Abstract
Apomyoglobin folds by a sequential mechanism in which the A, G, and H helix regions undergo rapid collapse to form a compact intermediate onto which the central portion of the B helix subsequently docks. To investigate the factors that frustrate folding, we have made mutations in the N-terminus of the B helix to stabilize helical structure (in the mutant G23A/G25A) and to promote native-like hydrophobic packing interactions with helix G (in the mutant H24L/H119F). The kinetic and equilibrium intermediates of G23A/G25A and H24L/H119F were studied by hydrogen exchange pulse labeling and interrupted hydrogen/deuterium exchange combined with NMR. For both mutants, stabilization of helical structure in the N-terminal region of the B-helix is confirmed by increased exchange protection in the equilibrium molten globule states near pH 4. Increased protection is also observed in the G-H turn region in the G23A/G25A mutant, suggesting that stabilization of the B-helix facilitates native-like interactions with the C-terminal region of helix G. These interactions are further enhanced in H24L/H119F. The kinetic burst phase intermediates of both mutants show increased protection, relative to wild type protein, of amides in the N-terminus of the B helix and in part of the E helix. Stabilization of the E helix in the intermediate is attributed to direct interactions between E helix residues and the newly stabilized N-terminus of helix B. Stabilization of native packing between the B and G helices in H24L/H119F also favors formation of native-like interactions in the GH turn and between the G and H helices in the ensemble of burst phase intermediates. We conclude that instability at the N-terminus of the B helix of apomyoglobin contributes to the energetic frustration of folding by preventing docking and stabilization of the E helix.
Keywords: NMR, quench flow pulse labeling, H/D exchange, kinetic and equilibrium
INTRODUCTION
Apomyoglobin (apoMb) is uniquely suited for elucidation of the detailed molecular events that occur during protein folding. The protein exhibits straightforward folding kinetics with well-defined folding intermediates and also forms an equilibrium molten globule intermediate at mildly acidic pH.1 Kinetics and hydrogen exchange pulse labeling experiments performed more than a decade ago showed that apoMb folds via an on-pathway burst phase intermediate with helical structure in the A, G, and H helix regions and in part of the B helix.2–4 Amide protons in the remainder of the B helix, and in the C helix, CD loop, and E helix become fully exchange-protected only during the slower stages of folding.2,5,6 The burst phase intermediate is heterogeneous in structure and constitutes a kinetic trap in which progression of folding is impeded by non-native packing between the G and H helices caused by translocation of helix H.7 Subsequent kinetic experiments with faster time resolution revealed that apoMb folds by a sequential pathway with two compact helical intermediates, U ⇆ Ia ⇆ Ib ⇆ N.4,8 Ia is fully formed in less than 300μs at 26°C and neutral pH,8 converts to Ib at a rate of 200s−1 which then progresses to the native state N at a rate of 20s−1.4,8 Recent ultra-fast hydrogen exchange pulse labeling experiments show that Ia contains folded helical structure in the A, G, and H helix regions, and that subsequent folding events involve docking of parts of the B helix followed by the C and E helices onto the AGH core to form the Ib intermediate.9 Folding of the B helix appears to play a key role in formation of Ib and progression to the native folded state.
We have previously addressed the role of the B helix in apoMb folding using a series of single-site mutants, where long hydrophobic side chains at positions 28, 29, 30, and 32 were changed to alanine.10 Hydrogen exchange pulse labeling studies revealed specific long-range contacts between the B/C and B/G helices in the burst phase kinetic intermediates formed after 6ms refolding time. In particular, substitution of I28 by alanine disrupted key hydrophobic contacts with the E and G helices and substantially decreased the folding rate.
An important stabilizing interaction between the B and G helices involves formation of a hydrogen bond between the side chains of H24 and H119; disruption of this hydrogen bond by protonation of H24 has been proposed as the trigger for acid-induced unfolding of apoMb to form the pH 4 molten globule.11 Substitution of H24 and H119 by valine and phenylalanine, respectively, stabilizes apoMb against acid unfolding.12 These observations suggest that interactions between the B and G helices are key to formation of the equilibrium molten globule. H24 remains protonated in the transition state for apoMb folding;13 thus the B/G helix contacts appear to be energetically frustrated, with the native H24/H119 hydrogen bond forming only very late in the folding process.
To obtain more detailed insights into the role of the B helix in apoMb folding, we have mutated its N-terminal region to enhance its intrinsic helical stability and promote hydrophobic packing interactions with the G helix. Two mutant proteins were prepared. In the first, G23 and G25 were both mutated to alanine, to stabilize helical structure at the N-terminus of the B-helix. In the second, H24 was mutated to leucine and H119 substituted by phenylalanine both to stabilize the N-terminus of the B helix and to promote long range packing interactions between the B and G helices. The resulting G23A/G25A and H24L/H119F apoMb mutants were studied using hydrogen exchange pulse labeling and interrupted H/D exchange experiments, incorporating the use of DMSO as an aprotic solvent for the detection of partially-exchanged species,6 to determine the effects of B helix stabilization and packing on the folding kinetics and on the structure of the kinetic and equilibrium molten globule intermediates.
RESULTS
Acid Unfolding of the Mutant Proteins
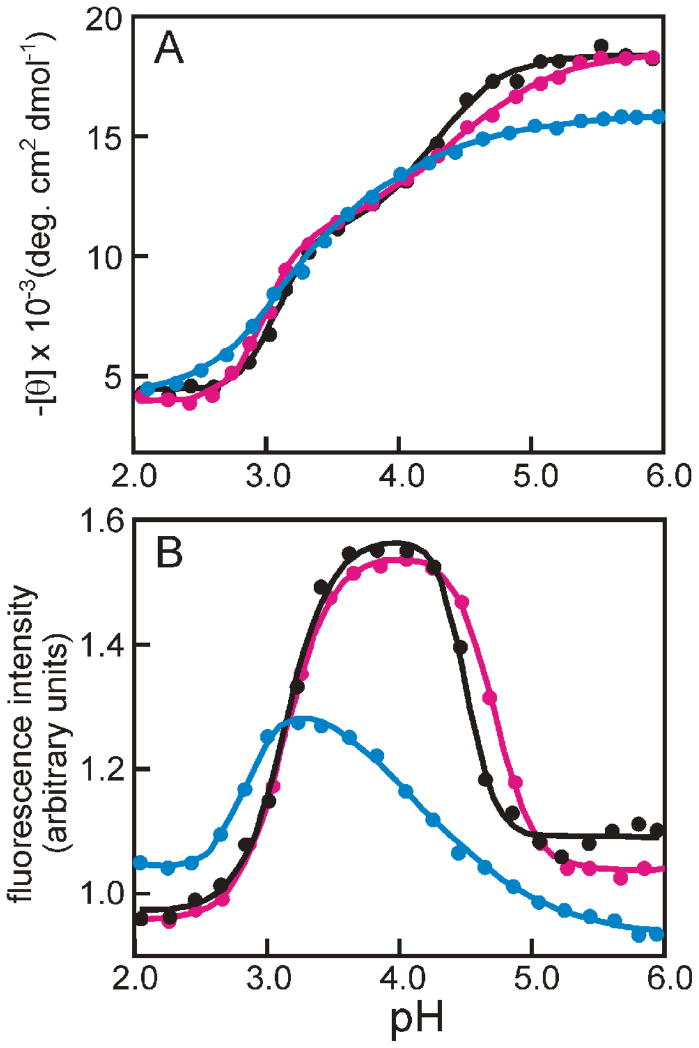
The conditions under which the population of the equilibrium intermediate is maximal were determined by measuring the pH dependence of tryptophan fluorescence and circular dichroism spectra (Fig. 1). For the G23A/G25A mutant, the CD and fluorescence spectra appear very similar to those of the wild type protein. The spectra of H24L/H119F are similar to those of H24V/H119F.11,12 The maximum ellipticity at pH 6 is significantly lower for both H24L/H119F (Fig. 1A) and H24V/H119F11 than for wild-type apoMb, and the maximum in the fluorescence intensity is observed at a pH of 3.2 for both mutants (Fig. 1B),11 compared to pH 3.8 for wild-type and G23A/G25A. The peak in fluorescence intensity corresponds to the maximum population of the equilibrium intermediate, because the tryptophan fluorescence is quenched both at lower pH where the protein unfolds and at higher pH where quenching occurs due to the close proximity of Lys79 and Met131 in the native apoMb structure. The decreased sensitivity of the H24L/H119F mutant towards acid denaturation probably results from the absence of the H24/H119 “pH-unfolding trigger” as well as additional stabilizing hydrophobic interactions between the L24 and F119 side chains.
Figure 1.
pH-dependent unfolding of apomyoglobin and mutants. A. helical content monitored at 222 nm in the CD spectrum. B. fluorescence emission intensity recorded with the excitation wavelength at 288 nm for wild-type (black), G23A/G25A (red), and H24L/H119F (blue). Solid curves were fitted to the data by using linear least-squares algorithm.
Urea-induced Unfolding of the Native and Intermediate States
The stability of the wild-type and mutant proteins towards urea denaturation was determined for both the equilibrium intermediate (pH 3–4) and native (pH 6) states by CD titrations (Table 1). For the native state at pH 6, the thermodynamic parameters are similar for wild-type apoMb and the G23A/G25A mutant, but both ΔG and Cm are much higher for H24L/H119F. For the molten globule intermediate, ΔG for the G23A/G25A and H24L/H119F mutants is significantly larger than for wild-type apoMb, and the Cm value is also higher for the H24L/H119F mutant. These results suggest a change in the structure of the equilibrium intermediate, particularly for H24L/H119F and to a lesser extent for G23A/G25A.
Table 1.
Kinetic and Thermodynamic Data for the G23A/G25A and H24L/H119F Mutants
| (a) Urea Titration | ||||
|---|---|---|---|---|
| Sample | pH | ΔG(H2O) (kcal/mol) | m (kcal/mol/M) | Cm (M) |
| wild type | 6.0 | −4.8 ± 0.2 | 1.50 ± 0.05 | 3.17 |
| G23A/G25A | 6.0 | −4.6 ± 0.1 | 1.36 ± 0.02 | 3.38 |
| H24L/H119F | 6.0 | −6.3 ± 0.2 | 1.51 ± 0.05 | 4.17 |
| wild type | 4.0 | −1.46 ± 0.03 | 1.02 ± 0.02 | 1.43 |
| G23A/G25A | 4.0 | −1.78 ± 0.04 | 1.15 ± 0.02 | 1.55 |
| H24L/H119F | 3.4 | −1.67 ± 0.02 | 0.91 ± 0.01 | 1.84 |
| (b) Stopped Flow CD (pH jump) | ||
|---|---|---|
| Sample | burst phase amplitude (%) | Rate constant observable phase (s −1) |
| wild type | 69 | 3.5 ± 0.1 |
| G23A/G25A | 76 | 4.6 ± 0.1 |
| H24L/H119F | 72 | 4.3 ± 0.1 |
Protection Factors of the Equilibrium Intermediates
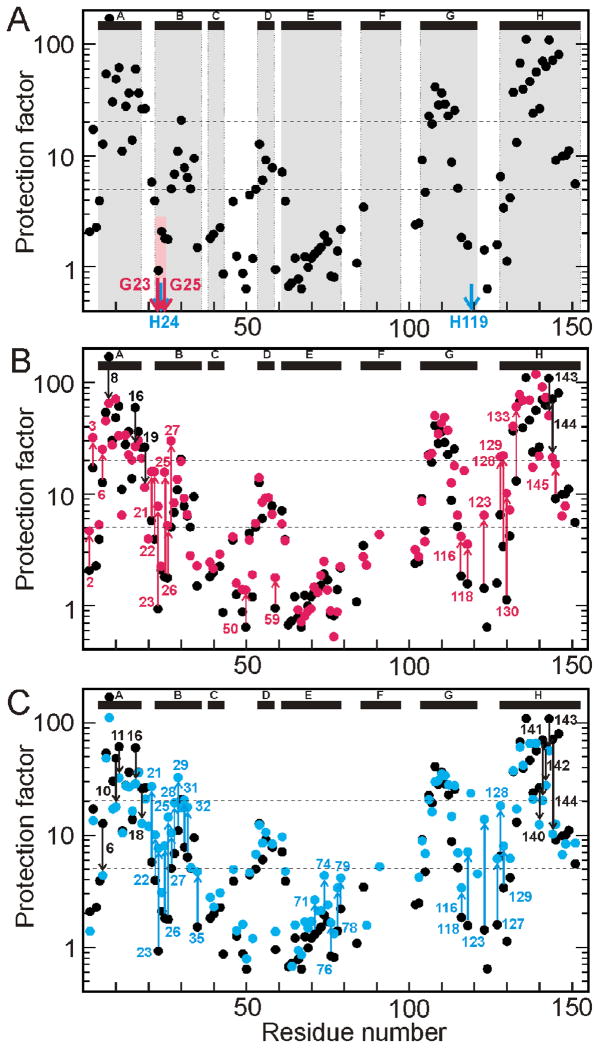
The amide protection factors for the equilibrium intermediate that forms under weakly acidic conditions were determined from interrupted H/D exchange measurements.6,14 As noted previously,6 the protection factors for residues in the central parts of the A, G, and H helices of wild-type apoMb are significantly higher (20 –100) than those near the termini of each helix (Fig. 2A). The presence of the two glycine residues at positions 23 and 25 in the B helix appears to destabilize the local helical structure in the equilibrium intermediate, since the protection factors for these residues (Fig. 2A) are lower than for residues closer to the C-terminus of the B-helix. Stabilization of the B helix by substitution of these glycine residues by alanine (G23A/G25A) or stabilization of packing between helices B and G (H24L/H119F) results in a significant increase in protection factor in the N-terminal region of the B helix (Figs. 2B, 2C). Increased protection is also observed at long range for both mutants. The GH turn region (residues 116–133, comprising the C-terminal end of the G-helix, the GH turn, and the N-terminal region of the H-helix) shows greater protection in G23A/G25A than wild type, suggesting the presence of contacts between the B, G and H helices in the equilibrium molten globule intermediate of the mutant protein. More extensive long-range effects are observed in the equilibrium intermediate of H24L/H119F. In addition to stabilization of amides in the GH turn region, including protection of residues 119 and 121 that are exchanged out in the wild type protein and G23A/G25A, increased exchange protection is observed for residues I28 - S35 in the C-terminal part of helix B and residues A71, A74, L76, K78, and K79 in the C-terminal half of the E-helix. It is of interest that several residues in the C-terminal part of the H helix display diminished protection (Fig. 2C).
Figure 2.
Protection factor as a function of residue number for the equilibrium (pH ~4) intermediates. A. wild-type, pH 4 (black), showing the mutation sites for the G23A/G25A mutant protein (red arrows) and the H24L/H119F mutant protein (blue arrows). The location of the helices in the wild-type protein in the fully-folded state are shown at the top of the panel, and their locations in relation to the protection factor data are shown as gray boxes. The position of the G23 and G25 within the B helix and their apparent effect on the protection factor of the wild-type protein at pH 4 is emphasized with a pink box. B. Superposition of the wild-type data from part A (black) with that for the G23A/G25A mutant protein, pH 4 (red). Residues showing a significant change in the mutant are labeled and the extent of the change indicated by arrows, red for increased protection in the mutant and black for decreased protection. C. Superposition of the wild-type data from part A (black) with that for the H24L/H119F mutant protein, pH 3.4 (blue). Residues showing a significant change in the mutant are labeled and the extent of the change indicated by arrows, blue for increased protection in the mutant and black for decreased protection. A significant change is defined as value of > 2 or < 0.5 in the ratio of the protection factors of mutant and wild type at that residue.
Time-course of Refolding from the Intermediate to the Native State
Refolding of apoMb from the pH ~2 unfolded state following rapid pH jump to pH 6 was monitored by stopped-flow CD. The resulting kinetic parameters are reported in Table 1. The burst phase amplitude and folding rate constants were increased somewhat for the mutant proteins.
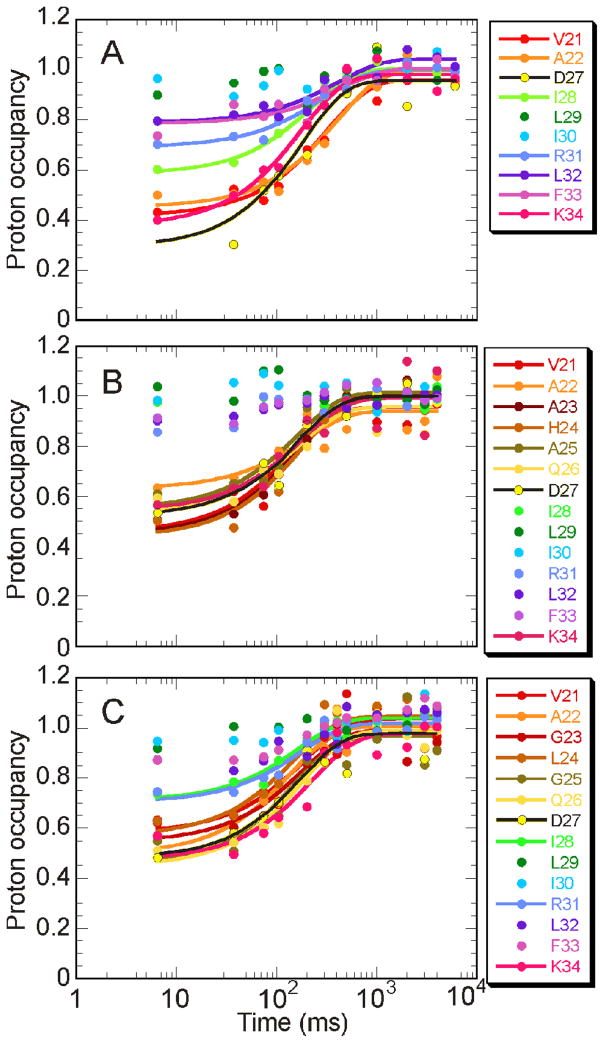
The time-course for exchange protection of amides by formation of stable helical structure was monitored by pH-pulse labeling (Fig. 3). More probes are available for the B-helix residues in the mutant proteins (Fig. 3B, C) compared to wild type (Fig. 3A), because the mutations stabilize the amides of residues G23–Q26 against exchange. Proton occupancies in the kinetic (burst phase) intermediate were determined from the HSQC cross peak intensity at the shortest folding time (6 ms). Consistent with the increase in the burst phase amplitude observed in the stopped-flow refolding experiments, the proton occupancy of residues in the B-helix region was increased in the burst phase intermediates of both mutant proteins.
Figure 3.
Time course of amide protection for the B-helix in A. wild type, B. G23A/G25A, and C. H24L/H119F. A transition curve of the proton occupancy has been fitted by using linear least-squares algorithm for residues whose proton occupancy at 6.4 ms is below 0.8.
The overall rate for the observable folding process, which corresponds to the transition from the kinetic intermediate to the native state, is similar for the wild type and mutant proteins. The measured rate constants (wild-type, 4.7 s−1; G23A/G25A, 5.3 s−1; H24L/H119F, 4.9 s−1) are consistent with the rates determined by stopped-flow CD (Table 1).
Proton occupancy in the burst phase intermediates
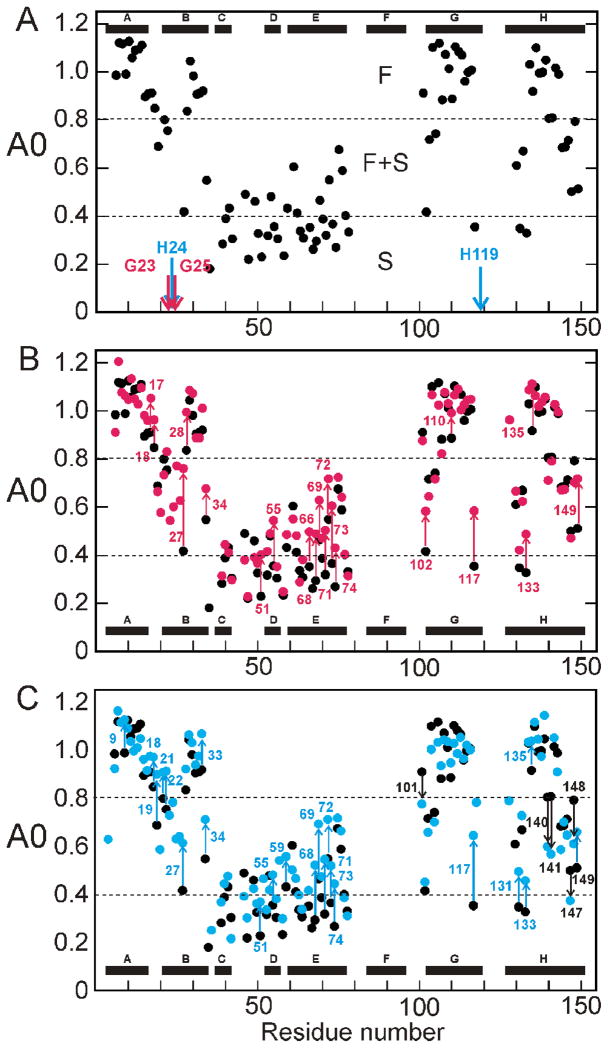
The protection of amides from exchange in the burst phase intermediate was analyzed in more detail using A0 values, calculated by plotting proton occupancy as a function of pH-pulse duration and extrapolating to zero pulse duration to compensate for the effects of exchange-out during the high-pH pulse.5 The results are shown in Fig. 4. For wild-type apoMb, the overall pattern of protection is consistent with that observed in the equilibrium intermediate except in the B-helix region, where the kinetic intermediate shows greater exchange protection.6 An A0 value close to 1 indicates that an amide is already fully protected in the burst phase intermediate; such amides are designated “fast folding” (F), and are observed in the central regions of the A, B, G, and H helices. Residues in the N-terminal regions of the B and G helices, the C-terminal region of the E helix, and both ends of the H helix display biphasic kinetics. These amides are partially protected in the burst phase (A0 values are between 0.4 and 0.8) and only become fully protected during the slow phase of refolding; such amides are designated “fast plus slow” (F+S).5 Residues with fractional values of A0 are located in regions that fluctuate between protected and unprotected states in the conformational ensemble of the burst phase intermediate.9 The remaining amides of the wild-type protein, including those belonging to residues in the C and D-helices, the CD-loop and the N-terminal part of the E-helix are not significantly protected in the burst phase, but are protected as the intermediate folds to the native state in the second, slower phase; such amides are designated “slow folding” (S).
Figure 4.
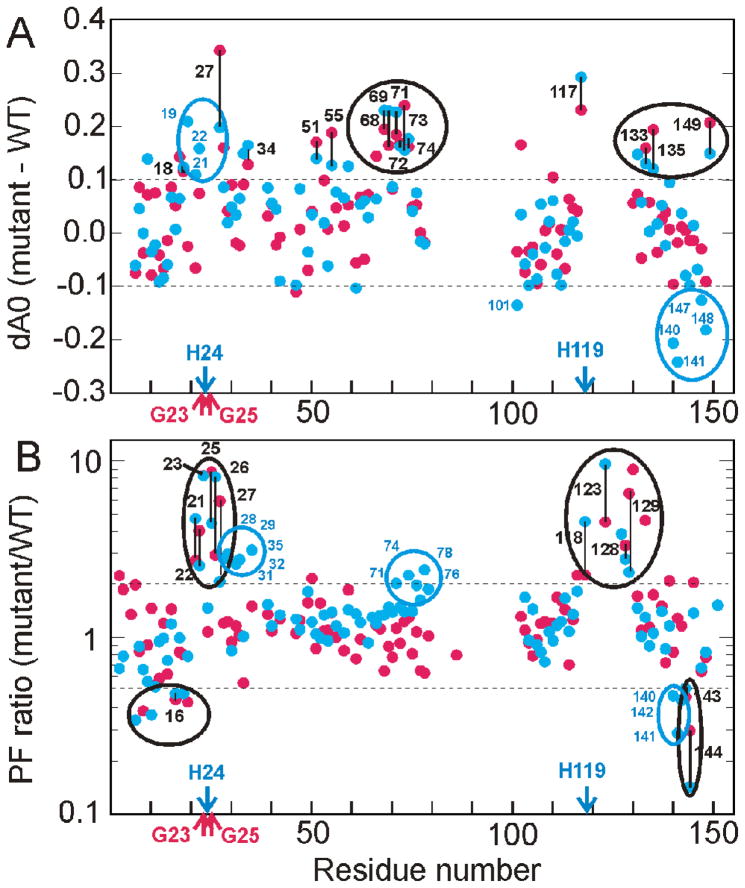
Proton occupancy (A0) for the burst phase intermediate as a function of residue number. A. wild type (black) showing the mutation sites for the G23A/G25A mutant protein (red arrows) and the H24L/H119F mutant protein (blue arrows). B. Superposition of the wild-type data from part A (black) with that for the G23A/G25A mutant protein. Residues showing a significant change in the mutant are labeled and the extent of the change indicated by arrows, red for increased protection in the mutant and black for decreased protection. C. Superposition of the wild-type data from part A (black) with that for the H24L/H119F mutant protein. Residues showing a significant change in the mutant are labeled and the extent of the change indicated by arrows, blue for increased protection in the mutant and black for decreased protection. A significant change is defined as dA0 (A0mutant – A0wild type) greater than 0.1 or less than −0.1.
Effect of mutations on the kinetic burst phase intermediates
The A0 values of many amides in the F+S and S groups are increased in the G23A/G25A and H24L/H119F mutant proteins relative to wild type, indicating stabilization of partly folded states in which the amides are protected from exchange (Fig. 4B,C). Not surprisingly, the N-terminal region of the B-helix, near the site of the mutations, is also stabilized in the kinetic intermediate. Many of the E-helix residues show larger A0 values in both mutants and some residues at the C-terminal end of the A-helix are also more protected compared to the wild type protein. It is also of note that residues 118 and 128, near the C-terminus of helix G and in the N-terminal region of helix H, respectively, are strongly protected in the burst phase for H24L/H119F but not for the other mutant or wild type protein.
The differences in A0 values between the wild type and the two mutant proteins are shown in Fig. 5A. It is interesting that amides of residues in the middle of the E-helix (V68 – A74) are stabilized significantly (A0 increased by 0.15–0.25) for both mutants, suggesting contacts between helix E and the N-terminus of helix B in the burst phase kinetic intermediate (Fig. 6A,B). Increased A0 values are also observed for several B-helix residues, two D-helix residues and two residues in the H-helix that are close to the contact site for the E helix in the fully folded protein (Fig. 6A,B). All other sites showing increased occupancy occur as single points. The proton occupancies of I101, at the N-terminus of helix G, and residues 140, 141, 147, and 148 in the C-terminal region of helix H are greatly decreased by the H24L/H119F mutation (Fig. 5A, 6B), while the A0 values for residues 128, 131, and 133 in the N-terminal region of the H helix increase substantially (Figs. 4C, 5A).
Figure 5.
Differences in the folding behavior of the G23A/G25A and H24L/H119F mutants. A. difference in values of the proton occupancy A0 between wild type and mutants (G23A/G25A, red, H24L/H119F blue). Values greater than 0.1 or less than −0.1 (dotted horizontal lines) for both mutants are circled in black (with vertical black lines connecting the corresponding values for the two mutants) or for the H24L/H119F mutant alone in blue. B. ratio of the protection factor between wild type and mutant (G23A/G25A, red H24L/H119F blue). Values greater than 2 or less than 0.5 (dotted lines) for both mutants are circled in black (with vertical black lines connecting the corresponding values for the two mutants) or for the H24L/H119F mutant alone in blue.
Figure 6.
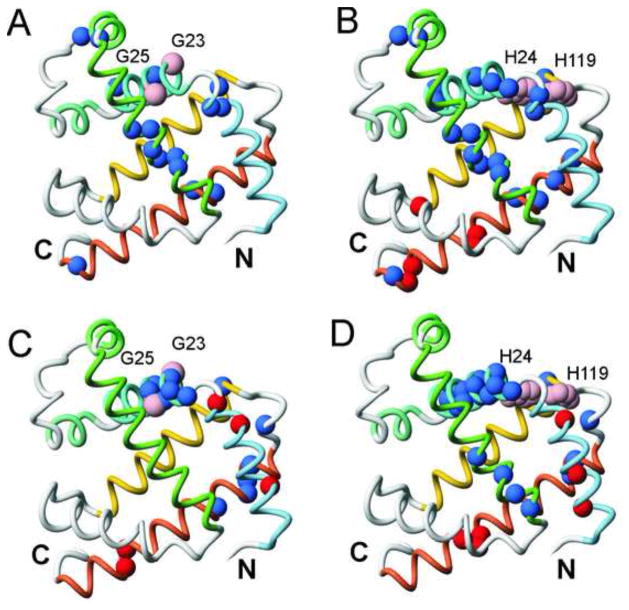
Mapping of the locations of significantly stabilized (blue) or destabilized (red) amides in the kinetic and equilibrium intermediates of the G23A/G25A and H24L/H119F mutants onto the backbone of the X-ray crystal structure of MbCO.30 Helices A-H are denoted by colored backbone: blue (A), turquoise (B), aquamarine (C), light green (D), green (E), gray (F), yellow (G) and coral (H). A. kinetic intermediate of G23A/G25A. The mutation sites are indicated with pink spheres at the CA positions of G23 and G25. B. kinetic intermediate of H24L/H119F. The locations of the two histidine side chains are indicated as pink spheres. C. equilibrium intermediate of G23A/G25A. D. equilibrium intermediate of H24L/H119F. Mutation sites indicated as in parts A and B. These figures were generated in MolMol.31
Effect of mutations on the equilibrium intermediates
The changes in protection factors in the equilibrium intermediates of the mutant proteins, relative to wild type, are plotted in Fig. 5B. Although the pattern is similar to that of the changes in A0 values (Fig. 5A), the effects appear more pronounced for the equilibrium intermediate, particularly in the B helix region. Both mutations greatly increase the protection factors of the residues near the N-termini of the B and H helices (Fig. 5B, 6C, D). For the H24L/H119F mutant protein, but not for the G23A/G25A mutant, small but significant increases in amide proton protection are also observed in the C-terminal halves of the B and E helices. Both equilibrium intermediates show decreased protection factors in the A helix as well as in the C-terminal region of the H helix, as observed also in the A0 values for the H24L/H119F kinetic intermediate.
DISCUSSION
The present experiments provide important new insights into the origins of energetic frustration in apomyoglobin folding. A major bottleneck arises from non-native packing of the G and H helices in the AGH burst phase intermediate; the H helix is translocated by approximately one helical turn towards its N-terminus to maximize burial of hydrophobic residues.7 The N-terminal region of the B helix appears to be an additional site of energetic frustration. Even though the central region of helix B forms native-like contacts with the G helix in the kinetic burst phase intermediate,10 residues 21–26 near the N-terminus of B are largely unstructured in the pH4 equilibrium molten globule15 and amides in this region are not exchange-protected in the burst phase.5,6 The G23A/G25A mutation was designed to determine explicitly the effect of stabilizing helical structure in the N-terminal region of the B helix, without affecting the H24-H119 hydrogen bonding interaction between the B and G helices. The H24L/H119F mutation was designed to assess the role of H24-H119 hydrogen bond formation in the folding process. H24 is protonated in the pH 4 molten globule intermediate and is the trigger for pH-induced unfolding of wild type apoMb.11 H24 is also protonated in the folding transition state; i.e., the H24-H119 hydrogen bond is formed only after passage through the transition state.13 In H24L/H119F, the hydrogen bond is replaced by a pH-independent stabilizing hydrophobic interaction. In this work, we chose to mutate H24 to leucine, instead of the valine substitution performed in previous work,12 to avoid potential destabilization of the B helix by incorporation of an unfavorable β-branched side chain.
The G23A/G25A mutation has little effect on the stability or helicity of the native apo protein, but significantly stabilizes the equilibrium molten globule against urea unfolding (Table 1), in accord with a previous report.16 The native state of the H24L/H119F apoMb is greatly stabilized against urea denaturation (ΔG −6.3 kcal.mol−1 versus −4.8 for wild type). However, the total helical content is significantly lower, probably due to the steric clashes in the native structure induced by the mutations.
Both the G23A/G25A and H24L/H119F mutants fold somewhat faster than wild type apoMb (Table 1). For H24L/H119F the increased folding rate appears to be due to an enhanced interaction between the B and G helices, resulting in a more efficient conformational search in this mutant. For G23A/G25A the increase appears to be due to the stabilization of helix in the N-terminal part of the B-helix.
Effect of B helix stabilization on the structure of the equilibrium intermediate
Our results show that, as expected, both the G23A/G25A and H24L/H119F mutations lead to stabilization of structure and increased H/D exchange protection factors in the N-terminal region of the B helix in the equilibrium molten globule intermediate (Fig. 2). Interestingly, long-range effects of the increased helical structure in G23A/G25A are observed: several residues near the C-terminus of the G helix, in the GH turn, and near the N-terminus of helix H show increased protection in the mutant, suggesting the presence of native-like contacts between the B and G helices that lead to stabilization of the GH region. More extensive long-range effects on the equilibrium intermediate structure are observed for H24L/H119F, which induces stabilization in the C-terminal regions of the E and B helices as well as in the GH region (Fig. 6D). Thus, it appears that stabilization of helical structure in the N-terminal part of the B helix (in both the G23A/G25A and H24L/H119F mutants) allows the B helix to make more stable contacts with the AGH core, while the additional B-G interaction provided by the hydrophobic contacts between L24 and F119 further enhances B helix docking in the equilibrium molten globule formed by the H24L/H119F mutant and promotes docking of part of the E-helix to the ABGH core. In contrast to the stabilizing effects described above, amide protons in parts of the A helix and the H helix show decreased protection factors in the equilibrium molten globule intermediates of H24L/H119F and, to a lesser extent, G23A/G25A.
Effect of B helix stabilization on the structure of the kinetic intermediate
For both mutant proteins, increased exchange protection is observed in the burst phase kinetic intermediates in much of the B helix, in the AB turn, the D helix, the C-terminal region of the E-helix, the C-terminal end of the G helix (especially in H24L/H119F), and in the N-terminal region of the H helix (Figs. 4, 5A). These results strongly suggest that stabilization of helix B and subsequent packing interactions with helix G promote packing and folding of regions of the D helix and E helix. Interestingly, residue 101 at the N-terminus of helix G and the amides of several residues in the C-terminal half of the H helix are less well protected in the kinetic intermediate of the H24L/H119F mutant than in the wild type protein; this effect is less pronounced for the G23A/G25A mutant. Previous studies have shown that the H helix in the burst phase intermediate of wild type apoMb is translocated by approximately one helical turn in the direction of its N-terminus to maximize burial of hydrophobic surface.7 The observed stabilization of amides at the N-terminus of the H helix and concomitant destabilization of the C-terminal region of H strongly suggests that there is an increased population of molecules in the ensemble in which the H helix is packed via more native-like contacts with helix G in the burst phase ensemble of H24L/H119F, and is no longer translocated as in the wild type intermediate. The pattern of protection and deprotection of amides observed in interrupted H/D exchange experiments (Figs.2 and 5B) suggests that the H helix also makes more native-like interactions with helix G in the ensemble of structures formed by the equilibrium molten globules of H24L/H119F and, to a lesser extent, G23A/G25A.
It should be noted that even though the B helix is less stably folded in the burst phase intermediate of wild type apoMb, and appears to be completely unstructured in its N-terminal region, the C-terminal region of helix B still makes stabilizing interactions with helix G. Evidence for a native-like B-G helix interaction in the wild-type kinetic intermediate comes from the observation that the mutation L32A in the B-helix significantly lowers proton occupancies for several residues in the G helix that are in close proximity to L32 in the native myoglobin structure.10
Stabilization of the E-helix in the mutant proteins
Compared to the wild type protein, greater exchange protection is observed in the E-helix region in the kinetic burst phase ensemble of both mutants (Fig. 4B,C) and in the equilibrium molten globule of H24L/H119F (Fig. 2C). The observed stabilization of the E helix region is of particular interest since correct packing and folding of the E helix is required before the transition state for folding can be reached.7
The substantial increase in burst phase proton occupancies for residues V68-A74 in both G23A/G25A and H24L/H119F indicates stabilization of contacts between this region of the E helix and the ABGH core. This could arise in several ways, but most likely derives directly from stabilization of helical structure between residues V21 and D27 in the N-terminal region of helix B in the mutant proteins. Several residues in this region of the B helix (V21, A22, G25, Q26, and L29) pack against the N-terminal half of helix E (L61, K62, G65, V66, and L69) in the native myoglobin structure. Thus, the observed stabilization of structure between residues 68–74 may originate from native-like interactions between the stabilized B helix and the N-terminal region of E, leading to initiation and propagation of helical structure towards the C-terminus of helix E that is further stabilized by hydrophobic packing against the AGH core. In particular, the side chain of L69 packs into a pocket formed by V17, V21, G25, and L29 in the native fold, so that stabilization of the N-terminus of the B helix and the neighboring AB region could promote native docking of L69. Alternatively, stabilization of the residue 68–74 region of helix E might result from transient non-native interactions directly with the N-terminal region of helix B. Indeed, for the wild type protein, previous mutagenesis studies have provided evidence of disorder in the packing of the E helix in the burst phase ensemble and have revealed the presence of non-native interactions in which L72 docks directly into the hydrophobic pocket at the interface of helices A, B, and G.7
It is also possible that stabilization of helix E arises not through direct contacts with the N-terminus of helix B, but rather as a consequence of changes in the A/B/G/H core caused by the mutations. We have noted above that there appears to be a population of molecules in the burst phase ensemble of the H24L/H119F mutant in which the GH turn is stabilized and in which the packing between the G and H helices is native-like. Contacts between the C-terminal region of helix E and the “native” G-H helical hairpin might then be responsible for stabilization of helix between V68 and A74. However, this explanation is considered unlikely. The H helix appears to be non-natively packed in the burst phase intermediate of G23A/G25A, similar to the wild type protein, yet the proton occupancies of the E helix residues are increased to the same extent as in the H24L/H119F mutant. This points towards a mechanism other than formation of native-like E/H contacts as the origin of stabilized helical structure in the E helix.
Finally, one might ask why, if correct folding and packing of the E helix is required prior to the folding transition state,7 stabilization of E helix structure in the G23A/G25A and H24L/H119F mutants led to only a modest (20–30%) increase in the folding rate? Clearly, other sites of energetic frustration remain that lead to a heterogeneous conformational ensemble in the burst phase intermediate and hinder proper docking of the E helix. A major source of frustration is undoubtedly associated with translocation of the H helix in the AGH core, which impedes native-like docking of helix E. Also, as noted above, it is possible that stabilization of E helix structure in the mutants occurs through direct but non-native interactions with the N-terminus of helix B; these would not be expected to promote formation of the transition state ensemble and would not accelerate folding. A further site of frustration of E helix folding is the distal histidine (H64), a residue that is of functional importance in holomyoglobin to enhance O2 binding and to discriminate against competing ligands such as CO. Substitution of H64 with a non-polar phenylalanine greatly stabilizes helical structure in the entire E helix region in both the burst phase conformational ensemble and in the equilibrium molten globule intermediate and results in a 2-fold increase in the apoMb folding rate.17,18 The frustration caused by the distal histidine has been attributed to the energetic cost of desolvation and deprotonation of the imidazole group required for packing in the hydrophobic core.18 It will be of interest in future work to determine whether the effects of B helix stabilization and mutation of H64 to Phe are additive or synergistic.
MATERIALS AND METHODS
Preparation of proteins
Recombinant sperm-whale apoMb was over-expressed in Escherichia coli BL21-DE3 cells with a pET17b vector containing the Mb gene.19 Site-directed mutagenesis was performed using the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene). The 15N-labeled wild-type and mutant proteins for both pH-pulse labeling and interrupted H/D exchange experiments and the 15N,13C-double-labeled wild-type and mutant proteins for resonance assignments were produced in M9 minimal medium containing 13C-glucose and 15N-ammonium sulfate. Proteins were purified by a previously described method.7 The same extinction coefficient (15,900 M−1cm−1 at 280 nm, pH6.1) was used for determination of the protein concentration for both wild-type and mutants proteins.20
CD and fluorescence studies
CD spectra were collected on an Aviv 62DS spectrometer at 25°C. The ellipticity of the proteins (10 μM) in 10 mM sodium acetate buffer was monitored at 222 nm as functions of the pH and concentration of urea.
Emission spectra derived from the fluorescence of the two tryptophan residues at positions 7 and 14 were recorded on a Fluorolog-3 fluorescence spectrometer (Jobin Yvon-Horiba) at 25°C. The spectra of proteins (2 μM) in 10 mM sodium acetate buffer were collected by scanning from 300 nm to 400 nm. The excitation wavelength was set at 288 nm with the bandwidth of both excitation and emission at 2 nm. The maximum intensities of the Trp fluorescence were plotted as a function of pH. Individual samples were prepared for each data point in both the CD and fluorescence titrations.
Stopped-flow CD
The change in ellipticity at 222 nm following initiation of folding was monitored on a DX-17MV Applied Photophysics stopped flow CD instrument at 8°C. The CD signal was collected at 225 nm after pH-jump from pH 2.2 (wild-type and G23A/G25A) or 2.0 (H24L/H119F) to pH 6.0 with a slit width of 2 nm. The final concentration of protein was adjusted to 10 μM after 1 to 5 dilution with 30 mM sodium acetate buffer. The dead time of the instrument was about 20 ms.
Pulse-labeling H/D exchange
Hydrogen exchange pulse labeling was used to identify transient protection of amide protons in helical structure formed during refolding.6 Pulse labeling experiments were performed at 8°C with a Biologic QFM5 Quench Flow instrument (Grenoble, France). Folding of apoMb (2.0 mg/ml at pH 2.2 for wild-type and G23A/G25A, and at pH 2.0 for H24L/H119F) in H2O was initiated by pH jump to pH* 6.0 using a 7.5-fold volume of 20 mM sodium acetate buffer in D2O. (pH* refers to the pH measured in a D2O solution, uncorrected for the deuterium isotope effect). The pulse labeling was performed using 100 mM 3-[cyclohexylamino]-1-propanesulfonic acid buffer in D2O, pH* 10.3. Experiments at the shortest refolding time (6.4 ms) were repeated for a series of pulse durations (12, 20, 35, 65 and 95 ms) to determine the proton occupancy of the burst phase intermediate (A0) by extrapolation to a pulse duration of zero.5 After the high-pH pulse, the pH was immediately dropped to pH* 6.0 with 300 mM 3-(N-morpholino) propane-sulfonic acid buffer in D2O. To determine the time course of folding, refolding times were varied between 6.4 ms – 8 s with a pulse duration at pH* 10.3 of 35 ms. The solution containing the pH-pulse labeled protein was flushed into a tube containing 0.1 % (v/v) trifluoroacetic acid solution in D2O. This solution (final pH* 2.5) was immediately frozen in liquid N2 followed by lyophilization. The lyophilized protein was kept in the −80 C freezer until NMR data collection.
Interrupted H/D exchange
For studies of amide proton exchange from the equilibrium intermediate, a solution of protein (1.5 mg/ml) in 2 mM sodium acetate buffer in H2O at pH 4.0 (wild-type and G23A/G25A) or pH 3.4 (H24L/H119F) was mixed with a 10-fold volume of the same buffer in D2O at 4°C. H/D exchange was allowed to proceed for times that varied between 40 s and 24 h at 4°C, after which the pH* was decreased to 2.5 using 0.1 % (v/v) trifluoroacetic acid in D2O. The solution was immediately frozen in liquid N2 and lyophilized and the lyophilized protein stored at −80°C before NMR data collection.
NMR spectroscopy
15N-1H heteronuclear single quantum correlation (HSQC) spectra were recorded at 30°C on a Bruker DRX600 spectrometer equipped with a cryoprobe. Lyophilized protein samples from the H/D exchange experiments were dissolved in a mixture of 99.6 % (v/v) d6-DMSO (99.96 % deuterated, Cambridge Isotope Laboratories, Andover, MA) and 0.4 % (v/v) D2O (99.8 % deuterated, CIL) to give a final concentration of about 0.2 mM. NMR data acquisition was started 8 min after the addition of the solvent, with spectral widths of 9,615 Hz and 2,048 points in the 1H dimension and 1,538 Hz and 256 points in the 15N dimension. The small amount of D2O was added to the DMSO solvent in order to correct for exchange during the NMR measurements, caused by variable amounts of residual water in the DMSO solvent.6 For each sample, HSQC spectra were acquired every 20 min and the changes in intensity of each cross peak, due to exchange with the added D2O, were fitted to an exponential decay curve. The proton occupancy and protection factor for each amide was calculated by extrapolation of the exponential curve to zero time, as described in detail elsewhere.6 The intensities in each HSQC spectrum were normalized relative to the peak volume of the methyl proton signals observed in 1D proton NMR spectra acquired at the same time as the HSQC measurements. Protection factors were calculated using published values for the intrinsic H/D exchange rate.21
Backbone resonance assignments in the DMSO solutions were made using 3D HNCO,22 HNCACO,23 HNCA,22 HNCOCA,22 HNCACB,24,25 and CBCA(CO)NH26 spectra recorded at 30°C on a Bruker DRX800 spectrometer as described elsewhere.6 In order to minimize changes in the position of HSQC cross peaks due to differences in buffer components and salt concentration, the 15N,13C-double-labeled protein samples for assignments were prepared in the same way as the 15N-labeled samples that were used for the pH-pulse labeling and interrupted H/D exchange experiments. The NMR data were collected in the indirect dimension with the TPPI-States method,27 processed using NMRPipe,28 and analyzed by NMRView.29
Acknowledgments
We thank Linda Tennant for excellent technical assistance, and John Chung for valuable help with NMR spectroscopy.
This work was supported by grant DK34909 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Wright PE, Baldwin RL. The folding process of apomyoglobin. In: Pain RH, editor. Mechanisms of Protein Folding. Oxford University Press; Oxford: 2000. pp. 309–329. [Google Scholar]
- 2.Jennings PA, Wright PE. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 3.Tsui V, Garcia C, Cavagnero S, Siuzdak G, Dyson HJ, Wright PE. Quench-flow experiments combined with mass spectrometry show apomyoglobin folds through an obligatory intermediate. Protein Sci. 1999;8:45–49. doi: 10.1110/ps.8.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamin M, Baldwin RL. Two forms of the pH 4 folding intermediate of apomyoglobin. J Mol Biol. 1998;276:491–504. doi: 10.1006/jmbi.1997.1543. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura C, Dyson HJ, Wright PE. The apomyoglobin folding pathway revisited: structural heterogeneity in the kinetic burst phase intermediate. J Mol Biol. 2002;322:483–489. doi: 10.1016/s0022-2836(02)00810-0. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura C, Dyson HJ, Wright PE. Enhanced picture of protein-folding intermediates using organic solvents in H/D exchange and quench-flow experiments. Proc Natl Acad Sci USA. 2005;102:4765–4770. doi: 10.1073/pnas.0409538102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimura C, Dyson HJ, Wright PE. Identification of native and non-native structure in kinetic folding intermediates of apomyoglobin. J Mol Biol. 2006;355:139–156. doi: 10.1016/j.jmb.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 8.Uzawa T, Akiyama S, Kimura T, Takahashi S, Ishimori K, Morishima I, Fujisawa T. Collapse and search dynamics of apomyoglobin folding revealed by submillisecond observations of α-helical content and compactness. Proc Natl Acad Sci USA. 2004;101:1171–1176. doi: 10.1073/pnas.0305376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzawa T, Nishimura C, Akiyama S, Ishimori K, Takahashi S, Dyson HJ, Wright PE. Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR. Proc Natl Acad Sci USA. 2008;105:13859–13864. doi: 10.1073/pnas.0804033105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishimura C, Wright PE, Dyson HJ. Role of the B helix in early folding events in apomyoglobin: evidence from site-directed mutagenesis for native-like long range interactions. J Mol Biol. 2003;334:293–307. doi: 10.1016/j.jmb.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 11.Geierstanger B, Jamin M, Volkman BF, Baldwin RL. Protonation behavior of histidine 24 and histidine 119 in forming the pH 4 folding intermediate of apomyoglobin. Biochemistry. 1998;37:4254–4265. doi: 10.1021/bi972516+. [DOI] [PubMed] [Google Scholar]
- 12.Barrick D, Hughson FM, Baldwin RL. Molecular mechanisms of acid denaturation. The role of histidine residues in the partial unfolding of apomyoglobin. J Mol Biol. 1994;237:588–601. doi: 10.1006/jmbi.1994.1257. [DOI] [PubMed] [Google Scholar]
- 13.Jamin M, Geierstanger B, Baldwin RL. The pKa of His-24 in the folding transition state of apomyoglobin. Proc Natl Acad Sci USA. 2001;98:6127–6131. doi: 10.1073/pnas.111157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughson FM, Wright PE, Baldwin RL. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990;249:1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- 15.Eliezer D, Chung J, Dyson HJ, Wright PE. Native and non-native structure and dynamics in the pH 4 intermediate of apomyoglobin. Biochemistry. 2000;39:2894–2901. doi: 10.1021/bi992545f. [DOI] [PubMed] [Google Scholar]
- 16.Kiefhaber T, Baldwin RL. Intrinsic stability of individual α helices modulates structure and stability of the apomyoglobin molten globule form. J Mol Biol. 1995;252:122–132. doi: 10.1006/jmbi.1995.0479. [DOI] [PubMed] [Google Scholar]
- 17.Garcia C, Nishimura C, Cavagnero S, Dyson HJ, Wright PE. Changes in the apomyoglobin folding pathway caused by mutation of the distal histidine residue. Biochemistry. 2000;39:11227–11237. doi: 10.1021/bi0010266. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzinger S, Mohana-Borges R, Kroon GJA, Dyson HJ, Wright PE. Structural characterization of partially folded intermediates of apomyoglobin H64F. Protein Sci. 2008;17:313–321. doi: 10.1110/ps.073187208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings PA, Stone MJ, Wright PE. Overexpression of myoglobin and assignment of the amide, Cα and Cβ resonances. J Biomol NMR. 1995;6:271–276. doi: 10.1007/BF00197808. [DOI] [PubMed] [Google Scholar]
- 20.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J Magn Reson. 1992;96:432–440. [Google Scholar]
- 23.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J Magn Reson. 1992;99:201–207. [Google Scholar]
- 25.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J Magn Reson. 1993;101:201–205. [Google Scholar]
- 26.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 27.Marion D, Ikura M, Tschudin R, Bax A. Rapid recording of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. J Magn Reson. 1989;85:393–399. [Google Scholar]
- 28.Delaglio F, Grzesiek S, Vuister GW, Guang Z, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:604–613. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 30.Kuriyan J, Wilz S, Karplus M, Petsko GA. X-ray structure and refinement of carbon-monoxy (Fe II)-myoglobin at 1.5 Å resolution. J Mol Biol. 1986;192:133–154. doi: 10.1016/0022-2836(86)90470-5. [DOI] [PubMed] [Google Scholar]
- 31.Koradi R, Billeter M, Wüthrich K. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]