Abstract
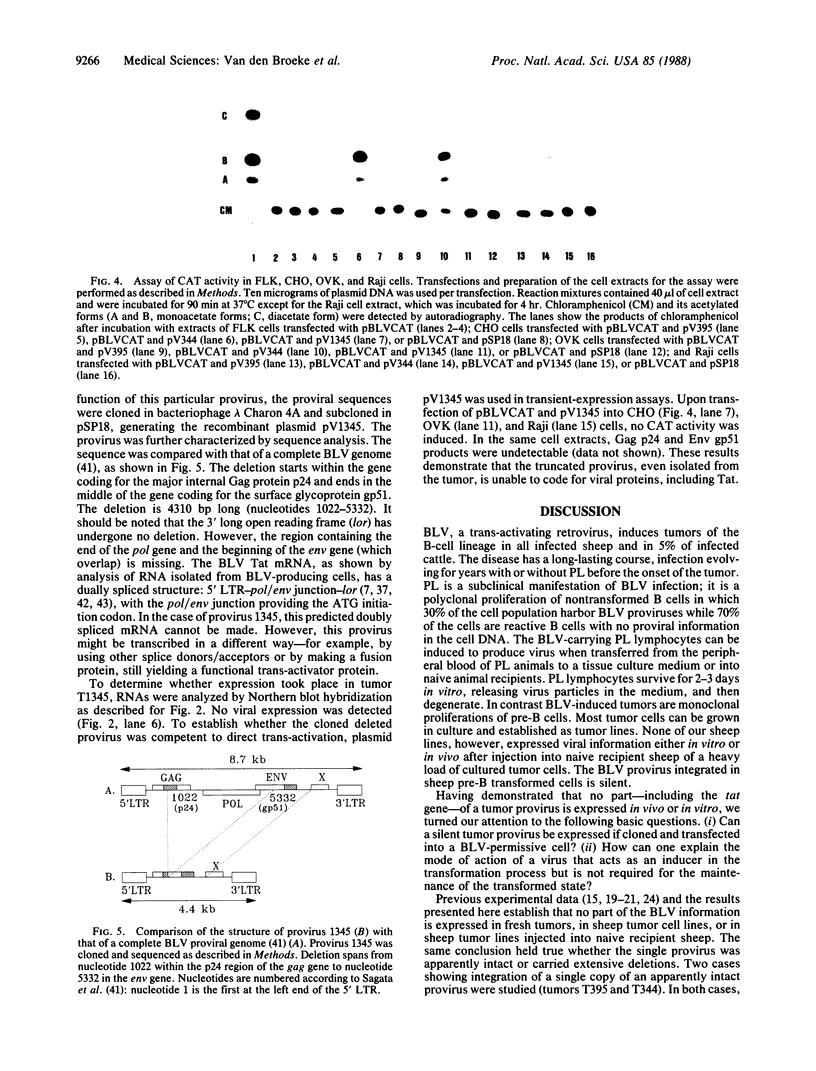
To investigate the role of proviral integration and expression in cellular transformation induced by bovine leukemia virus (BLV), three BLV-induced tumors harboring a single proviral copy were selected upon restriction and hybridization analysis. Tumors 344 and 395 were shown to contain a full-size proviral copy, whereas in tumor 1345 the provirus appeared to be heavily deleted. RNA gel blot hybridization with an antisense RNA probe showed no transcription of the viral sequences in the fresh tumors or in sheep tumor cells growing in vitro. The proviruses were cloned and transfected in mammalian cell lines. Transient-expression experiments revealed that the complete proviruses were still able to express the trans-activating protein (Tat) as well as structural proteins, demonstrating that the nonexpression of a provirus in a tumor cell does not necessarily imply a structural alteration of the viral information. In contrast, sequence analysis of the provirus with a large deletion and transient-expression assays proved that this truncated provirus, isolated from a tumor, was unable to code for viral proteins. These data indicate that expression of viral genes, including tat, is not required for the maintenance of the transformed state.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baliga V., Ferrer J. F. Expression of the bovine leukemia virus and its internal antigen in blood lymphocytes. Proc Soc Exp Biol Med. 1977 Nov;156(2):388–391. doi: 10.3181/00379727-156-39942. [DOI] [PubMed] [Google Scholar]
- Burny A., Cleuter Y., Kettmann R., Mammerickx M., Marbaix G., Portetelle D., Van den Broeke A., Willems L., Thomas R. Bovine leukaemia: facts and hypotheses derived from the study of an infectious cancer. Cancer Surv. 1987;6(1):139–159. [PubMed] [Google Scholar]
- Clarke M. F., Trainor C. D., Mann D. L., Gallo R. C., Reitz M. S. Methylation of human T-cell leukemia virus proviral DNA and viral RNA expression in short- and long-term cultures of infected cells. Virology. 1984 May;135(1):97–104. doi: 10.1016/0042-6822(84)90120-x. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D. Bovine leukemia virus transcription is controlled by a virus-encoded trans-acting factor and by cis-acting response elements. J Virol. 1987 Aug;61(8):2462–2471. doi: 10.1128/jvi.61.8.2462-2471.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Caradonna S. J., Casey J. W. Bovine leukemia virus long terminal repeat: a cell type-specific promoter. Science. 1985 Jan 18;227(4684):317–320. doi: 10.1126/science.2981431. [DOI] [PubMed] [Google Scholar]
- Derse D., Casey J. W. Two elements in the bovine leukemia virus long terminal repeat that regulate gene expression. Science. 1986 Mar 21;231(4744):1437–1440. doi: 10.1126/science.3006241. [DOI] [PubMed] [Google Scholar]
- Derse D. trans-acting regulation of bovine leukemia virus mRNA processing. J Virol. 1988 Apr;62(4):1115–1119. doi: 10.1128/jvi.62.4.1115-1119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps J., Kettmann R., Burny A. Experiments with cloned complete tumor-derived bovine leukemia virus information prove that the virus is totally exogenous to its target animal species. J Virol. 1981 Nov;40(2):605–609. doi: 10.1128/jvi.40.2.605-609.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll D. M., Olson C. Bovine leukemia virus-associated antigens in lymphocyte cultures. Am J Vet Res. 1977 Nov;38(11):1897–1898. [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Grégoire D., Couez D., Deschamps J., Heuertz S., Hors-Cayla M. C., Szpirer J., Szpirer C., Burny A., Huez G., Kettmann R. Different bovine leukemia virus-induced tumors harbor the provirus in different chromosomes. J Virol. 1984 Apr;50(1):275–279. doi: 10.1128/jvi.50.1.275-279.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P., Kashmiri S. V., Ferrer J. F. Transcriptional control of the bovine leukemia virus genome: role and characterization of a non-immunoglobulin plasma protein from bovine leukemia virus-infected cattle. J Virol. 1984 Apr;50(1):267–270. doi: 10.1128/jvi.50.1.267-270.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn J., Matthes H. W., Gait M. J., Brenner S. A new selective phage cloning vector, lambda 2001, with sites for XbaI, BamHI, HindIII, EcoRI, SstI and XhoI. Gene. 1984 Dec;32(1-2):217–224. doi: 10.1016/0378-1119(84)90049-0. [DOI] [PubMed] [Google Scholar]
- Kettmann R., Cleuter Y., Gregoire D., Burny A. Role of the 3' long open reading frame region of bovine leukemia virus in the maintenance of cell transformation. J Virol. 1985 Jun;54(3):899–901. doi: 10.1128/jvi.54.3.899-901.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettmann R., Cleuter Y., Mammerickx M., Meunier-Rotival M., Bernardi G., Burny A., Chantrenne H. Genomic integration of bovine leukemia provirus: comparison of persistent lymphocytosis with lymph node tumor form of enzootic. Proc Natl Acad Sci U S A. 1980 May;77(5):2577–2581. doi: 10.1073/pnas.77.5.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettmann R., Deschamps J., Cleuter Y., Couez D., Burny A., Marbaix G. Leukemogenesis by bovine leukemia virus: proviral DNA integration and lack of RNA expression of viral long terminal repeat and 3' proximate cellular sequences. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2465–2469. doi: 10.1073/pnas.79.8.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettmann R., Deschamps J., Couez D., Claustriaux J. J., Palm R., Burny A. Chromosome integration domain for bovine leukemia provirus in tumors. J Virol. 1983 Jul;47(1):146–150. doi: 10.1128/jvi.47.1.146-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettmann R., Mammerickx M., Dekegel D., Ghysdael J., Portetelle D., Burny A. Biochemical approach to bovine leukemia. Acta Haematol. 1975;54(4):201–209. doi: 10.1159/000208076. [DOI] [PubMed] [Google Scholar]
- Kettmann R., Mammerickx M., Portetelle D., Grégoire D., Burny A. Experimental infection of sheep and goat with bovine leukemia virus: localization of proviral information on the target cells. Leuk Res. 1984;8(6):937–944. doi: 10.1016/0145-2126(84)90047-x. [DOI] [PubMed] [Google Scholar]
- Kettmann R., Marbaix G., Cleuter Y., Portetelle D., Mammerickx M., Burny A. Genomic integration of bovine leukemia provirus and lack of viral RNA expression in the target cells of cattle with different responses to BLV infection. Leuk Res. 1980;4(6):509–519. doi: 10.1016/0145-2126(80)90062-4. [DOI] [PubMed] [Google Scholar]
- Mammerickx M., Palm R., Portetelle D., Burny A. Experimental transmission of enzootic bovine leukosis to sheep: latency period of the tumoral disease. Leukemia. 1988 Feb;2(2):103–107. [PubMed] [Google Scholar]
- Mamoun R. Z., Astier-Gin T., Kettmann R., Deschamps J., Rebeyrotte N., Guillemain B. J. The pX region of the bovine leukemia virus is transcribed as a 2.1-kilobase mRNA. J Virol. 1985 May;54(2):625–629. doi: 10.1128/jvi.54.2.625-629.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbaix G., Kettmann R., Cleuter Y., Burny A. Viral RNA content of bovine leukemia virus-infected cells. Mol Biol Rep. 1981 May 22;7(1-3):135–138. doi: 10.1007/BF00778744. [DOI] [PubMed] [Google Scholar]
- Rice N. R., Simek S. L., Dubois G. C., Showalter S. D., Gilden R. V., Stephens R. M. Expression of the bovine leukemia virus X region in virus-infected cells. J Virol. 1987 May;61(5):1577–1585. doi: 10.1128/jvi.61.5.1577-1585.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Kettman R., Haseltine W. A. Activation of enhancer sequences in type II human T-cell leukemia virus and bovine leukemia virus long terminal repeats by virus-associated trans-acting regulatory factors. J Virol. 1986 Mar;57(3):738–744. doi: 10.1128/jvi.57.3.738-744.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Willems L., Kettmann R., Campbell K., Zaya R., Burny A., Haseltine W. A. The 3' region of bovine leukemia virus genome encodes a trans-activator protein. EMBO J. 1986 Oct;5(10):2585–2589. doi: 10.1002/j.1460-2075.1986.tb04538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagata N., Yasunaga T., Ikawa Y. Two distinct polypeptides may be translated from a single spliced mRNA of the X genes of human T-cell leukemia and bovine leukemia viruses. FEBS Lett. 1985 Nov 11;192(1):37–42. doi: 10.1016/0014-5793(85)80038-7. [DOI] [PubMed] [Google Scholar]
- Sagata N., Yasunaga T., Tsuzuku-Kawamura J., Ohishi K., Ogawa Y., Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci U S A. 1985 Feb;82(3):677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short M. K., Okenquist S. A., Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987 Apr;61(4):1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L., Bruck C., Portetelle D., Burny A., Kettmann R. Expression of a cDNA clone corresponding to the long open reading frame (XBL-I) of the bovine leukemia virus. Virology. 1987 Sep;160(1):55–59. doi: 10.1016/0042-6822(87)90043-2. [DOI] [PubMed] [Google Scholar]
- Willems L., Gegonne A., Chen G., Burny A., Kettmann R., Ghysdael J. The bovine leukemia virus p34 is a transactivator protein. EMBO J. 1987 Nov;6(11):3385–3389. doi: 10.1002/j.1460-2075.1987.tb02661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. G., Blattner F. R. Construction and characterization of the hybrid bacteriophage lambda Charon vectors for DNA cloning. J Virol. 1979 Feb;29(2):555–575. doi: 10.1128/jvi.29.2.555-575.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]