Abstract
We have previously shown that ovarian tumors express prostate-derived Ets transcription factor (PDEF). However, the precise role of PDEF in the prognosis of ovarian cancer is unknown. In our study, we report for the first time that expression of PDEF in tumor lesions of patients with ovarian cancer is associated with favorable prognosis. Evaluation of samples from 40 patients with ovarian cancer showed that early stage (IA) and borderline (IIB, III) ovarian tumors expressed higher levels of PDEF mRNA and protein and lower levels of survivin compared to late stage ovarian tumors (IIIC and IV, p < 0.05). Normal ovarian tissues expressed the highest levels of PDEF mRNA and protein when compared to tumor tissues (p < 0.05). A Log-Rank test showed that overall survival of patients with PDEF-positive and survivin-negative ovarian tumors was significantly longer than those with PDEF-negative and survivin-positive tumors (p < 0.01). Forced expression of PDEF in PDEF-negative ovarian tumor cells inhibited tumor cell growth, induced apoptosis, downregulated survivin expression and its promoter activity. Furthermore, treatment of ovarian cancer cells with vitamin D or a selenium compound resulted in reexpression of PDEF, downregulation of survivin, induction of apoptosis and inhibition of tumor cell growth when compared to untreated controls (p < 0.05). Together, these observations showed an inverse correlation between PDEF and survivin expression and suggested that increased PDEF expression along with reduced survivin was associated with prolonged survival of patients with ovarian cancer.
Keywords: PDEF, survivin, ovarian cancer, vitamin D3, selenium
Ovarian cancer is the second most commonly diagnosed gynecologic malignances in women. In the United States alone, ~23,000 new cases of ovarian cancers are diagnosed every year and among them around 16,000 deaths occur.1 The high mortality rate of ovarian cancer is due to difficulties with early detection. Most women (75%) continue to be diagnosed with advanced stages (IIIC–IV) of the disease. Current available therapies for ovarian cancer fail to control tumor progression in most patients. Despite significant advances in surgery and cytotoxic chemotherapy over the last two decades, the overall 5-year survival rate for these patients is only 30%.2 Thus, identification of new molecular markers/targets for ovarian cancer is important in order to improve early detection of the disease and develop new therapeutic regimens.3,4
Prostate derived Ets transcription factor (PDEF) is a member of Ets transcription factor family. This family controls multiple biological functions including cell proliferation, differentiation, apoptosis, angiogenesis, transformation and invasion.5,6 Whether PDEF is a marker of good or poor prognosis is a matter of controversy. While some reports showed expression of PDEF in some normal epithelial cells and its upregulation in malignant cells,3,7,8 other reports showed PDEF loss during tumorigenesis.5–12 Our previous findings in breast cancer10 and present report in ovarian cancer are consistent with other reports showing PDEF loss during tumorigenesis.5–6,11,12 We and others reported PDEF expression in normal breast tissues as we all in normal prostate, ovary, colon and salivary gland tissues but PDEF loss in invasive prostate and breast tumors.5–10 Recently, we demonstrated PDEF-mediated downregulation of survivin expression and inhibition of breast cancer cell growth in vitro and in vivo.10 Our pilot study demonstrated the role of PDEF protein as a breast tumor suppressor gene in vivo. PDEF protein was also shown to act as tumor suppressor in vitro that inhibits migration and invasion in breast cancer cell lines.6,11 Reexpression of PDEF in invasive breast cancer cell lines inhibited cell growth, migration, and invasion.6,11 Phenotypic changes in breast tumors, induced by PDEF, were also shown to be associated with downregulation of the antiapoptotic protein survivin and metastasis activator urokinase-type plasminogen activator (uPA).10,11 More recently, it was shown that downregulation of PDEF expression increased expression of mesenchymal genes such as vimentin and N-cadherin and enhanced invasiveness of prostate cancer cells.12 On the other hand, one report showed the lack of expression of PDEF in normal ovarian tissues and its expression in 27–35% of ovarian cancers using immunohistochemistry (IHC).13 We performed Western blot and real-time RT-PCR analyses in addition to IHC using a highly specific PDEF antibody10 and obtained consistent results by all 3 methods. Forced PDEF expression studies were also performed to further evaluate the role of PDEF in tumorigenicity. We also evaluated PDEF and survivin expression after treatment of ovarian cancer cell lines with vitamin D3 (VD3) or a selenium compound, methylseleninic acid (MSA), as anticancer drugs.
Material and methods
Cell lines, tissue procurement and reagents
The human normal ovarian cell line (Hose) and ovarian tumor lines (Skov3, Skov6, Ov432, P11 (2008) and A2780) were grown in medium supplemented with 10% heat inactivated FCS, 100 U/ml of penicillin and 0.1 µg/ml of streptomycin in a 5% CO2 incubator. The Skov3, Skov6 and Ov432 cells were grown in DMEM, whereas Hose, P11 (2008) and A2730 were grown in RPMI 1640. Ten normal human ovarian tissues and 40 epithelial ovarian tumor samples from patients ranged from 41 to 84 (median 60) years old were obtained from the tissue procurement facility at Roswell Park Cancer Institute (RPCI) under IRB approved protocols. Tumor samples were staged on the basis of histology and were classified according to International Federation of Gynecology and Obstetrics (Stage IA, n = 1; Stage IIB, n = 5; Stage III, n = 4; Stage IIIC, n = 26; Stage IV, n = 4). These samples were used in IHC and Western blot. These tumor samples consisted of papillary serous adenocarcinoma 70% (28 out of 40), clear cell 5% (2 out of 40), endometrioid 7.5% (3 out of 40), mucinous 2.5% (1 out of 40) and undifferentiated 15% (6 out of 40). Although tumor contents of tumor tissues were not determined in these samples, we also performed IHC using tissue micro array (TMA) containing four nonneoplastic single spots of four patients and 50 duplicated spots from 25 ovarian tumors (1.0 mm in diameter) (Stage IA, n = 12; Stage IIA, n = 2; Stage IIIA, n = 2; Stage IIB, n = 2; Stage IIIB, n = 2; Stage IIIC, n = 20; Stage IV, n = 10) obtained from AccuMax Array Company (ISU ABXIS). These TMA tumours consisted of serous adenocarcinoma 20 % (10 out of 50), mucinous adenocarcinoma 20% (10 out of 50), clear cell carcinoma 20% (10 out of 50), transitional cell carcinoma 20% (10 out of 50) and endometrioid 20 % (10 out of 50). We obtained consistent results using tumor tissues or TMA. PDEF antibody was prepared in our laboratory10 and used at a 1:500 concentration. Survivin antibody (FL-142) was purchased from Santa Cruz. Actin antibody and HRPO-conjugated goat anti rabbit antibodies were purchased from Sigma (St. Louis, MO). Lipofectamine™ 2000 reagents were purchased from Invitrogen (Grand Island, NY). VD3 compounds analog EB1089 was provided from Leo Pharmaceutical Products (Ballerup, Denmark). MSA was purchased from Wako Chemical (Richmond, VA).
Western blot analysis
Ovarian cancer cells were washed with PBS and lysed using lysis buffer [50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 2mM EDTA, 0.1% SDS, 1% NP40] containing 10 µg/ml phenylmethyl sulfonyl fluoride, 1% of protease and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO) at 4°C for 30 min. Ovarian tissues (10 normal and 40 tumor samples) were homogenized in lysis buffer and then kept at 4°C for 30 min. Cell extracts were cleared by centrifugation at 12,000g for 30 min at 4°C and protein concentration was determined using a BCA kit (Pierce, Rockford, IL). Samples were separated on 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to Immobilon-P membranes (Millipore, Bedford, MA). The membrane was blocked in TBS-T buffer [20 mM Tris/HCl (pH 7.5), 0.137 M NaCl, and 0.05% Tween-20] containing 5% skim milk at room temperature for 2–3 hr. The membranes were incubated with PDEF or survivin antibodies diluted (1:500) in TBS-T overnight at 4°C. After washing with TBS-T, the membrane was incubated in 5% skim milk in TBS-T buffer containing a secondary antibody (1:5000), for 45–60 min at room temperature with shaking. Proteins of interest were detected using a HRPL kit (National Diagnostics/LPS, Rochester, NY) or a Chemo-luminescent Reagent Plus kit (Perkin Elmer, Waltham, MA) and visualized by autoradiography after various exposure times (usually 20–120 sec). For normalization of protein loading, the same membranes were stripped with stripping buffer (100 mM 2-mercaptoethanol, 2% sodium dodecyl sulphate, 62.5 mM Tris-HCl, pH 6.7) and used for Western blot with a monoclonal antibody against actin at 1:1000 using the same procedure.
IHC analysis
Immunohistochemical studies were performed on 10 cases of normal ovarian tissues and 40 cases of ovarian tumors as well as four normal tissues and 50 duplicate spots from 25 tumors in a TMA. Tissues were probed with purified rabbit anti-human PDEF antibody. Immunoreactivity and specificity of this antibody has previously been determined in our laboratory.10 The paraffin-embedded sections of normal and ovarian tumor tissues were dewaxed, rehydrated, and endogenous peroxidase activity was blocked using 3% H2O2 for 10 min. Slides were microwaved in citrate buffer antigen retrieval solution (Vector Laboratories) and washed with PBS for 5 min. Slides were then blocked with 10% normal goat serum in PBS for 30 min and incubated for over night with PDEF antibody diluted in PBS (1:500). The slides were washed 3 times using PBS, incubated with biotinylated secondary antibody (goat anti-rabbit diluted in 1:100 in PBS contain 1.5% normal goat serum) for 30 min. Immunohistochemical staining was performed using avidin–biotin complex (ABC) method (ABC Kit, Vector Laboratories, Burlingame, CA). A positive reaction was detected using 3-diaminobezidine (Vector Laboratories). Slides were counterstained with light green and the cells with nuclear PDEF staining were considered as positive. The IHC slides were evaluated semi-quantitatively based on the intensity and percentage of staining. The staining intensity was classified as strong (3+), moderate (2+), weak (1+) or negative (−) and the percentage of staining was divided into 3 categories: >10% positive cells, <10% positive cells and 0% positive cells. Finally, the cases were considered negative if there was no staining or <10% staining. On the other hand, cases were considered positive when >10% tumor cells stained positive. No staining was observed using isotype control Ig.
RNA extraction and RT-PCR
The tissues characteristics used for RNA extraction are summarized in Table I. To preserve the quality of RNA, normal ovary and ovarian tumor tissue specimens obtained from surgery were immediately immersed in the “RNA later” solution (Ambion, Austin, TX). Total RNA was extracted from cells or tissues using Tri Reagent™ following the instruction of manufacturer (MRC, Cincinnati, OH). Concentration of RNA was quantified spectrophotometrically. To test the quality of RNA, 5 µg of isolated total RNA from each sample was separated on a 1% agarose gel containing 0.45 M formaldehyde. Intact bands for 18S and 28S rRNAs without degradation were used as a criterion for assessing the quality of isolated total RNAs from normal ovarian, ovarian cancer tissues and cell lines (data not shown). The mRNA expression for PDEF, survivin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was determined using a one-step RT-PCR kit (Qiagen, Valencia, CA) as previously described.7 The following primers used in RT-PCR reactions: 5′-ATGGGCAGCGCCAGCCCGGGTC-3′ (forward) and 5′-TCAGATGGGGTGCACGAACTGGT-3′ (reverse) for PDEFPCR products (1008 bp), 5′-GAGGCTGGC TTCATCCACTG-3′ (forward) and 5′- CAGCTGCTCGATGGC ACGGC-3′ (reverse) for survivin PCR products (299 bp), and 5′-GCTTCCCGTTCTCAGCCTTGAC-3′ (forward) and 5′-ATG GGAAGGTGAAGGTCGGAG-3′ (reverse) for GAPDH PCR products (195 bp, internal control). PCR products were separated on a 1.8% agarose gel containing 0.01% ethidium bromide.
TABLE I.
Patients’ tumor characteristics and survival by PDEF and survivin status
| Patients group Total = 40 | PDEF protein | Survivin protein | Relapse | Survival |
|---|---|---|---|---|
| Well differentiated (early stage), n = 13 | ++/+++ | −/+ | 8/13 (62%) | 8/13 (62%) |
| Moderately differentiated (borderline), n = 17 | + | + | 13/17 (76%) | 10/17 (59%) |
| Undifferentiated (late stage), n = 10 | − | ++/+++ | 8/10 (80%) | 2/10 (20%) |
Quantitative analysis of PDEF expression in normal ovarian tissues and tumor lesions by real-time PCR
Real-time PCR was performed to quantitate the level of expression of PDEF mRNA in ovarian normal and tumor tissues at different stages, as previously described.7 Briefly, 2 µg of total RNA from each tissue was mixed with 1 µl (100 pmol) of oligodT and incubated at 70°C for 10 min. The reaction mixture was chilled on ice and centrifuged, and then 200 µM each deoxynucleotide triphosphate, 2 U/µl RNase inhibitor, and 2 U/µl reverse transcriptase (Promega) in a total volume of 10 µl was added. Reverse transcription was performed at 42°C for 1 hr and enzyme inactivation at 95°C for 10 min. After the reverse transcription, the reaction mixture was 5-fold diluted with diethyl pyrocarbonate-treated water. Real-time PCR amplification was performed on an ABI Prism 7700 sequence detector system (PE Applied Biosystems) using the Taq MAN universal PCR mix following the instruction of manufacturer. The amplification was performed in Micro Amp Optical tubes (PE Bio System, Foster city, CA) using primers and probes designed for specific detection of PDEF. PDEF-specific sense primer 5′-GTGAGGAGAGCTGGACCGAC-3′ and antisense primers 5′-GGGCTGAGTCCTCAATTTTGAAG-3′ and a Taq Man-labeled probe 5′-CGAGGTGGACTCATCATGCTCCGG-3′ were used. A 167-bp PDEF-specific sequence was amplified by these primers that flanked the probe sequence. The reaction mixture contained cDNA synthesis from 500 ng of total RNA as template, 5 pmol each primers, 7.5 pmol Taqman fluorescent label probe and 1× Taqman universal PCR master mix (PE Applied Biosystems) in a total volume of 25 µl. Each tumor sample was tested in duplicate, with results indicating excellent reproducibility. To analysis real-time PCR data, PDEF-negative ovarian tumors along with blank (without template) which show threshold PCR cycle value of 26 or higher were considered as negative controls. In contrast normal ovarian and ovarian tumors at different stages with 25 or below a threshold PCR cycle value were considered PDEF positive. These criteria were used to classify the ovarian tumors into PDEF-positive or PDEF-negative categories. The fold level of PDEF expression was calculated using 2N formula. The N is the subtraction of the number of PCR cycle of PDEF positive tumors from the PCR cycle of negative control.
The effect of ectopic expression of PDEF on cell growth of ovarian cancer
To examine the PDEF-mediated growth suppression, the P11 (2008) ovarian cancer cell (3 × 105) were seeded in 6-well plates in the 2 ml of RPMI medium without antibiotic 1 day prior to transfection. The cells were transfected with pcDNA3.1 PDEF vector constructed previously10 or mock control pcDNA3.1 with different concentration (2 and 4 µg) using Lipofectamine 2000 following the instruction of manufacturer (Invitrogen). Thirty-six hours after transfection, cell viability was determined by trypan blue exclusion staining. Cell count by using trypan blue exclusion showed that PDEF positive cells had a lower viability and a reduced total number of cells compared to PDEF negative cells (data not shown). The level of PDEF and survivin expression were evaluated using 1-step PCR and Western blot assay as described earlier.
MTT proliferation assay
Equal numbers of 104 P11 cells were seeded in 96-well plates and transfected with Lipofectamine 2000 as instructed by manufacturer (Invitrogen). MTT (3-[4,5-dimethylthiazol-2-yl]-2.5-diphenyltetrazolium bromide) was added to a final concentration 0.5 mg/ml 48 hr after transfection. The cells were incubated for 4 hrs and then lysed with a cell lysis buffer (20% SDS, 50% N,N-dimethylformamide, pH 4.7). Spectrophotometric absorbance from each sample was measured at 570 nm using an ultramicroplate reader.
Induction of apoptosis
A cell death detection ELISA assay (Roche, Indianapolis, IN) was performed for evaluation of apoptosis. Briefly, equal numbers of 3 × 104 P11 cells were seeded in a 48-well plate and transfected with Lipofectamine 2000 following the instruction of manufacturer (Invitrogen). Thirty-six hours after transfection, medium was removed and cells were washed with PBS. Cells were then lysed in 200 µl lysis buffer (supplied in the kit) for 30 min at room temperature, centrifuged at 200g and then 20 µl supernatants were dispensed into streptavidin-coated 96-well microtiter plates in duplicates. The DNA-histon complex (supplied in the kit) was used as positive control. The reaction was followed by adding 80 µl of immunoreagents. The immunoreagent consisted of a mixture of anti-histone biotin and anti-DNA-HRP directed against various histones (H1, H2A, H2B, H3 and H4) and antibodies to nucleosome single-stranded and double-stranded DNA. The plate was incubated at room temperature for 2 hr while shaking gently. The unbound components were removed by washing 3 times with 250 µl incubation buffer. One hundred microliters of HRP substrate [2, 2′-azino-di-(3-ethylbenzthiazoline sulfonate) diammonium salt, ABTS] was added to each well and the plate were placed on a shaker at 250 rpm for color development. Measurements were made at 405 nm against an ABTS solution as blank (reference wavelength 490 nm) using an ultramicroplate reader (Bio-Tek Instruments).
Cotransfection of survivin promoter and luciferase reporter assay constructs
To determine whether ectopic expression of PDEF downregulates survivin promoter activity, ovarian cancer cell line P11 (2008) were seeded in 24-well plates (5 × 104 cells per well) and grown to about 50–60% confluence. Cells were cotransfected with the survivin promoter luciferase constructs (pLuc-6270)14 and pRL-TK (TK promoter-Renilla luciferase construct as internal control) along with the PDEF expression vector (pcDNA3.1-PDEF) or control vector (pcDNA3.1) using Lipofectamine™ 2000 Plus according to the manufacturer’s recommendation (Invitrogen). Briefly, 450 ng of pLuc-survivin construct, 10 ng of pRL-TK and 50 ng of pcDNA3.1 PDEF or pcDNA3.1 were added in 50 µl serum-free medium (RPMI 1640) in a 1.5 ml tube for each well using 24-well plates. After incubation at room temperature for 30 min, the DNA-Lipofectamine mixture was added to each well containing 200 µl of RPMI 1640. The DNA-Lipofectamine complex was replaced by complete medium containing 10% fetal bovine serum after incubation for 2–3 hr. Cells were lysed 36–48 hr after transfection, and luciferase activity was assessed using a dual luciferase reporter assay system (Promega, Madison, WI). Briefly, cell lysates (20 µl/well) were used for measurement of luciferase activity in a luminometer by first mixing the cell lysates (20 µl) with 20 µl luciferase assay reagent for measuring firefly luciferase activity and subsequently adding 20 µl of Stop-Glo reagent for measuring Renilla luciferase activity. Data were normalized to Renilla luciferase activity (internal control) as arbitrary units.10,14
Treatment of ovarian cancer cell lines with VD3 or selenium compounds
Ovarian cancer cell lines P11 (2008) and A2780 were seeded at a density of 2 × 105 cells/60-mm dish. The cells were allowed to attach to the plate overnight and then treated with 100 nM VD3 concentration for 72 hr15 or with different concentrations of 4, 10, or 30 µM MSA as previously described.16 Cell growth/death was assessed using MTT assay and trypan blue exclusion.10,17 Cells were lysed and expression of PDEF and survivin were evaluated by Western blot.10 To evaluate the induction of apoptosis by VD3 and MSA, 3 × 104 ovarian cancer cells were seeded in a 48-well plate and treated with VD3 or MSA. Untreated cells were considered as negative controls. The treated and untreated cells were lysed with 200 µl of lysis buffers/well (supplied in the kit) for 30 min at room temperature, centrifuged and 20 µl of supernatants were dispensed into streptavidin-coated 96-well plates in duplicate wells. Induction of apoptosis was determined using cell death detection ELISA assay kit as described earlier.
Patient’s characteristics
The records of 40 patients with epithelial ovarian cancer who were surgically staged by gynecologic oncologist at RPCI between 1992 and 2000 were reviewed. The review included out-patient and in-patient treatments, including surgery and chemotherapy. The Institutional Review Boards approved the study design. All pathology specimens were reviewed at RPCI and tumors were classified according to WHO criteria.18 Study outcomes included overall survival and time to progression, each measured from the time of definitive surgery. Progression was defined as objective evidence of recurrence, because all therapy was given in the adjuvant setting. The expression levels of PDEF mRNA quantitated by real time PCR after normalization with GAPDH as an internal control. In addition, the level of PDEF and survivin protein expression determined by Western blot ImageQuant5.2 Software (Molecular Dynamics). Relative intensities of PDEF and survivin expressions, after normalization with actin as an internal control, were calculated and recorded as 100% at the highest levels of expression.
Statistical analysis
Results were analyzed using Student’s t test. A value of p < 0.05 was considered statistically significant. Survival probability was estimated by Kaplan–Meier method19,20 and Log-Rank test was used to estimate statistical significance of survivability of patients with PDEF-positive and survivin-negative tumors versus the PDEF-negative and survivin-positive tumors.
Results
Expression of PDEF in ovarian cancer is inversely correlated with the stage of the disease
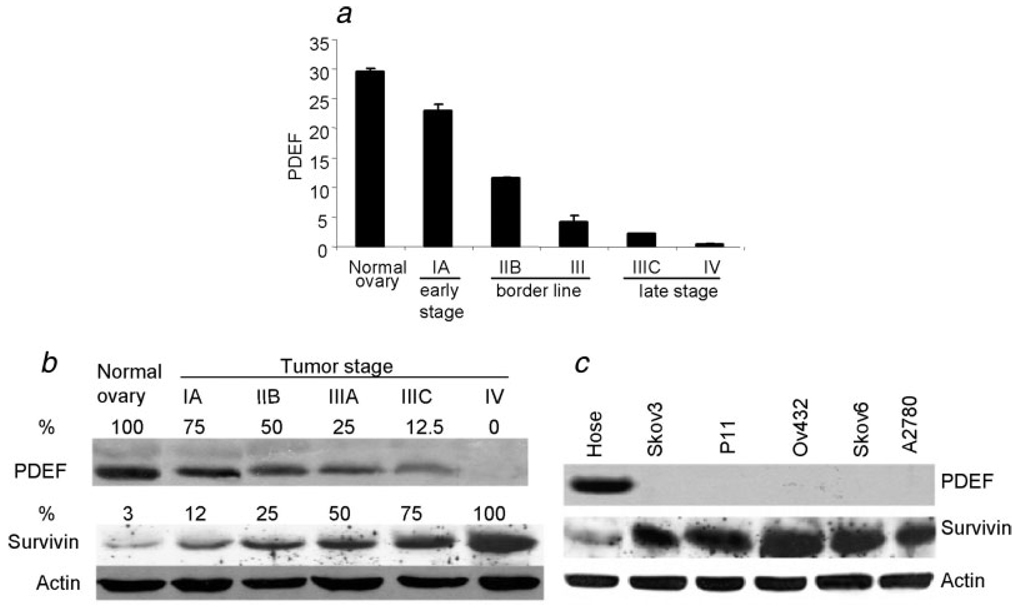
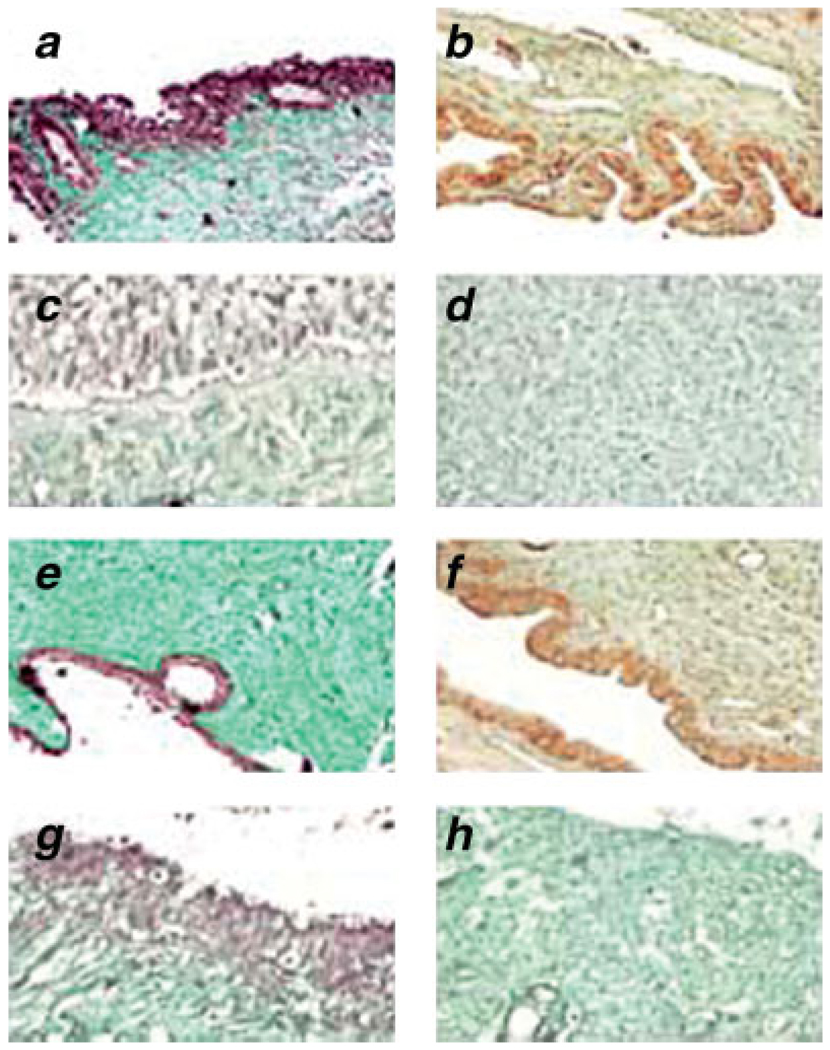
In order to perform quantitative analysis of PDEF expression in normal ovarian tissues and tumors at different stages, we performed real-time RT-PCR. Quality of total mRNA was verified in normal and tumor tissues. The highest level of PDEF mRNA was detected in normal tissues while it was downregulated and eventually lost as cancer progressed from early to late stages (p = 0.00003; Fig. 1a). Western blot analysis confirmed downregulation and eventual loss of PDEF protein as disease progressed from early to late stage cancer (Fig. 1b). Importantly, downregulation/loss of PDEF protein was associated with upregulation of survivin through early to late stage tumors (Fig. 1b). Western blot analysis of a normal epithelial cell, Hose, and the ovarian tumor lines showed total loss of PDEF protein expression and its inverse correlation with the expression of survivin (Fig. 1c). IHC analysis of normal ovarian tissues or early and late stage ovarian tumors showed strong nuclear staining for PDEF in normal tissues (Fig. 2a), reduced expression of PDEF in early (Fig. 2b) or borderline (Fig. 2c) tumors, and complete loss of PDEF in late stage tumors (Fig. 2d). The IHC on TMA cores also produced similar results (Figs. 2e–2h). The distribution intensity of PDEF staining of nonneoplastic (benign) vs. ovarian tumor with different tumour-nodemetastasis (TNM) stage in ovarian tom our samples and TMA are shown in Table II and Table III, respectively.
FIGURE 1.
Reduced expression of PDEF during tumour progression is inversely correlated with the expression of survivin. Total RNA and whole cell protein were extracted from normal and cancerous ovarian tissues and analyzed by quantitative real-time RT-PCR and Western blot, respectively. (a) Quantitative real-time RT-PCR analysis of PDEF expression in normal and ovarian tumour lesions with different disease stages. Representative data are presented from triplicate experiments using 10 normal tissues and tumour samples (Stage IA, n = 1; Stage IIB, n = 5; Stage III, n = 4; Stage IIIC, n = 26; Stage IV, n = 4). Mean ± SD derived from 3 independent experiments. Sample size was indicated in “Material and methods” section. (b) Western blot analysis of lysates of normal ovary and tumour lesions for the expression of PDEF and survivin. Actin was used as internal control for normalization of the results to determine relative % expression of the proteins in each sample. The highest expression was set to 100%. Representative results are presented from 3 independent experiments. Samples size was indicated in the “Material and methods” section. (c) Western blot analysis of PDEF and survivin expression in normal ovarian (Hose) and cancerous cell lines. Actin was used as an internal control.
FIGURE 2.
Loss of PDEF protein expression in ovarian cancer lesions compared to normal ovarian tissues. PDEF expression was analyzes using IHC with specific anti PDEF antibody. Representative nuclear staining for PDEF expression from the total of 10 normal (a) and 40 malignant tumours (b–d) as well as tissue microarray data of 4 benign (e) and 50 tumour samples (f–h) are shown. All images are shown at ×200 of the original magnification.
TABLE II.
Distribution of intensity of PDEF staining in TMA
| PDEF score | Benign (%) n = 4 |
TNM stage (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| IA | IIA | IIIA | IIB | IIIB | IIIC | IV | ||
| n = 12 | n = 2 | n = 2 | n = 2 | n = 2 | n = 20 | n = 10 | ||
| Negative (−) | 0 | 0 | 0 | 0 | 0 | 0 | 80 | 100 |
| Weak (1+) | 0 | 35 | 0 | 50 | 50 | 50 | 10 | 0 |
| Moderate (2+) | 25 | 65 | 100 | 50 | 50 | 50 | 10 | 0 |
| Strong (3+) | 75 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
TABLE III.
Distribution of intensity of PDEF staining in 40 ovarian tumors using IHC
| PDEF score | Normal (%) n = 10 |
TNM stage (%) | ||||
|---|---|---|---|---|---|---|
| IA | IIB | III | IIIC | IV | ||
| n = 1 | n = 5 | n = 4 | n = 26 | n = 4 | ||
| Negative (−) | 0 | 0 | 0 | 0 | 23 | 100 |
| Weak (1+) | 0 | 0 | 40 | 50 | 58 | 0 |
| Moderate (2+) | 0 | 100 | 60 | 50 | 19 | 0 |
| Strong (3+) | 100 | 0 | 0 | 0 | 0 | 0 |
Expression of PDEF in ovarian tumors is associated with prolonged survival in patients with ovarian cancer
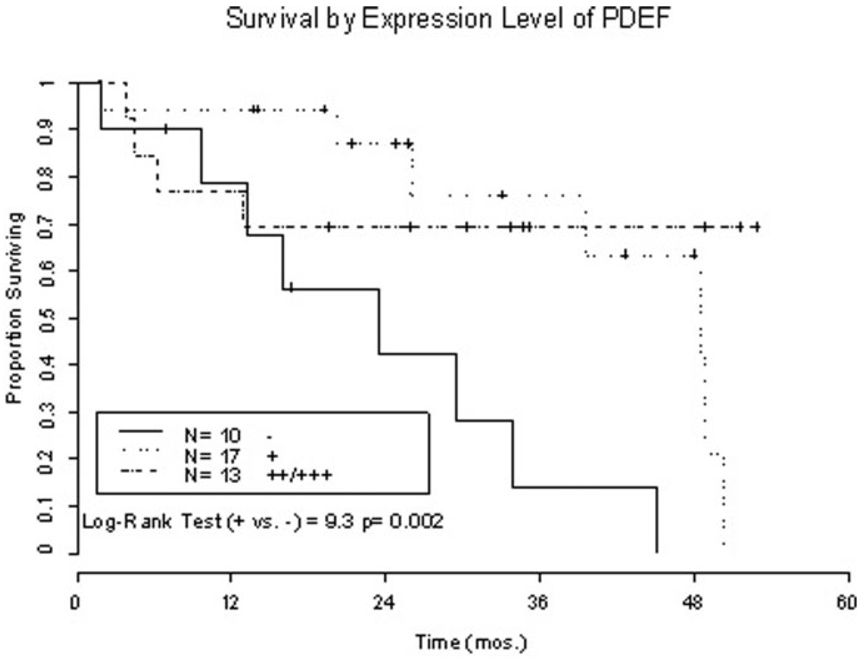
To evaluate the clinical relevance of PDEF expression in ovarian tumor lesions to the patient’s survival, two groups of ovarian cancer patients with or without expression of PDEF in their tumors were compared. Data from Kaplan–Meir analyses and two-tailed Log-Rank test indicated that survivability of patients with PDEF positive ovarian cancers was significantly longer than those without PDEF expression (p = 0.002, Fig. 3). When fitting the data to the Cox model, the relative risk estimate was 0.28 for the low PDEF group (+) and 0.19 for the high PDEF group (++/+++) in comparison with patients without PDEF expression in their tumor lesions (Wald Test [2] = 9.6, p = 0.008). Adjusting for disease stages, the relative risk estimates were unchanged from the crude estimates and remained statistically significant (adjusted relative risk of high and low PDEF expression vs. PDEF negativity is 0.19 and 0.28, respectively).
FIGURE 3.
PDEF expression is associated with longer survival of ovarian cancer patients. Kaplan–Meir analysis of survivability of ovarian cancer patients expressing PDEF in their tumour lesions (++: n = 7;+++: n = 6) vs. those without PDEF expression. Two-tailed Log-Rank test indicated that patient survivability is significantly different in comparison with PDEF negativity vs. PDEF positivity (p = 0.002). When fitting the data to the Cox model the relative risk estimate was 0.28 for the low PDEF groups (+) and 0.19 for the high PDEF groups (++/+++) in comparison with patients without PDEF expression in their tumour lesions (Wald Test [2] =9.6, p = 0.008).
The survival rate in patients with PDEF negative tumors was only 20% (2 out of 10) while patients with PDEF-positive tumors had a survival rate of 58–62% (Table I, p = 0.002). There is also a trend towards an inverse correlation between PDEF expression and disease progression/recurrence. Among patient who survived those with PDEF negative and survivin positive (++/+++) showed higher rates of relapse (80%) compared to those with PDEF positive (++/+++) and survivin negative (−/+) tumors (57%).
Forced expression of PDEF inhibits cell growth, downregulates endogenous survivin, induces apoptosis and reduces survivin promoter activity.
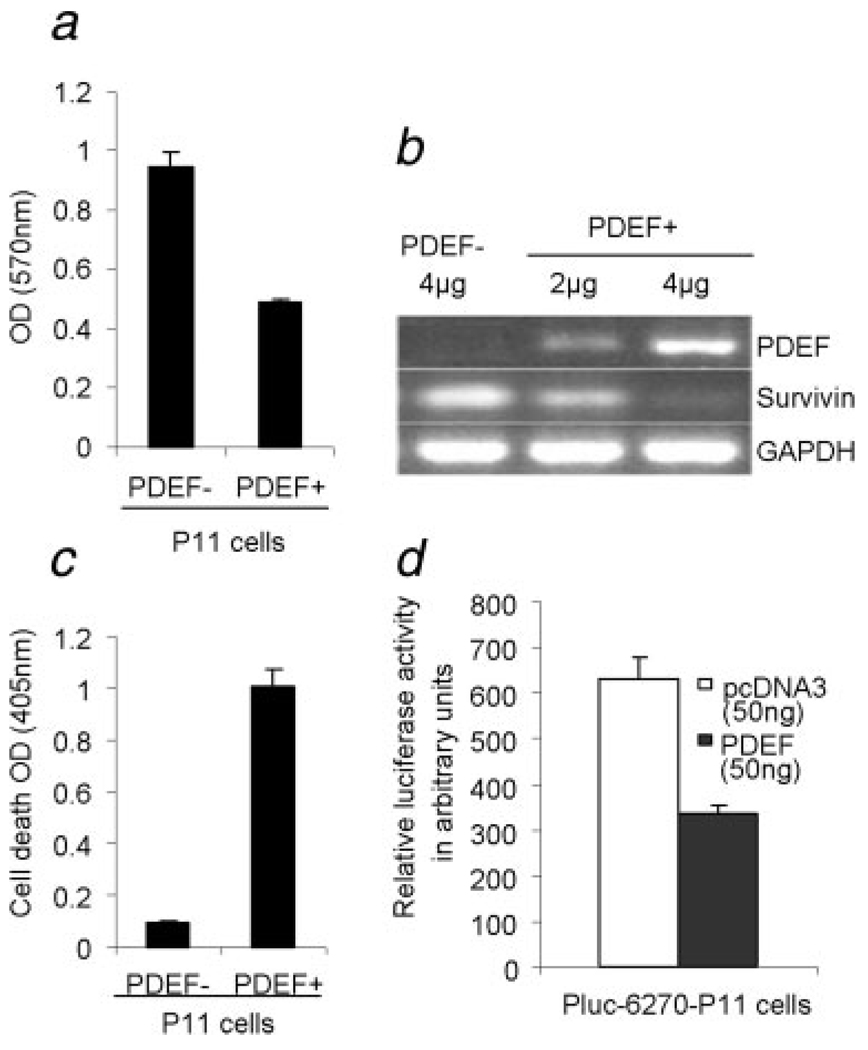
To determine the role of PDEF expression in ovarian tumor cell growth and regulation of the expression of survivin, the PDEF-negative ovarian cell line, P11 (2008), was transfected with PDEF using pcDNA3.1 PDEF construct or mock control pcDNA3.1 vector (2 and 4 µg vectors). Expression of PDEF in the cell line, using the 4 µg vector, was ~40% of that in HOSE cell line (data not shown). Proliferation of these tumor lines in vitro was then determined by MTT assay. As shown in Figure 4a, forced expression of PDEF, when using the 4 µg vector for transient transfection, resulted in a significant inhibition of P11 cell proliferation after 48 hr when compared to mock P11 cells (p = 0.00007). We also made similar observations while using additional cell lines, MDA-MB231 10 and PC3 (manuscript submitted). In addition, RT-PCR analysis revealed higher expression of PDEF and loss of survivin expression (Fig. 4b) leading to apoptosis of PDEF positive cells (Fig. 4c, p = 0.00002). Rates of cell growth and apoptosis in the 2 µg construct lied between the 4 µg construct and PDEF cells (data not shown). Cotransfection of survivin promoter along with PDEF significantly decreased survivin promoter-driven luciferase activity (Fig. 4d, p = 0.00004).
FIGURE 4.
Ectopic expression of PDEF inhibits cell growth, induces apoptosis, downregulates survivin promoter activity and endogenous survivin expression in a highly invasive P11 (2008) ovarian cancer cell line. (a) MTT assay performed for detection of proliferation pcDNA3.1 PDEF P11 cells (PDEF+) and mock pcDNA3.1 P11 cells (PDEF−). (b) Ectopic expression of PDEF (using 4 µg pcDNA3.1 PDEF) efficiently inhibited endogenous survivin expression. The relative expression of PDEF and survivin after normalization to GAPDH internal control are shown. Total mRNA extracts were collected 48 hr posttransfection and analyzed by 1-step RT-PCR (Qiagen). (c) Detection of apoptosis in pcDNA3.1 PDEF P11 cells (PDEF+) and mock pcDNA3.1 P11 by cell death detection ELISA assay. High level of apoptosis was induced when 4 µg of pcDNA3.1 PDEF vector was used for transfection (p = 0.00002). (d) Luciferase assay showing that ectopic expression of PDEF in the PDEF-negative P11 ovarian cancer cell line significantly decreased survivin promoter-driven luciferase activity. Means ± SD derived from 3 independent assays (p = 0.00004).
Reactivation of PDEF after treatment with VD3 (analogs EB 1089) and a selenium compound, MSA
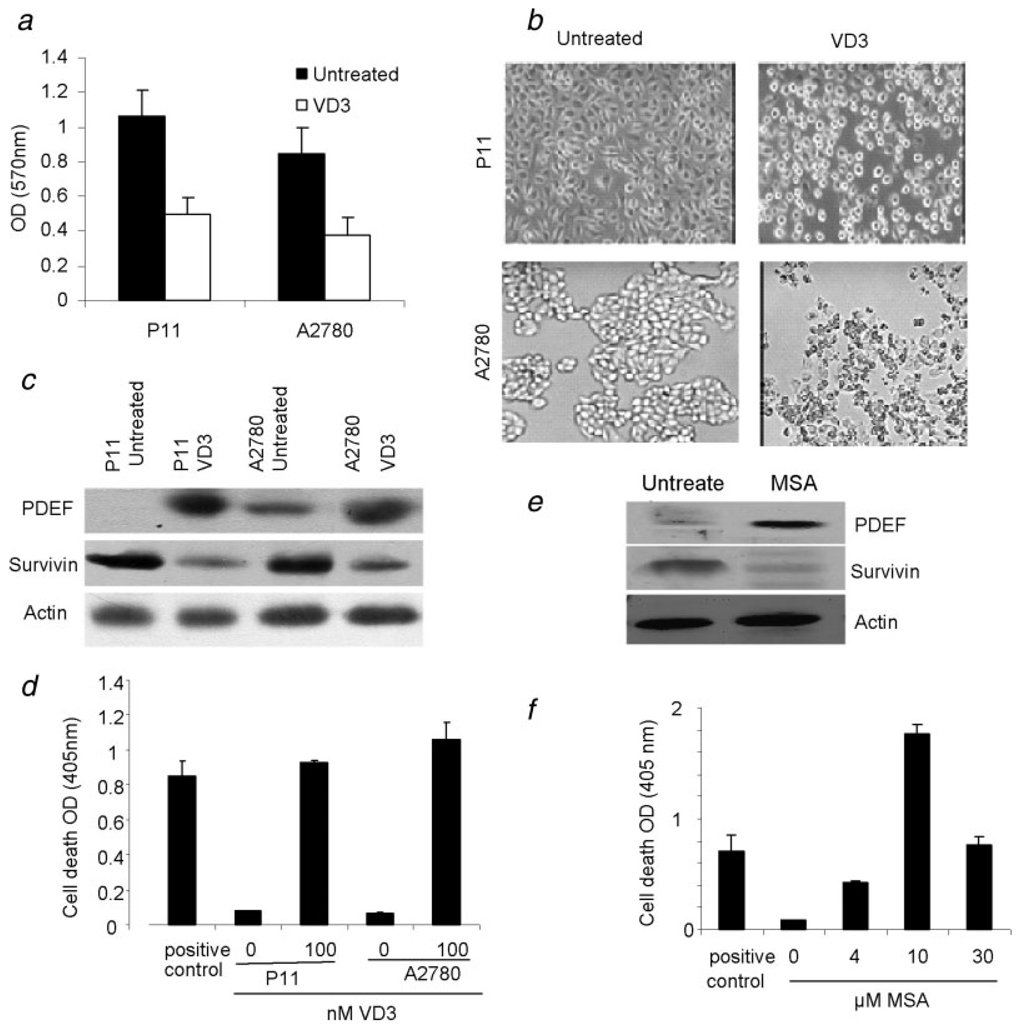
Previous studies indicated that VD3 and selenium compound (MSA) did inhibit cell proliferation and induced apoptosis in vitro and in vivo in many cell types including ovarian cancer cell lines.21–25 However, mechanisms by which these compounds manifest their antitumor function is not fully understood. We wondered whether such function VD3 and MSA may be via restoration of PDEF expression. Therefore, we cultured the PDEF-negative ovarian tumor lines, P11 and A2780, in the presence or absence of VD3 analogs EB 1089 or MSA for 72 hr. As shown in Figure 5a, presence of VD3 analogs resulted in a significant inhibition of cell proliferation (p = 0.006). Interestingly, presence of VD3 analogs (100 nM) changed cell morphology (Fig. 5b), induced PDEF expression and downregulated survivin expression (Fig. 5c). Changes in cell morphology were evident from changes in cell projections and reduced cell-cell adhesion which were not associated with cell migration or invasiveness because presence of VD3 analogs and MSA induced apoptosis in tumor cells, as determined by DNA fragmentation assay using a cell death detection ELISA assay kit (Roche, Indianapolis, IN). As shown in Figure 5d, the highest level of apoptosis was detected in ovarian cancer cell lines P11 and A2780 after treatment with VD3 (p < 0.0006). Similar results were obtained when 10 µM MSA were used (Figs. 5e and 5f).
FIGURE 5.
Reexpression of PDEF in PDEF-negative ovarian tumour lines by treatment with the vitamin D3 analogs EB1089 and MSA. VD3 enhanced PDEF expression and reduced survivin expression in the highly invasive P11 and A2780 ovarian cancer cell lines. (a) MTT assay for detection of proliferation in ovarian tumour lines in the presence (open box) or absence (black box) of 100 nM VD3 after 72 hr. Mean OD values are shown from 3 independent experiments for each cell line (p = 0.006). (b) Treatment of P11 cells after 72 hr treatment with VD3 caused changes in cell morphology as evidenced by reduction in cell projections and cell–cell adhesion. Representative images from 3 independent experiments are shown at ×200 magnifications using light microscopy. (c) Western blot analysis of ovarian tumour lines P11 and A2780 before and after treatment with 100 nM VD3 for 72 hr. Representative data from 3 independent experiments are shown. (d) Highest levels of apoptosis were detected in ovarian cancer cell lines P11 and A2780 after treatment with VD3. Mean OD values from three independent experiments are shown. (e) Western blot analysis of PDEF and survivin expression in P11 cell lysates before or after treatment with 10 µM MSA. Representative data from 3 independent experiments are shown. Actin was used as internal control. (f) Treatment of ovarian cancer cell line P11 with different concentration of MSA induced apoptosis, as determined by DNA fragmentation assay using cell death detection ELISA. Mean OD values are shown from 3 independent experiments.
Discussion
There has been little change in ovarian cancer incidence and mortality over the past 5 decades, and unfortunately, there are a number of significant barriers to progress in its treatment. These include poor understanding of the underlying biology of this disease, inadequate screening tools as well as few early warning signs. The 5-year survival for patients with Stage I disease can exceed 90%, but it is less than 25% for women with advanced-stage of the disease.2 These statistics underscore the need for identification of the molecular targets/markers for a better screening and staging of ovarian cancer.
The Ets family transcription factors regulate transcription of a number of gene that are involved in cellular proliferation, development, angiogenesis, differentiation, apoptosis, transformation and tumor invasion.26 PDEF is a new member in the Ets family proteins that acts as a tumor suppressor reducing motility, invasion and metastasis in breast and prostate cancers.6–12 Forced expression of PDEF in MDA-MB-231 breast cancer cell line mediated in part by a G0–G1 cell cycle arrest.6 We showed that forced expression of PDEF was associated with not only inhibition of cell growth and proliferation but also induction of apoptosis in the arrested cells. We report for the first time that PDEF is highly expressed in normal ovarian tissues while its expression tends to be reduced or lost during ovarian cancer progression. Loss of PDEF expression was associated with upregulation of survivin in both ovarian cancer tissues and ovarian tumor cell lines. Moreover, PDEF expression was inversely correlated with ovarian tumor stages. Patients with early tumor stages expressed higher level of PDEF in their tumors while those with late stages cancer lost the expression of PDEF in their tumors. The clinical relevance of PDEF expression was determined by showing that upregulation of PDEF along with downregulation of survivin was associated with favorable prognosis in patients with ovarian cancer. Downregulation of survivin expression by forced expression of PDEF provided additional evidence for inverse correlation between PDEF and survivin expression and suggested that the loss of PDEF expression and upregulation of survivin contribute to the ovarian cancer progression and malignancy. It is known that survivin is involved in promoting cancer cell proliferation and inhibition of apoptosis.27,28 As currently available diagnostic tools are inadequate for accurately staging ovarian cancer patients, determination of the quantitative expression of PDEF and survivin at the same time may represent a better prognostic marker for early detection of the disease. This may facilitate more accurate staging of patients and selection of treatment modalities that avoid unnecessary treatment related toxicities for patients with good prognosis. Moreover, as novel targets, they may help to develop various options to prevent ovarian cancer progression.
Contrary to our observation and other reports,6–11 a recent report demonstrated loss of PDEF expression in all normal ovarian tissues but its expression in only 33% of ovarian tumors.13 However, they did not determine whether forced expression of PDEF might enhance tumorigenicity. The discrepant results might be due to the specificity of antibody and the methods used for PDEF detection, so that inconsistent results were obtained.13 In addition, verification of PDEF expression by using Western blot and real-time PCR analyses is necessary to confirm the consistency of findings while using different methods.
Ets transcription factors bind to a core motif of “GGAA/T,” which is usually flanked with purine-rich DNA sequences.6 Consistence with pervious report10 we found that PDEF inhibited survivin expression and its promoter activity in ovarian tumor cells. Although there is no Ets binding sites (EBSs) in the core promoter region of survivin, inspection of the 2.8-kb DNA promoter region of survivin revealed 28 EBS with a perfect match to the core motif recognized by Ets transcription factors (unpublished observation). The presumptive EBSs present in the 2.8-kb promoter are also within the promoter-luciferase (6270) promoter. Thus, PDEF as a transcription factor may bind to these sites and inhibit survivin expression. However, further investigation is needed to confirm this hypothesis and evaluate the molecular mechanism by which PDEF downregulates survivin transcription.
Previous studies indicated that VD3 analog EB1089and MSA inhibit cell proliferation and induce apoptosis in vitro and in vivo in many cell types including ovarian cancer cell lines.15–17,21–25,29 However, the underlying mechanisms by which these compounds exert their effects are not well understood. We found that treatment of ovarian cancer cells with VD3 or MSA induced PDEF expression, which in turn, resulted in downregulation of the antiapoptotic protein survivin and subsequent induction of apoptosis in tumor cells. These findings suggest that cancer prevention and tumor inhibition by VD3 or MSA is associated with the activation of PDEF. Moreover, our findings are consistent with the observations suggesting that PDEF acts as a tumor suppressor and metastasis inhibitor, and that survivin expression in cancer is directly associated with cancer stage, drug/radiation resistance, shorter patient survival and oncogenesis.27,28
In summary, our observations of inverse correlation between PDEF and survivin expression suggest that expression of PDEF along with loss or downregulation of survivin provides favorable prognostic marker for ovarian cancer patients.
Acknowledgements
The authors thank Dr. Candance Johnson for her support of this work and Dr. Zahra Fayazi for technical assistance in our study. The authors also thank the members of the tissue procurement facility for their support.
Grant sponsor: NIH; Grant number: CA109481; Grant sponsor: Susan Komen Foundation; Grant number: BCTR63806; Grant sponsor: American Cancer Society; Grant number: IRG-02-197-06.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Friedlander ML. Prognostic factors in ovarian cancer. Semin Oncol. 1998;25:305–314. [PubMed] [Google Scholar]
- 3.Ghadersohi A, Chitta K, Greco WR, Harvey S, Winston J, Slocum H, Odunsi K, Sood AK. Tumour antigens and markers for breast and ovarian cancers. Front Biosci. 2002;7:48–57. doi: 10.2741/ghader. [DOI] [PubMed] [Google Scholar]
- 4.Shridhar V, Lee J, Pandita A, Iturria S, Avula R, Staub J, Morrissey M, Calhoun E, Sen A, Kalli K, Keeney G, Roche P, et al. Genetic analysis of early-versus late-stage ovarian tumour. Cancer Res. 2001;61:5895–5904. [PubMed] [Google Scholar]
- 5.Nozawa M, Yomogida K, Kanno N, Nonomura N, Miki T, Okuyama A, Nishimune Y, Nozaki M. Prostate-specific transcription factor hPSE is translated only in normal prostate epithelial cells. Cancer Res. 2000;60:1348–1352. [PubMed] [Google Scholar]
- 6.Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, Watson DK. Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res. 2003;63:4626–4631. [PubMed] [Google Scholar]
- 7.Ghadersohi A, Sood AK. Prostate epithelium-derived Ets transcription factor mRNA is overexpressed in human breast tumours and is a candidate breast tumour marker and a breast tumour antigen. Clin Cancer Res. 2001;7:2731–2738. [PubMed] [Google Scholar]
- 8.Gunawardane RN, Sgroi DC, Wrobel CN, Koh E, Daley GQ, Brugge JS. Novel role for PDEF in epithelial cell migration and invasion. Cancer Res. 2005;65:11572–11580. doi: 10.1158/0008-5472.CAN-05-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsujimoto Y, Nonomura N, Takayama H, Yomogida K, Nozawa M, Nishimura K, Okuyama A, Nozaki M, Aozasa K. Utility of immunohistochemical detection of prostate-specific Ets for the diagnosis of benign and malignant prostatic epithelial lesions. Int J Urol. 2002;9:167–172. doi: 10.1046/j.1442-2042.2002.00444.x. [DOI] [PubMed] [Google Scholar]
- 10.Ghadersohi A, Dalin P, Fayazi Z, Hicks DG, Winston JS, Li F. Prostate-derived Ets transcription factor (PDEF) downregulates survivin expression and inhibit breast cancer cell growth in vitro and xenograft tumour formation in vivo. Breast Cancer Res Treat. 2007;102:19–30. doi: 10.1007/s10549-006-9314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner DP, Moussa O, Sauane M, Fisher PB, Watson DK. Prostatederived ETS factor is a mediator of metastatic potential through the inhibition of migration and invasion in breast cancer. Cancer Res. 2007;67:1618–1625. doi: 10.1158/0008-5472.CAN-06-2913. [DOI] [PubMed] [Google Scholar]
- 12.Gu X, Zerbini LF, Out HH, Bhasin M, Yang Q, Joseph MG, Grall F, Onatunde T, Correa RG, Libermann TA. Reduced PDEF expression increases invasion and expression of mesenchymal genes in prostate cancer cell. Cancer Res. 2007;67:4219–4226. doi: 10.1158/0008-5472.CAN-06-3689. [DOI] [PubMed] [Google Scholar]
- 13.Rodabaugh KJ, Mhawech-Fauceglia P, Groth J, Lele S, Sood AK. Prostate-derived Ets Factor is overexpressed in serous epithelial ovarian tumours. Int J Gynecol Pathol. 2007;26:10–15. doi: 10.1097/01.pgp.0000225386.41244.bd. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Ling X, Pan D, Apontes P, Song L, Liang P, Altieri DC, Beerman T, Li F. Molecular mechanism of inhibition of survivin transcription by the GC-rich sequence selective DNA-binding antitumour agent, hedamycin: evidence of survivin downregulation associated with drug sensitivity. J Biol Chem. 2005;280:9745–9751. doi: 10.1074/jbc.M409350200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershberger PA, Modzelewski RA, Shurin ZR, Rueger RM, Trump DL, Johnson CS. 1,25-Dihydroxycholecalciferol (1,25-D3) inhibits the growth of squamous cell carcinoma and down-modulates p21(Waf1/Cip1) in vitro and in vivo. Cancer Res. 1999;59:2644–2649. [PubMed] [Google Scholar]
- 16.Zhao H, Whitfield LM, Xu T, Botstein D, Brooks JD. Diverse effects of methylseleninic acid on the transcriptional program of human prostate cancer cells. Mol Biol Cell. 2004;15:506–519. doi: 10.1091/mbc.E03-07-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clement IP. Lessons from basic research in selenium and cancer prevention. J Nutr. 1998;128:1845–1844. doi: 10.1093/jn/128.11.1845. [DOI] [PubMed] [Google Scholar]
- 18.Serov SF, Scully RE, Sobin LH. Histological typing of ovarian tumours. World Health Organization; International classification of tumours. 1973
- 19.Kaplan EL, Meir P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–486. [Google Scholar]
- 20.Peto R, Pike MC, Armitage P, Breslow NE, Cox DR, Howard SV, Mantel N, Mcpherson K, Peto J, Smith PG. Design and analysis of randomized clinical trails requiring prolonged observation of each patient. Br J Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azrak RG, Frank CL, Ling X, Slocum HK, Li F, Foster BA, Rustum YM. The mechanism of methylselenocysteine and docetaxel synergistic activity in prostate cancer cells. Mol Cancer Ther. 2006;5:2540–2548. doi: 10.1158/1535-7163.MCT-05-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Ling X, Huang H, Brattain L, Apontes P, Wu J, Binderup L, Brattain M. Differential regulation of survivin expression and apoptosis by vitamin D3 compounds in two isogenic MCF-7 breast cancer cell sublines. Oncogene. 2005;24:1385–1395. doi: 10.1038/sj.onc.1208330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Getzenberg RH, Light BW, Lapco PE, Konety BR, Nangia AK, Acierno JS, Dhir R, Shurin Z, Day RS, Trump DL, Johnson CS. Vitamin D inhibition of prostate adenocarcinoma growth and metastasis in the Dunning rat prostate model system. Urology. 1997;50:999–1006. doi: 10.1016/S0090-4295(97)00408-1. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Jiang F, Li P, Li C, Qiuping Ma, Nicosia SV, Bai W. Growth suppression of ovarian cancer xenografts in nude mice By Vitamin D analogue EB1089. Clinical Cancer Res. 2005;11:323–328. [PubMed] [Google Scholar]
- 25.Johnson CS, Hershberger PA, Trump DL. Vitamin D-related therapies in prostate cancer. Cancer Metastasis Rev. 2002;21:147–158. doi: 10.1023/a:1020836226594. [DOI] [PubMed] [Google Scholar]
- 26.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 27.Li F. Role of survivin and its splice variants in tumourigenesis. Br J Cancer. 2005;92:212–216. doi: 10.1038/sj.bjc.6602340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F. Survivin Study: what is the next wave? J Cell Physiol. 2003;197:8–29. doi: 10.1002/jcp.10327. [DOI] [PubMed] [Google Scholar]
- 29.Vijayakumar S, Mehta RR, Boerner PS, Packianathan S, Mehta RG. Clinical trials involving vitamin D analogs in prostate cancer. Cancer J. 2005;11:362–373. doi: 10.1097/00130404-200509000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J. 1999;2:305–311. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]