA3 stabilizes BACE and enhances β-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma H, et al. Involvement of β-site APP cleaving enzyme 1 (BACE1) in amyloid precursor protein–mediated enhancement of memory and activity-dependent synaptic plasticity. Proc. Natl. Acad. Sci. USA. 2007;104:8167–8172. doi: 10.1073/pnas.0609521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laird FM, et al. BACE1, a major determinant of selective vulnerability of the brain to amyloid-β amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J. Neurosci. 2005;25:11693–11709. doi: 10.1523/JNEUROSCI.2766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, et al. Bace1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- 22.Willem M, et al. Control of peripheral nerve myelination by the β-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- 23.Engstrom PG, et al. Complex loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 25.Wahlestedt C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov. Today. 2006;11:503–508. doi: 10.1016/j.drudis.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 26.Su Y, Ryder J, Ni B. Inhibition of Aβ production and APP maturation by a specific PKA inhibitor. FEBS Lett. 2003;546:407–410. doi: 10.1016/s0014-5793(03)00645-8. [DOI] [PubMed] [Google Scholar]
- 27.Thakker DR, Hoyer D, Cryan JF. Interfering with the brain: use of RNA interference for understanding the pathophysiology of psychiatric and neurological disorders. Pharmacol. Ther. 2006;109:413–438. doi: 10.1016/j.pharmthera.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Borghi R, et al. The increased activity of BACE1 correlates with oxidative stress in Alzheimer’s disease. Neurobiol. Aging. 2006;28:1009–1014. doi: 10.1016/j.neurobiolaging.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Tamagno E, Bardini P, Guglielmotto M, Danni O, Tabaton M. The various aggregation states of β-amyloid 1–42 mediate different effects on oxidative stress, neurodegeneration, and BACE-1 expression. Free Radic. Biol. Med. 2006;41:202–212. doi: 10.1016/j.freeradbiomed.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Harkany T, et al. Mechanisms of β-amyloid neurotoxicity: perspectives of pharmacotherapy. Rev. Neurosci. 2000;11:329–382. doi: 10.1515/revneuro.2000.11.4.329. [DOI] [PubMed] [Google Scholar]
- 31.Yatin SM, et al. Temporal relations among amyloid β-peptide–induced free-radical oxidative stress, neuronal toxicity, and neuronal defensive responses. J. Mol. Neurosci. 1998;11:183–197. doi: 10.1385/JMN:11:3:183. [DOI] [PubMed] [Google Scholar]
- 32.Meyer-Luehmann M, et al. Rapid appearance and local toxicity of amyloid-β plaques in a mouse model of Alzheimer’s disease. Nature. 2008;451:720–724. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh DM, et al. The role of cell-derived oligomers of Aβ in Alzheimer’s disease and avenues for therapeutic intervention. Biochem. Soc. Trans. 2005;33:1087–1090. doi: 10.1042/BST20051087. [DOI] [PubMed] [Google Scholar]
- 34.Link CD, et al. Gene expression analysis in a transgenic Caenorhabditis elegans Alzheimer’s disease model. Neurobiol. Aging. 2003;24:397–413. doi: 10.1016/s0197-4580(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 35.Ray S, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat. Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 36.Chishti MA, et al. Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J. Biol. Chem. 2001;276:21562–21570. doi: 10.1074/jbc.M100710200. [DOI] [PubMed] [Google Scholar]
- 37.Li F, et al. Increased plaque burden in brains of APP mutant MnSOD heterozygous knockout mice. J. Neurochem. 2004;89:1308–1312. doi: 10.1111/j.1471-4159.2004.02455.x. [DOI] [PubMed] [Google Scholar]
- 38.McConlogue L, et al. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP transgenic mice. J. Biol. Chem. 2007;282:26326–26334. doi: 10.1074/jbc.M611687200. [DOI] [PubMed] [Google Scholar]
- 39.Zhong Z, et al. Levels of β-secretase (BACE1) in cerebrospinal fluid as a predictor of risk in mild cognitive impairment. Arch. Gen. Psychiatry. 2007;64:718–726. doi: 10.1001/archpsyc.64.6.718. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Zhou W, Tong Y, He G, Song W. Control of APP processing and Aβ generation level by BACE1 enzymatic activity and transcription. FASEB J. 2006;20:285–292. doi: 10.1096/fj.05-4986com. [DOI] [PubMed] [Google Scholar]
- 41.Cirrito JR, et al. Synaptic activity regulates interstitial fluid amyloid-β levels in vivo. Neuron. 2005;48:913–922. doi: 10.1016/j.neuron.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 42.Emilsson L, Saetre P, Jazin E. Alzheimer’s disease: mRNA expression profiles of multiple patients show alterations of genes involved with calcium signaling. Neurobiol. Dis. 2006;21:618–625. doi: 10.1016/j.nbd.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Brooks WM, et al. Gene expression profiles of metabolic enzyme transcripts in Alzheimer’s disease. Brain Res. 2007;1127:127–135. doi: 10.1016/j.brainres.2006.09.106. [DOI] [PubMed] [Google Scholar]
- 44.Rossner S, Sastre M, Bourne K, Lichtenthaler SF. Transcriptional and translational regulation of BACE1 expression–implications for Alzheimer’s disease. Prog. Neurobiol. 2006;79:95–111. doi: 10.1016/j.pneurobio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor β-secretase in Alzheimer’s disease. Ann. Neurol. 2002;51:783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh H, et al. AMPAR removal underlies Aβ-induced synaptic depression and dendritic spine loss. Neuron. 2006;52:831–843. doi: 10.1016/j.neuron.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St Laurent G, III, Wahlestedt C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Vassar R. The β-secretase, BACE: a prime drug target for Alzheimer’s disease. J. Mol. Neurosci. 2001;17:157–170. doi: 10.1385/JMN:17:2:157. [DOI] [PubMed] [Google Scholar]
- 49.Kumar P, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.