Summary
Although it was observed that inhibition of the antiapoptotic protein survivin expression in lung cancer cells induces apoptosis, the expression and role of survivin variants (survivin-2B and survivin-ΔEx3) in lung cancer have not yet been characterized. We analyzed 24 non-small-cell lung cancer (NSCLC) samples by semi-quantitative RT-PCR. Surprisingly, our results revealed that high-level expression of survivin-2B is significantly associated with the patient category of “no relapse and alive” (p-value < 0.0001). In contrast, high-level expression of survivin-ΔEx3 is highly associated with the patient category of “relapse and dead” (p-value < 0.0001). Consistent with this observation, exogenous expression of survivin-2B in A549 lung cancer cells inhibited cell growth, disrupted the mitochondria potential, and induced apoptotic cell death, while expression of survivin-ΔEx3 protected the mitochondria potential and facilitated cell survival. These findings provide evidence that survivin-2B and survivin-ΔEx3 play opposite roles in disease relapse and NSCLC cell survival, which is likely through the differential modulation of mitochondrial potential. Thus, controlling the differential expression of survivin-2B and survivin-ΔEx3 may represent novel approaches for cancer therapeutics in NSCLC.
Keywords: Survivin-2B, Survivin-ΔEx3, Non-small-cell lung cancer
1. Introduction
Lung cancer is the leading cause of cancer-related death in the United States [1–3]. Non-small-cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancers. Despite recent advances in surgery, radiation and medical treatments, the 5-year survival rate of patients with NSCLC remains among the lowest of all major human cancers [1–3]. Developing better molecular markers for the prediction of disease progression and relapse will be crucial for the improvement of medical management of this disease.
The survivin gene has four exons, and the alternative splicing of its pre-mRNA can produce three different mRNAs, which encode three distinct survivin proteins (survivin, survivin-2B and survivin-ΔEx3) [4]. In addition, a new survivin variant, survivin-3B was recently reported [5], although its function is unknown.
Studies with soft tissue sarcoma [6] and gastric cancer [7] showed that survivin-2B and survivin-ΔEx3 are expressed in these cancers in addition to survivin. However, the function of survivin-2B and survivin-ΔEx3 in these cancers is uncertain. Recently, it was reported that the expression ratio of survivin-ΔEx3/survivin is significantly higher in malignant brain tumors than in their benign counterparts [8]. Expression of survivin-ΔEx3 was dominant in malignant brain tumors, whereas survivin-2B expression was dominant in benign ones [8]. These observations suggest that survivin-ΔEx3 and survivin-2B may have differential functions in tumorigenesis and/or cell viability. Thus, investigation of the role of survivin-2B and survivin-ΔEx3 in disease outcome for cancer patients may have significant consequences to develop novel approaches for cancer diagnosis and therapeutics. Moreover, the expression of survivin-2B and survivin-ΔEx3 in many cancers including lung cancer is even not determined.
Here, we employed both clinical lung cancer tissues and an established lung cancer cell line to study the expression and function of survivin-2B and survivin-ΔEx3 in disease outcome, and regulation of cell growth and apoptosis. We hypothesized that in addition to survivin, lung cancer also expresses survivin-2B and survivin-ΔEx3, which may play a role in modulation of cancer progression and patient survival through affecting cancer cell growth and viability. Consistent with this hypothesis, our studies suggest that monitoring the expression of survivin-2B and survivin-ΔEx3 in NSCLC may provide a new method to predict the disease outcome. Control of the expression of survivin-2B and survivin-ΔEx3 may also represent novel approaches for cancer therapeutics.
2. Materials and methods
2.1. Lung cancer tissue selection
Cancer tissues used in this study had to be resected from the diagnosed primary NSCLC in patients as their initial treatment modality. Lung cancerous or adjacent normal tissues were isolated through surgery by a pathologist and immediately frozen in liquid nitrogen, and stored in a −80 °C freezer. In addition, the selected tissues had anonymized clinicopathological information (Table 1). Thus, 24 lung cancer tissues and 10 corresponding normal lung tissues matching the tissue selection criteria were obtained from the Translational Research Tissue Support Resource of Pathology, Roswell Park Cancer Institute (RPCI). This study was performed following an Institutional Review Board (IRB)-approved protocol for the investigation of molecular markers in lung cancer.
Table 1.
Selected clinicopathological informationa
| Variables | Status | Patient no. |
|---|---|---|
| Smoke | Never | 0 |
| Ever | 24 | |
| 0 | 10 | |
| PS | 1 | 10 |
| 2 | 4 | |
| AD | 8 | |
| Histology | SCC | 10 |
| ASCC | 2 | |
| LCC | 4 | |
| W | 2 | |
| Grade | M | 4 |
| P | 12 | |
| U | 6 | |
| Stage | Unknown | 24 |
| Pre- chemotherapy |
Yes | 10 |
| No | 14 | |
| Pre- radiotherapy |
Yes | 0 |
| No | 24 |
PS: performance status; AD: adenocarcinoma; SCC: squamous cell carcinoma; ASCC: adenosquamous cell carcinoma; LCC: large cell carcinoma; W: well differentiated; M: moderately differentiated; P: poorly differentiated; U: undifferentiated.
2.2. RNA isolation, reverse transcription (RT) and semi-quantitative polymerase chain reaction (semiQPCR)
Total RNAs from both normal and cancerous lung tissues were isolated by TRI REAGENT (Molecular Research Center, Cincinnati, OH) following the manufacturer’s instruction. Briefly, 30–60 mg of tissues were homogenized in 0.5–1 ml TRI REAGENT by a PowerGen 125 homogenizer with a 7 mm × 95 mm ST generator (Fisher) for three times, 30 s each in maximum speeds. Each homogenate was stored at room temperature for 5–10 min and then frozen on dry ice. Then each homogenate was thawed and vortexed for 15–20 s at the maximal speed after adding 0.05–0.1 ml BCP phase separation reagents (MRC, Cincinnati, OH). Following a 2–15 min interval at room temperature, supernatant was collected by centrifugation. Total RNA was then isolated by isopropanol precipitation from the supernatant and washed with 75% ethanol. After drying for 5–10 min in air, RNA was resuspended in 8–15 µl RNase-free water. One microliter was used for determination of RNA concentration.
RT reaction was performed using Reverse Transcription System (#A3500, Promega, Madison, WI) following the manufacturer’s instruction. Briefly, total 20 µl of RT reactions contained 1 µg RNA, 4 µl MgCl2 (25 mM), 2 µl dNTP (10 mM), 2 µl 10 × RT buffer, 20 units of RNasin ribonuclease inhibitor, 0.5 µg random primers and 15 units of AMV reverse transcriptase. After the RT reaction was carried out in a themocycler with parameters in turn, 25 °C × 10 min, 42 °C × 60 min, 95 °C × 5 min and 4 °C hold, the reaction was diluted to 100 µl with water for PCR. Ten microliters of diluted RT reactions were used for semiQ-PCR in all experiments.
2.2.1. SemiQ-PCR reaction
Total 50 µl in 0.5 ml thin-wall PCR tube contained 10 µl of diluted RT reaction (above), 5 µl of 10 × PCR buffer, 3.5 µl of MgCl2 (25 mM), 1 µl of dNTP (10 mM), 200–500 nM of primers of each (see definition below) and 1.25 units of JumpStart Taq DNA Polymerase (Sigma).
2.2.2. SemiQ-PCR parameter
A Mastercycler Gradient (Eppendorf) was used to carry out PCR reaction. The PCR parameters are pre-heated at 94 °C for 1 min; followed by 24–30 PCR cycles (see definition below) of 94 °C × 20 s, 54 °C × 30 s and 72 °C × 1 min, and then the PCR reaction was held at 4 °C. PCR reactions were separated on a 1.5% agarose gel along with 100 bp DNA marker. PCR products were visualized under ultraviolet light (236 nm) after ethidium bromide staining. The picture was taken by a digital image system and saved in TIFF files.
2.2.3. Primer concentration
Survivin-related primers were kept at 500 nM each in all PCR reactions. GAPDH primers were kept at 200 nM, when mixed with survivin-related primers in one PCR reaction (GAPDH PCR product is an internal control at this situation), or kept at 400 nm when used separately (GAPDH PCR product is used as a parallel control in this situation).
2.2.4. Primer sequence information
Hsv5′P1 (5′-GAGGCT GGCTTC ATCCAC TG-3′) and 2B-2 (5′-GTTCCT CTCTCG TGATCC G-3′) were used to amplify survivin-2B PCR products (183 bp). SVV+ (5′-TCAAGG ACCACC GCATCT CTAC-3′) and Ex3 (5′-TGGTTT CCTTTG CATGGG G-3′) were used to amplify survivin-ΔEx3 PCR products (196 bp). GAPDH3 (5′-CCTTCA TTGACC TCAACT ACA-3′) and GAPDH8 (5′-GGCCAT CCACAG TCTTCT G-3′) were used to amplify the GAPDH PCR products (467 bp, control). All primers were designed in different exons. Any PCR product generated from genomic DNA contamination (if any) would be easily discriminated.
2.2.5. PCR cycle numbers
Thirty cycles were used for amplification of survivin-2B and survivin-ΔEx3. Twenty-four cycles were used for exclusive amplification of GAPDH PCR products as a parallel control. Otherwise, the cycle numbers for GAPDH amplification were 30 when GAPDH PCR product was used as an internal control.
2.3. Tumor sorting and semiQRT-PCR data analysis
The corresponding 24 patients with NSCLC were classified into four categories based on the available anonymized clinical patient information: (1) no relapse and alive (n = 6), (2) no relapse and dead (n = 7), (3) relapse and alive (n = 0) and (4) relapse and dead of disease (n = 11). The expression level of survivin-2B and survivin-ΔEx3 determined by the semiQRT-PCR experiments was quantitated using the Personal Densitometer SI and ImageQuant5.2 Software (Molecular Dynamics). The quantitative data were then analyzed using Microsoft Excel 2001. The correlation of disease outcomes in each category with the relative expression level of survivin-2B and survivin-ΔEx3 was analyzed as percentages, respectively, and plotted as histograms by Microsoft Excel 2001.
2.4. Molecular cloning of survivin-2B and survivin-ΔEx3
The cDNAs for the open reading frame of survivin-2B and survivin-ΔEx3 were amplified by RT-PCR and cloned into pcDNA3HA vector. Briefly, total RNA was isolated from the lung cancer cells with TRI REAGENT (Molecular Research Center). The RT reaction was performed with total RNA as template using a Reverse Transcription System (Promega) as described above. HsE1ATG-5′ (5′-GGAATTCC ATG GGT GCC CCG ACG TTG-3′, EcoR I underlined) and HsKX-EX3′P2 (5′-G GGG TAC CTCGAG CTA AGA CAT TGC TAA GGG GC-3′, Xho I underlined) were used for PCR amplification of survivin-2B (625bp) and survivin-ΔEx3 (437 bp) with 35 cycles. PCR products were separated on a 1.5% agarose gel. The corresponding cDNA fragments on the gel were isolated using GENECLEAN II Kit (BIO101, Carlsbad, CA). The purified cDNA fragments of survivin-2B (625bp) or survivin-ΔEx3 (437 bp) were digested by EcoR I and Xho I, and cloned into the pcDNA3HA vector in frame at the EcoR I and Xho I sites. The obtained HA-tagged expression vectors were confirmed by sequencing. Subsequently, the cDNAs of survivin-2B or survivin-ΔEx3 in pcDNA3HA were also subcloned into the pEGFPc1 (Clontech) in frame at the EcoR I and Xba I sites in cis-orientation.
2.5. Cell growth assay
Cell viability was used as a reflection for the effect of survivin variants on cell growth. A tetrazolium salt, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), was used as a colorimetric substrate for measuring cell viability (MTT assay). When cells are injured there is an alteration of cellular redox activity, rendering the cells unable to reduce the dye. MTT was added to a final concentration 0.5 mg/ml 72 h after transfection of survivin-2B or survivin-ΔEx3 vectors (for transfection, see below). Cells were continuously incubated in a 5% CO2 incubator at 37 °C for 4 h with MTT and then lysed with a cell lysis buffer (20% SDS, 50% N,N-dimethylformamide, pH 4.7), 100 µl per well, for 4 h in the incubator. Subsequently, cell absorbance in the relevant wells was measured at 570 nm with an Ultra Microplate Reader (Bio-Tek Instruments). Results are reported as the mean ± S.D. from six measurements at each point.
2.6. Expression vector transfection
Human A549 lung cancer cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 100 units/ml of penicillin and streptomycin (Invitrogen Co., Grand Island, NY) and 10% fetal bovine serum in a 5% CO2 incubator at 37 °C. Cells were transfected with pEGFPc1-surv-2B, pEGFPc1-surv-ΔEx3 or with pEGFPc1 empty vector by Lipofectamine 2000 (Invitrogen), respectively, as follows. A549 cells (1 × 105) were seeded on the round glass coverslips coated with 2% gelatin (Sigma, MO) in 12-well plates 1 day prior to transfection. On the following day, 0.8 µg of plasmid DNAs and 2 µl of lipofectamine 2000 transfection reagents were added to a 1.5 ml tube containing 100 µl of serum-free DMEM medium, respectively. The transfection solution for each well was prepared by mixing these two solutions and incubated at room temperature for 20–30 min. Then, the DNA-lipofectamine 2000 complex was added onto cells (at ~50% confluence) in each well containing 1 ml complete medium. The transfection medium was replaced with fresh complete medium 4 h after transfection.
2.7. Mitochondria potential determination
The mitochondria potential was determined using the JC-1 mitochondrial membrane potential detection kit (Cell Technology, MN) following the manufacturer’s recommendations. Forty-eight hours after transfection, JC-1 reagent was diluted to 1× with warmed DMEM medium immediately prior to the experiment. The medium in 12-well plates was removed and replaced with 500 µl diluted 1× JC-1 reagents. Fifteen minutes after incubation at 37 °C in 5% CO2 incubator, the 1× JC-1 solution was removed. Cells in each well were washed once with 1ml 1× assay buffer (supplied in the kit). Glass coverslips containing cells were mounted on glass slides with Gel/Mount™ solution (Biomedia, Foster City, CA). Cells were observed under a Zeiss Axiovert 100 M digital fluorescence microscope. In healthy cells, the JC-1 dye stains mitochondria bright red, but in apoptotic cells, the mitochondrial membrane potential collapses, and the JC-1 dye does not accumulate within the mitochondria (no color). Images were captured using Zeiss LSM510 v2.8and processed with Photoshop Element Software.
2.8. DNA fragmentation cell death detection
Cells was transfected with or without pEGFPc1 vector or pEGFPc1-survivin-2B vector. DNA fragmentation assay was performed using Cell Death Detection ELISAPlus assay kit (Roche, Indianapolis, IN) following the protocol recommended by manufacturer 36 h after transfection. Briefly, 3 × 104 cells per well were seeded in a 48-well plate and transfected with survivin-2B, or empty vector, as described. Medium in each well was removed and the cells washed with PBS 36 h after transfection. Cells were then lysed with 200 µl lysis buffers per well (supplied in the kit). Cell lysates were centrifuged at 200 × g for 10 min after incubation for 30 min at room temperature. Twenty microliters of aliquots from the supernatant were dispensed into streptavidin-coated 96-well microliter plates followed by addition of 80 µl of immuno-reagents. The immuno-reagent consisted of a mixture of anti-histone biotin and anti-DNA-HRP directed against various histones and antibodies to both single-stranded DNA and dsDNA, which are major constituents of the nucleosomes. After 2 h incubation at room temperature with gently shaking, unbound components were removed by washing with 250 µl of 1× incubation buffer. One hundred microliters of HRP substrate (2,2′-azino-di-(3-ethylbenzthiazoline sulfonate [6]) diammonium salt, ABTS) was added to each well, and the plate placed on a shaker at 250 rpm for color development. Measurements were made at 405 nm against an ABTS solution as a blank (reference wavelength ~490 nm) using an Ultra Microplate Reader (Bio-Tek Instruments).
2.9. Propidium iodide staining and flow cytometry analysis
Cells were transfected with and without the pEGFPc1-survivin-2B expression vector or pEGFPc1-empty vector as described above. After harvesting the cells by trypsinization and washing with ice-cold PBS, cells from each well (~1 × 105 to 10 × 105) were resuspended in 5 ml ice-cold 70% ethanol and kept on ice for a minimum of 30 min to fix the cells. Cells were then suspended in 0.5 ml PBS containing 25 µg/ml propidium iodide (PI), 0.2% Triton X-100 and 40 µg/ml RNase A, and incubated for at least 30 min at 4 °C. Cells were then analyzed using flow cytometry (FACScan, Becton Dickinson, San Jose, CA) by gating the transfected green cells by counting 10,000 cells per sample. Flow cytometric data were analyzed using WinList software (Verity Software House Inc., Topsham, ME). For each group, triplicate assays were performed.
2.10. Statistical analyses of clinical data
Wilcoxon Rank Sum test was used to evaluate whether the expression level distribution is the same between any two disease-outcome categories. A two-tail significance level was calculated and significance was considered to be indicated by a p-value of 0.05 or less [9].
3. Results
3.1. Survivin-2B and survivin-ΔEx3 are differentially expressed in lung cancer tissues
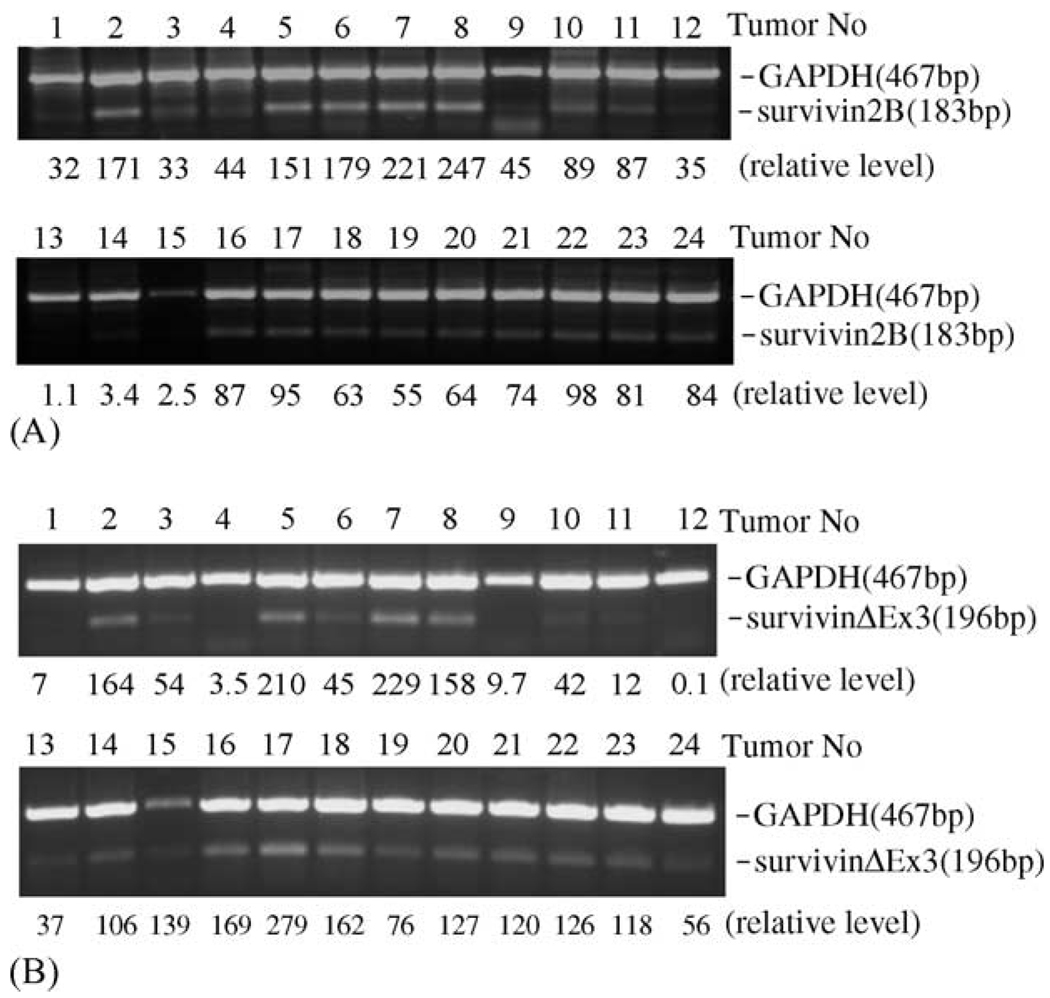
To study whether survivin-2B and survivin-ΔEx3 are expressed in lung cancer, 24 lung cancer tissues and 10 normal lung tissues were analyzed using survivin-2B- and survivin-ΔEx3-specific primers by reverse transcription-coupled semi-quantitative PCR (RT-semiQPCR). The results indicated that survivin-2B and survivin-ΔEx3 are differentially expressed in lung cancer tissues (Fig. 1). We also found that, while the expression of survivin is a predominant transcript, its expression is positively associated with survivin-ΔEx3 expression (not shown). The relative expression level for survivin-2B (Fig. 1A) and survivin-ΔEx3 (Fig. 1B) in the 24 lung cancer tissues was quantitated by densitometry, and is indicated at the bottom of the corresponding tumors in each lane after normalization to GAPDH expression level. We did not find the expression of either survivin-2B or survivin-ΔEx3 in 10 normal lung tissues (not shown).
Fig. 1.
Survivin-2B and survivin-ΔEx3 are differentially expressed in lung cancer tissues. The expression of survivin-2B (A) and survivin-ΔEx3 (B) was determined by semi-QRT-PCR using total RNAs as template isolated from 24 lung cancer tissues. PCR products were separated by agarose gel containing ethidium bromide and visualized under ultraviolet light (236 nm). The picture was taken under a digital image system and saved as Tiff files for further processing with Canvas software. The relative level of survivin-2B and survivin-ΔEx3 expression is indicated after normalization to the internal control of GAPDH expression level.
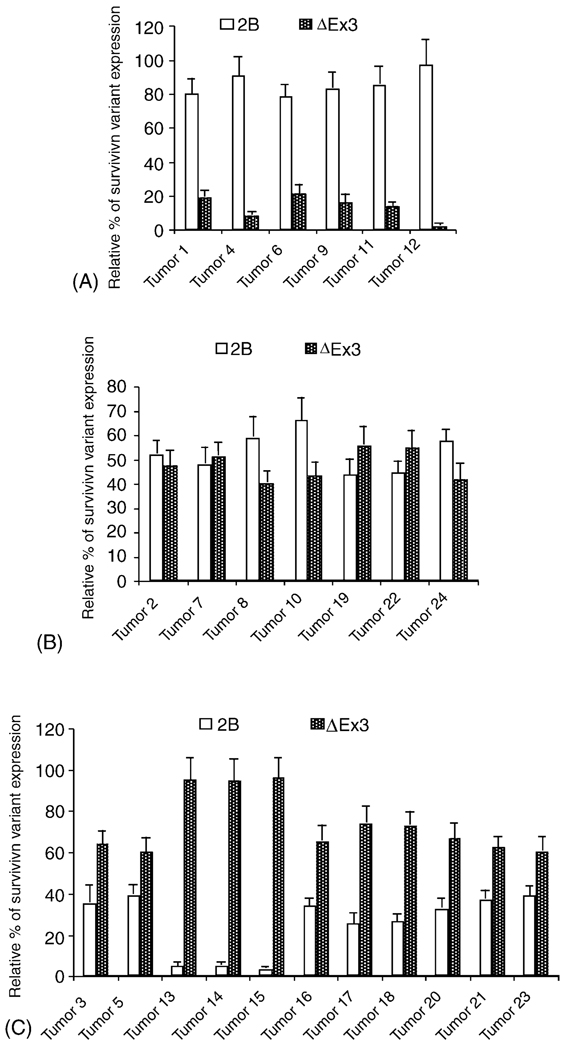
3.2. Expression of survivin-2B and survivin-ΔEx3 is inversely associated with disease outcome
Next, we conducted three pair-wise comparisons. We found that a predominant expression of survivin-2B relative to survivin-ΔEx3 is significantly associated with patient “no relapse and alive” categories (Fig. 2A) (p-values < 0.0001). In contrast, a predominant expression of survivin-ΔEx3 relative to survivin-2B is significantly associated with the patient category of “relapse and dead” (2C) (p-value < 0.0001). However, the relative expression level for survivin-2B and survivin-ΔEx3 is comparable in the lung cancer tissues from the patient category of “no relapse and dead” (Fig. 2B). These findings strongly argue that the expression of survivin-2B in lung tumor cells is a favorable factor for patient, while expression of survivin-ΔEx3 in lung tumor cells is an unfavorable factor for patients.
Fig. 2.
Expression of survivin-2B (2B) and survivin-ΔEx3 (ΔEx3) shows an opposite association with tumor relapse and patient survival. The patients corresponding to the 24 lung cancer samples were classified, on the basis of the clinical patient information, into four categories: (A) no disease relapse and alive (n = 6), (B) no disease relapse and dead (n = 7), (C) disease relapse and dead (n = 11), and (D) disease relapse and alive (n = 0). The expression level of survivin-2B and survivin-ΔEx3 is shown as percentages, respectively, and exported into a histogram by Microsoft Excel 2001. Data are the mean ± S.D. derived from three independent semi-QRT-PCRs.
3.3. Survivin-2B and survivin-ΔEx3 differentially affect cell survival and growth
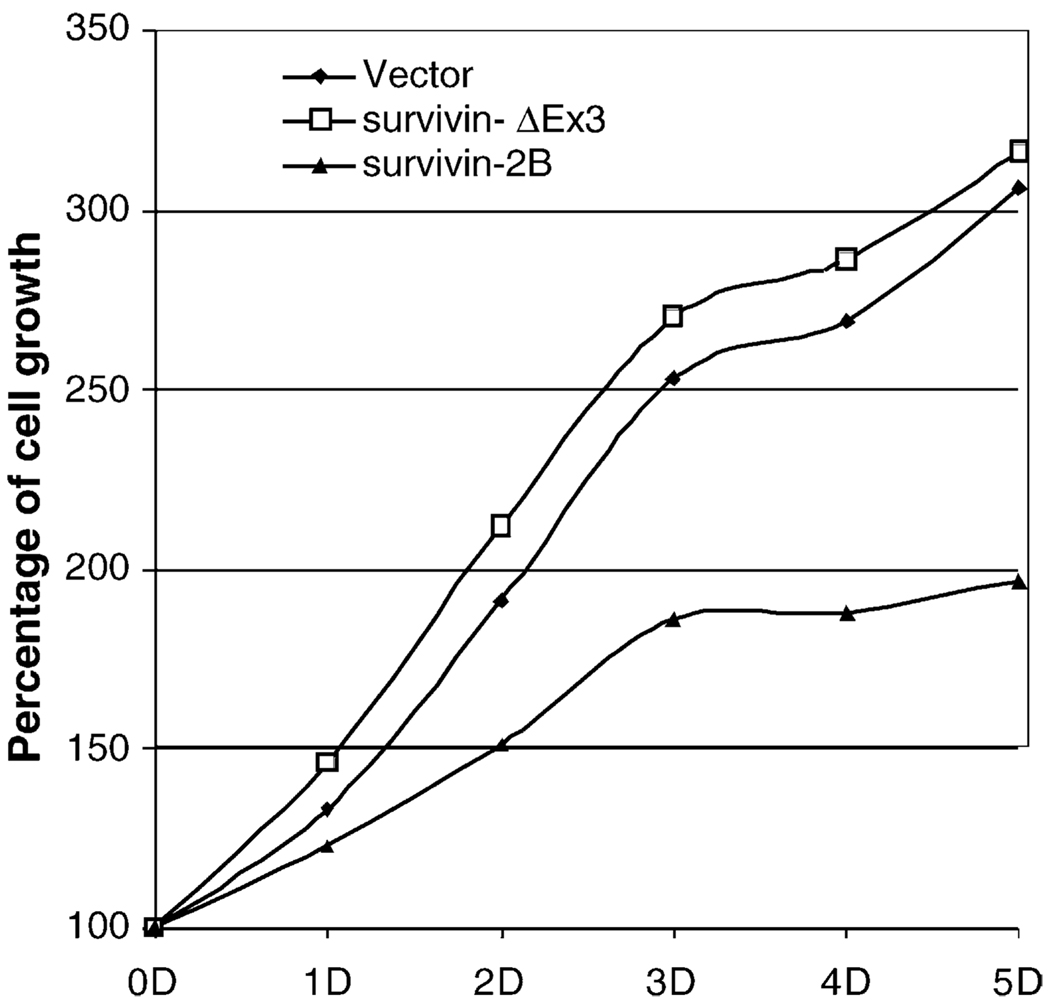
To back up the above observation, we first determined the role of survivin-2B and survivin-ΔEx3 in the regulation of lung cancer cell growth and viability using MTT assay. The results indicated that forced expression of survivin-2B inhibits cell growth while expression of survivin-ΔEx3 does not (Fig. 3), indicating their different roles in the maintenance of cell survival and growth.
Fig. 3.
Survivin-2B and survivin-ΔEx3 differentially regulate lung cancer cell growth/viability. A549 lung cancer cells were transfected with expression vectors for survivin-2B and survivin-ΔEx3 or empty vector using Lipofectamine 2000 (Invitrogen), respectively. Cell growth was determined by MTT assay over the time (day, D). As shown, exogenous expression of survivin-2B inhibited cell growth while survivin-ΔEx3 did not inhibit cell growth. (Note: variations are within 15% in each points.)
3.4. Survivin-2B and survivin-ΔEx3 differentially affect mitochondria potential
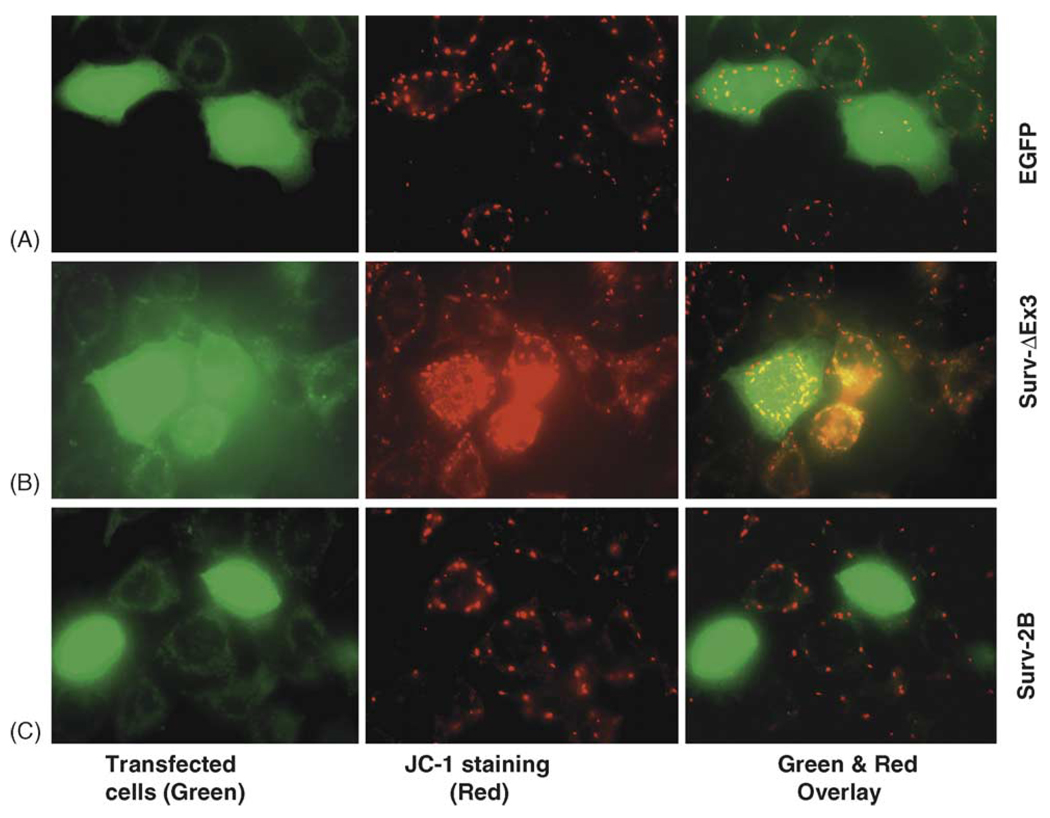
Next, we determined the effect of survivin-2B and survivin-ΔEx3 on mitochondrial potential. We found that enhanced expression of survivin-2B (green cells) disrupted the mitochondrial membrane potential (an early hallmark of apoptosis) indicated by the loss of JC-1 dye accumulation (no red) in the mitochondria (Fig. 4C). In contrast, exogenous expression of survivin-ΔEx3 protected the mitochondrial membrane potential revealed by the enhanced JC-1 dye accumulation (red) in the mitochondria (Fig. 4B) in comparison with the empty vector (pEGFPc1)-transfected cell control (Fig. 4A).
Fig. 4.
Role of survivin-2B and survivin-ΔEx3 in protecting mitochondrial potential. (A) A549 cells were transfected as shown. Forty-eight hours after transfection, the mitochondria potential was determined using the JC-1 mitochondrial membrane potential detection kit (Cell Technology, MN) following the manufacturer’s recommendations. In healthy cells, the JC-1 dye accumulated in the mitochondria as bright red, but in apoptotic cells the mitochondrial membrane potential collapses, and the JC-1 dye could not accumulate within the mitochondria (no color). Images were captured under a fluorescence microscope (Zeiss LSM510 v2.8) and processed with Photoshop Element Software.
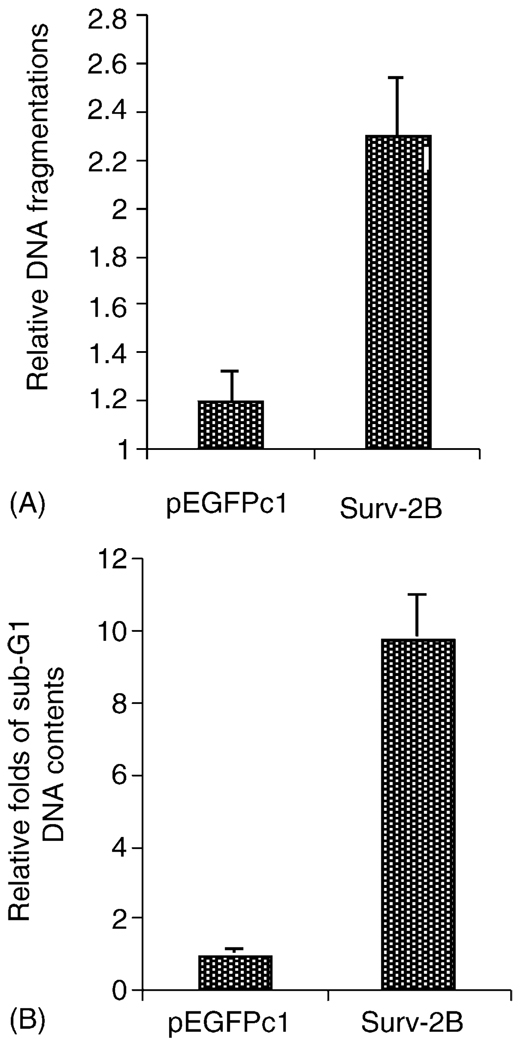
3.5. Survivin-2B induces death in lung cancer cells
We further investigated the role of survivin-2B in the induction of lung cancer cell death using both Cell Death Detection ELISA (Roche) and cell-flow cytometry. These experiments showed that transfection-mediated expression of survivin-2B strikingly induced DNA fragmentation (a hallmark of apoptosis, Fig. 5A) and sub-G1 DNA content increase (cell death, Fig. 5B). These results further indicated a pro-apoptotic role of survivin-2B in cell survival.
Fig. 5.
Role of survivin-2B in the induction of lung cancer cell death. (A) Cells were transfected with survivin-2B or empty vector (pEGFPc1) and lysed for determination of cell death/DNA fragmentation using the Cell Death Detection ELISAPlus kit (Roche) 36 h after transfection. As shown, exogenous expression of survivin-2B significantly increased DNA fragmentation in comparison with the empty vector-transfected control. Bars are the means ± S.D. derived from three independent assays. (B) Cells were transfected same as in (A) and analyzed using cell-flow cytometry by gating transfected green cells 36 h after transfection. The sub-G1 DNA contents in each condition were shown as folds (empty vector control as 1). Each bar is the mean ± S.D. derived from a representative experiment in triplicate.
4. Discussion
Survivin has been recognized as a novel molecular marker/target for cancer prognosis and therapeutics, and the role of survivin in lung cancer progression and prognosis has been extensively characterized [10]. However, there are some inconsistencies with respect to survivin as a prognostic marker in lung cancer. Studies have shown that patients whose cancer cells lacked survivin expression had a significantly better overall survival than patients whose cancer cells expressed survivin [11,12]. The frequency of venous invasion was significantly higher in the patients with elevated survivin expression in their cancer cells [12]. There was no significant correlation between survivin expression and cigarette smoking, histologic subtype, tumor differentiation, tumor size, or the presence of mediastinal lymph node metastases in surgical specimens [11]. Studies from other reports indicate that (cytoplasmic) survivin expression correlated with tumor stages [13,14], but not patient survival. Survivin expression was almost always present in early-stage NSCLC, indicating a possible role of survivin in carcinogenesis [13]. These studies suggest that other factors may play a role in lung cancer pathology. Here, we investigated the expression and role of survivin variants, survivin-2B and survivin-ΔEx3, in lung cancer. We expected that the results obtained from this study may extend the current vision regarding the role of survivin variants in lung cancer, and provide new perspectives for cancer therapeutics. It is currently unknown whether survivin-2B and survivin-ΔEx3 are expressed in lung cancer tissues, and whether the expression of these variants plays any role in disease outcome for lung cancer patients.
In this study, 24 lung cancer tissues were analyzed by semi-quantitative RT-PCR. The results demonstrate that survivin-2B and survivin-ΔEx3 are differentially expressed in lung cancer tissues (Fig. 1A and B), and that a predominant expression of survivin-2B relative to survivin-ΔEx3 is significantly associated with the patient category of “no relapse and alive” (Fig. 2A) (p-value < 0.0001). A predominant expression of survivin-ΔEx3 relative to survivin-2B was significantly associated with the patient category of “relapse and dead” (Fig. 2C) (p-value < 0.0001). Additionally, we also found that while the expression of survivin-2B in well and moderately differentiated lung tumors tends to be higher in comparison with the level of survivin-ΔEx3 expression, the difference is not statistically significant (not shown). To our knowledge, this is the first report indicating the potential of survivin-2B and survivin-ΔEx3 level to predict disease outcome in lung cancer patients. In spite of a differential expression of survivin-2B and survivin-ΔEx3 in lung cancer tissues (Fig. 1), we found that the level of survivin-ΔEx3 positively correlated with survivin expression (not shown). Others have noted that survivin-ΔEx3 is similar to survivin with respect to its antiapoptotic function [4].
To further validate the role of survivin-2B and survivin-ΔEx3 in the regulation of lung cancer cell viability, we performed additional experiments to evaluate the role of survivin-2B and survivin-ΔEx3 in cell survival and death (Fig. 3–Fig 5). These experiments revealed that ectopic expression of survivin-2B inhibited cell growth (Fig. 3), disrupted the mitochondria potential (Fig. 4) and induced the production of hypodiploid cells (DNA fragmentation, Fig. 5), while ectopic expression of survivin-ΔEx3 appeared to increase mitochondria potential (Fig. 4), implying a possible role of survivin-ΔEx3 in the maintenance of cell viability. Together, these studies not only strongly support the results derived from the analyses of clinical lung cancer tissues and patient information shown in Fig. 1 and Fig 2, but also provide a sound molecular mechanism to support the finding in Fig. 1 and Fig 2. This report extends our current knowledge of the role of survivin variants in lung cancer, and provides a potential mechanism for survivin-2B and survivin-ΔEx3 to exert their effects on disease outcome in lung cancer patients.
In summary, we assessed the expression and function of survivin-2B and survivin-ΔEx3 in 24 lung cancer tissues as well as in an established lung cancer cell line. We conclude that survivin-2B and survivin-ΔEx3 are differentially expressed in lung cancer tissues, and that survivin-2B and survivin-ΔEx3 play different roles in the modulation of cancer cell viability, disease relapse, and patient survival in NSCLC. Intervention of survivin-2B and survivin-ΔEx3 expression may represent a novel approach for cancer therapeutics.
Acknowledgements
This work was sponsored in part by Elsa U. Pardee Foundation (Midland, MI) and Concern Foundation (Beverly Hill, CA) to F.L., and by NIH R01 grants to L.Y. (CA9564301), M.Z. (CA82323) and F.L. (CA109481), and shared resources of an NCI Comprehensive Cancer Center Support Grant CA16056.
Abbreviations
- NSCLC
non-small-cell lung cancer
- semi-QRT-PCR
semi-quantitative reverse transcription-coupled polymerase chain reaction
References
- 1.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 2.Klittich WS, Caro JJ. Lung cancer screening: will the controversy extend to its cost-effectiveness? Am J Respir Med. 2002;1:393–401. doi: 10.1007/BF03257166. [DOI] [PubMed] [Google Scholar]
- 3.Soria JC, Kim ES, Fayette J, Lantuejoul S, Deutsch E, Hong WK. Chemoprevention of lung cancer. Lancet Oncol. 2003;4:659–669. doi: 10.1016/s1470-2045(03)01244-0. [DOI] [PubMed] [Google Scholar]
- 4.Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59:6097–6102. [PubMed] [Google Scholar]
- 5.Badran A, Yoshida A, Ishikawa K, Goi T, Yamaguchi A, Ueda T, et al. Identification of a novel splice variant of the human anti-apoptopsis gene survivin. Biochem Biophys Res Commun. 2004;314:902–907. doi: 10.1016/j.bbrc.2003.12.178. [DOI] [PubMed] [Google Scholar]
- 6.Kappler M, Kohler T, Kampf C, Diestelkotter P, Wurl P, Schmitz M, et al. Increased survivin transcript levels: an independent negative predictor of survival in soft tissue sarcoma patients. Int J Cancer. 2001;95:360–363. doi: 10.1002/1097-0215(20011120)95:6<360::aid-ijc1063>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 7.Krieg A, Mahotka C, Krieg T, Grabsch H, Muller W, Takeno S, et al. Expression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progression. Br J Cancer. 2002;86:737–743. doi: 10.1038/sj.bjc.6600153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada Y, Kuroiwa T, Nakagawa T, Kajimoto Y, Dohi T, Azuma H, et al. Transcriptional expression of survivin and its splice variants in brain tumors in humans. J Neurosurg. 2003;99:738–745. doi: 10.3171/jns.2003.99.4.0738. [DOI] [PubMed] [Google Scholar]
- 9.Hollander M, Wolfe DA. Nonparametric statistical methods. 2nd ed. Wiley and Sons; 1999. [Google Scholar]
- 10.Li F. Survivin study: what is the next wave? J Cell Physiol. 2003;197:8–29. doi: 10.1002/jcp.10327. [DOI] [PubMed] [Google Scholar]
- 11.Monzo M, Rosell R, Felip E, Astudillo J, Sanchez JJ, Maestre J, et al. A novel anti-apoptosis gene: re-expression of survivin messenger RNA as a prognosis marker in non-small-cell lung cancers. J Clin Oncol. 1999;17:2100–2104. doi: 10.1200/JCO.1999.17.7.2100. [DOI] [PubMed] [Google Scholar]
- 12.Ikehara M, Oshita F, Kameda Y, Ito H, Ohgane N, Suzuki R, et al. Expression of survivin correlated with vessel invasion is a marker of poor prognosis in small adenocarcinoma of the lung. Oncol Rep. 2002;9:835–838. [PubMed] [Google Scholar]
- 13.Falleni M, Pellegrini C, Marchetti A, Oprandi B, Buttitta F, Barassi F, et al. Survivin gene expression in early-stage non-small cell lung cancer. J Pathol. 2003;200:620–626. doi: 10.1002/path.1388. [DOI] [PubMed] [Google Scholar]
- 14.Kren L, Brazdil J, Hermanova M, Goncharuk VN, Kallakury BV, Kaur P, et al. Prognostic significance of anti-apoptosis proteins survivin and bcl-2 in non-small cell lung carcinomas: a clinicopathologic study of 102 cases. Appl Immunohistochem Mol Morphol. 2004;12:44–49. doi: 10.1097/00129039-200403000-00009. [DOI] [PubMed] [Google Scholar]