Abstract
Conjugation of biotin and fluorophore tags is useful for assaying covalent protein modification. Oxidative bioactivation of selective estrogen receptor modulators (SERMs) yields reactive quinoid electrophiles that covalently modify proteins; bioactivation is associated with carcinogenic and chemopreventive effects. Identification of the protein targets of electrophilic metabolites is of general important for xenobiotics. Four methodologies, using SERM derivatives and biotin/fluorophore tags, were compared for purification and quantification: (1) covert oxidatively activated tags (COATags; SERM conjugated to biotin); (2) dansylTags (SERM conjugated to fluorophore); and azidoTags (SERM azide derivatives) in a 2-step conjugation to biotin, either using (3) Staudinger ligation; or (4) click chemistry. All synthetic derivatives retained the estrogen receptor ligand characteristics of the parent SERMs. Model proteins with bioactivation by tyrosinase in buffer or cell lysates and liver proteins with in situ bioactivation in rat primary hepatocytes were studied by immunoassay and fluorescence. Comparison showed: the azidoTag/Staudinger method was sensitive but nonspecific; the azidoTag/click methodology had low sensitivity; and, the dansylTag methodology failed to detect modified proteins in hepatocytes. The COATag methodology was adjudged superior, detecting 5 ng of modified protein in vitro and identifying protein targets in hepatocytes. In metabolism studies in rat liver microsomes, the azide group was metabolically labile, one contributing factor in not selecting an azidoTag methodology in the highly oxidative environments required for bioactivation. For study of the protein targets of electrophilic metabolites formed by in situ oxidative bioactivation, the COATag is both sensitive and specific, and does not appear to suffer from poor cell permeability.
Keywords: xenobiotics, protein modification, biotin tags, quinone electrophiles, proteomics
INTRODUCTION
Selective estrogen receptor modulators (SERMs) are widely used in the clinic for the treatment and prevention of breast cancer, osteoporosis, and other indications in postmenopausal women (1). All SERMs and their active metabolites share common polyaromatic phenolic scaffolds that are redox active in vitro and in vivo (2, 3). Formation of o-quinones, quinone methides, and di-quinone methides has been reported from the oxidative metabolism of SERMs (4-7). As reactive electrophiles, SERM quinoids can potentially covalently modify cellular proteins with varied selectivity. Protein families that are involved in estrogen metabolism, ER signaling pathways, and redox sensing are theoretically susceptible to modification by SERM quinoids. It has been reported that metabolic enzymes, such as GST-P1 and P450 isoforms, can be inhibited by raloxifene and other SERMs (7-9); modification of ER and histone H3 by estrogens and quinoids has been reported (10-13); and sensor proteins such as Keap1 involved in cellular defense are also likely targets (14). It is increasingly clear that damage or covalent modification of cellular proteins is related to both toxicity and cytoprotective mechanisms, which impacts the balance between carcinogenesis versus chemoprevention for a variety of small molecules that form electrophilic, reactive intermediates (14). The correlation between the structural properties of these electrophiles and their biological targets is not yet clear, however, it is important to identify cellular targets of xenobiotics to understand the long-term biological consequences of exposure.
Identification of protein targets of electrophiles remains a technical challenge that is limited by a lack of efficient strategies for labeling targets in a physiological environment. Previous studies have widely used isotopically labeled electrophiles to analyze modified proteins (15-18); animals or cells were treated with radioactive electrophiles and were further analyzed by 1D/2D-PAGE and/or LC-MS/MS (17, 19). Antibodies that can recognize adducts formed from electrophiles such as 4-hydroxynonenal (4-HNE) and butylated hydroxytoluene (BHT) have also been used for protein identification (15, 16, 18), however, these techniques suffer from low coverage of adducted proteins and cross-reactivity. A more recent approach using tagged electrophiles is highlighted by the use of 4-HNE tagged with biotin to identify protein targets of 4-HNE (20). Whereas 4-HNE and commercially available tagged electrophiles (i.e. iodoacetamide) are inherently reactive, SERMs require oxidative bioactivation. Therefore, a novel methodology was developed, coined the covert oxidatively activated tag (COATag), in which a biotin linker is tagged to the partial structure of the xenobiotic. A COATag is not inherently reactive towards cellular proteins but requires bioactivation. The proof of principle study used a COATag of the clinical benzothiophene SERM raloxifene (Figure 1), and oxidative enzymes to activate the SERM to a reactive electrophile (7). In combination with mass spectroscopy, the COATag methodology revealed the selective modification of protein targets in liver microsomes (7), furthermore, raloxifene and desmethylarzoxifene (DMA) COATags were employed in uterine microsomes to demonstrate bioactivation by peroxidases (21). Of the potential problems with the use of COATags in vivo or in intact cell systems, the potential cell impermeability caused by the hydrophilic biotin linker is common to all biotin conjugates (22-24).
Figure 1.
Di-quinone methide and o-quinone metabolites formed by bioactivation of raloxifene/DMA. COATag, azidoTag, and dansylTag conjugates used in this study.
Progress in chemical biology has offered new approaches for identifying the protein targets of reactive electrophiles in intact cells, using azide or alkyne tags, followed by conjugation with the complementary biotin linker (20). These techniques exploit the so-called bioorthogonal reactivity of the azide functionality to conjugate with biotin or with a fluorophore using the mild reaction conditions of the modified Staudinger ligation or click chemistry (25, 26). Wide usage has been reported in cellular protein labeling (26), activity-based protein profiling (27), and recently, identification of cellular targets of electrophiles (20). In addition to biotin conjugation, fluorophore conjugates including dansyl, BODIPY, and rhodamine have been reported (28, 29). However, in a few reports have problems such as high background response, poor enrichment of modified proteins, and cellular impermeability been highlighted (20, 22, 23, 30).
The objective is to identify cellular targets of electrophiles formed by metabolic bioactivation. In this paper, the scope and limitations of different SERM tagging strategies are explored using raloxifene or DMA as the drugs of interest; comparing preformed COATags and fluorophore tags with conjugation methodologies based upon the bioorthogonal azide group subject to either Staudinger ligation or click chemistry. Perhaps surprisingly, the original COATag methodology was seen as superior, at least in part because of the influence on chemical conjugation of the oxidative environment required for bioactivation.
MATERIALS AND METHODS
Reagents and materials
All solvents and chemicals were purchased from Fisher Scientific (Itasca, IL) or Sigma-Aldrich Chemical (St. Louis, MO) unless stated otherwise. UltraLink Immobilized Monomeric Avidin, MagnaBind streptavidin beads, HRP-streptavidin, ECL western blotting substrate, and Imperial™ protein stain kit were obtained from Pierce (Rockford, IL). Click-iT biotinylation kit for click chemistry and all 1D and 2D gel electrophorosis instruments and reagents including NuPAGE precast gels, XCell SureLock Mini-Cell, ZOOM IPGRunner System and ZOOM strips were purchased from Invitrogen (Carlsbad, CA). [1,5-Bis [2-(dimethylamino) ethyl] amino]-4,8-dihydroxyanthracene-9, 10 -dione] (DRAQ5™) was purchased from Axxora (San Diego, CA). Rat liver microsomes were prepared according to an existing procedure (6). Phosphinyl biotin for Staudinger ligation, compound 9 for the synthesis of dansyl DMA, and dansyl alkyne 11 for the study of click chemistry model reaction were prepared according to published procedures (21, 31, 32).
Instrumentation
LC-MS-MS was carried out using an Agilent (Palo Alto, CA) ion trap mass spectrometer equipped with a model 1100 HPLC system and electrospray ionization source. Proteomic MS analysis used a Dionex Ultimate 3000 HPLC system linked to a Thermo Finnigan LTQ-FT mass spectrometer equipped with a nanospray interface. 1H-NMR and 13C-NMR spectra were obtained with Bruker AVANCE 300, UltraShield 400 spectrometers.
Synthesis of Azido raloxifene and Dansyl DMA
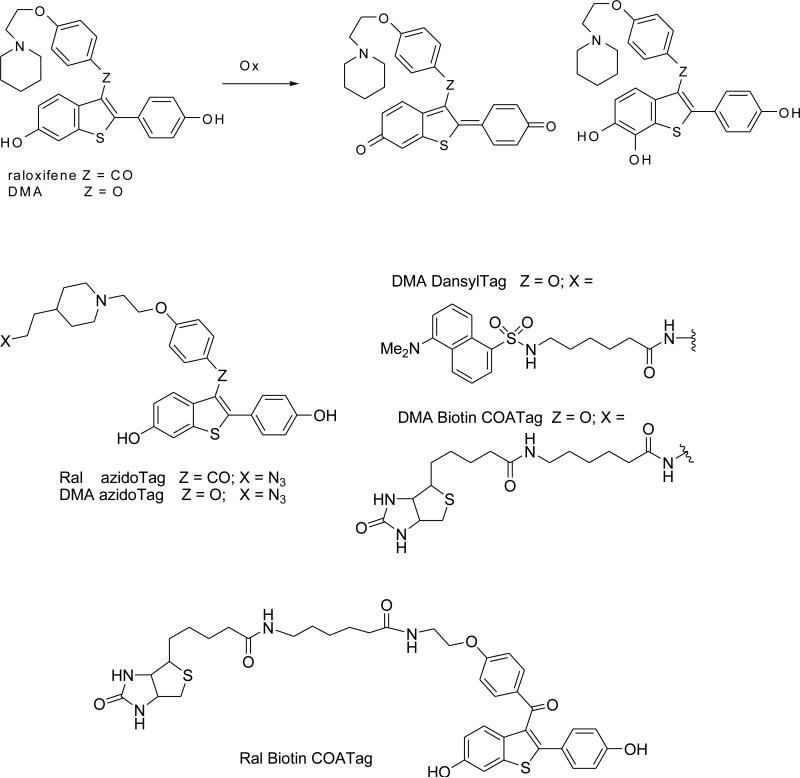
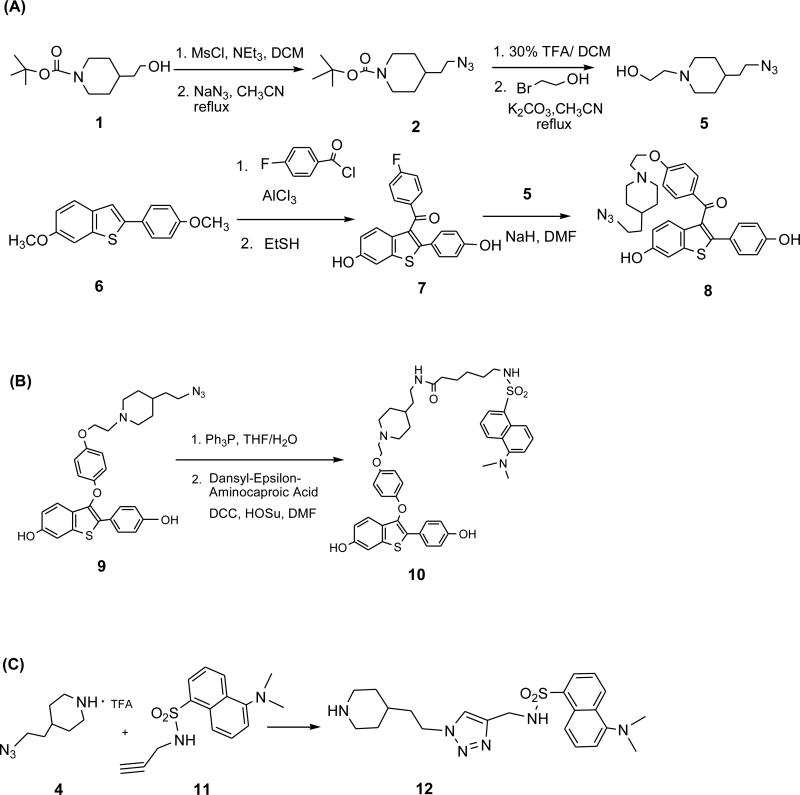
2-[1-(tert-Butoxycarbonyl)piperidin-4-yl]ethyl methanesulfonate (2)
Mesyl chloride (2.5 mL, 32.7 mmol) was added dropwise to compound 1 (5.0 g, 21.8 mmol) and NEt3 (6.0 mL, 43.6 mmol) in anhydrous THF (100 mL) cooled in an ice-water bath. The reaction mixture was stirred overnight at r.t. Most of the solvent was removed; residue was diluted with dichloromethane (200 mL); washed with brine; and, dried with anhydrous MgSO4. After filtration and concentration, the product was obtained as a white solid (5.2 g, 78%). 1H-NMR (CDCl3, 300 MHz): δ 4.28 (t, 2H, J = 6.1 Hz), 4.07-4.11 (m, 2H), 3.01(s, 3H) 2.64-2.73 (m, 2H), 1.66-1.71 (m, 5H), 1.45 (s, 9H), 1.11-1.19(m, 2H); 13C-NMR (CDCl3, 75MHz): δ 154.97, 79.57, 67.64, 43.93, 37.61, 35.79, 32.56, 31.92, 28.63.
2-(N-tert-Butoxycarbonyl-4-piperidinyl) ethyl azide (3)
Compound 2 (5.0 g, 16.3 mmol) dissolved in anhydrous acetonitrile (150 mL), NaN3 (4.3 g, 66 mmol) was added. The reaction mixture was refluxed overnight. Solvent was removed; the residue was diluted with dichloromethane (200 mL); washed with brine; and, dried with anhydrous MgSO4. After filtration and concentration, the product was obtained as a syrup with slight brown color (2.8 g, 80%). 1H-NMR (CDCl3, 300 MHz): δ 4.04-4.09 (m, 2H), 3.30 (t, 2H, J = 6.4 Hz), 2.62-2.70 (m, 2H), 1.49-1.66 (m, 5H), 1.43 (s, 9H), 1.05-1.14 (m, 2H); 13C-NMR (CDCl3, 75MHz): δ 154.91, 79.41, 48.90, 43.94, 35.34, 33.46, 31.94, 28.57.
4-(2-Azidoethyl) piperidine trifluoroacetate (4)
Compound 3 (1.2 g, 4.7 mmol) was dissolved in dichloromethane (10 mL), TFA (3.0 mL) was added and the reaction was stirred at r.t. for 1h. Most of the solvent was removed and a mixture of ether and water (v/v 1:1, 100 mL) was added. The aqueous phase was separated and washed with ether (30 mL). The aqueous solution was isolated and concentrated to give the product as white solid (890 mg, 70%). 1H-NMR (DMSO-d6, 300 MHz): δ 8.91(bs, 1H), 8.61(bs, 1H), 3.38 (t, 2H, J = 6.8 Hz), 3.22-3.26 (m, 2H), 2.75-2.92 (m, 2H), 1.77-1.81 (m, 2H), 1.55-1.71 (m, 1H), 1.44-1.51 (m, 2H), 1.20-1.36 (m,2H); 13C-NMR (DMSO-d6, 75MHz): δ 158.82, 158.40, 47.96, 43.01, 34.12, 30.57, 28.07.
[4-(2-Azidoethyl)- 1-piperidinyl] ethanol (5)
Compound 4 (1.2 g, 4.4 mmol) and 2-bromoethanol (333 μL, 4.5 mmol) were dissolved in anhydrous acetonitrile (20 mL), followed by addition of anhydrous K2CO3 (1.85 g, 13.5 mmol). The reaction mixture was refluxed overnight. After filtration and concentration, the residue was dissolved in ethyl acetate (100 mL) and washed with saturated NaCl aqueous solution. The organic phase was separated and concentrated to give a residue which was used in the subsequent step without further purification (610 mg, 68%). A small amount of sample was taken for proton NMR analysis to confirm the product identity. 1H-NMR (Acetone-d6, 400 MHz): δ 3.55 (t, 2H, J = 5.9Hz), 3.37 (t, 2H, J = 7.1Hz), 2.88-2.91 (m, 2H), 2.43 (t, 2H, J = 5.9Hz), 1.95-2.01 (m, 2H), 1.66-1.69 (m, 2H), 1.50-1.55 (m, 2H), 1.33-1.45 (m, 1H), 1.18-1.28 (m, 2H).
(4-Fluorophenyl)[6-hydroxy-2-(4-hydroxyphenyl)benzo[b]thien-3-yl] methanone (7)
Compound 6 (2.1 g, 7.7 mmol) and 4-fluorobenzoyl chloride (0.92 mL, 7.8 mmol) were dissolved in anhydrous dichloromethane (50 mL) and cooled in ice-water bath, followed by addition of AlCl3 (1.1 g). The reaction mixture was stirred at r.t. overnight. The reaction was cooled in an ice-water bath, ethanethiol (2.8 mL, 38.5 mmol) was added, and the mixture stirred at r.t. for 6 h. The reaction mixture was diluted with dichloromethane (200 mL) followed by addition of 20% HCl (5 mL) and H2O (50 mL) with vigorous stirring. The organic phase was separated and further washed carefully with saturated aqueous NaHCO3 and brine. After concentration, the product was purified by column chromatography (hexanes/ethyl acetate 1:1) to give a slightly orange solid (2.5 g, 88%). 1H-NMR (Acetone-d6, 400 MHz): δ 7.77-7.81(m, 2H), 7.55(d, 1H, J = 8.8Hz), 7.41(d, 1H, J = 2.0Hz), 7.21(d, 2H, J = 8.4Hz), 7.06 (t, 2H, J = 8.8Hz), 7.69 (dd, 1H, J = 8.8Hz, 2.0Hz), 6.73 (d, 2H, J = 8.8Hz); 13C-NMR (Acetone-d6, 100 MHz): δ 192.99, 167.49, 164.98, 158.86, 156.43, 144.30, 140.83, 135.16, 133.74, 133.28, 133.18, 131.25, 130.45, 125.46, 124.72, 116.34, 116.21, 115.99, 107.82.
[4-[2-[4-(2-Azidoethyl)-1-piperidinyl] ethoxy] phenyl][6-hydroxy-2-(4-hyroxy phenyl)benzo[b]thien-3-yl] methanone (8)
Compound 5 (590 mg, 2.9 mol) was dissolved in anhydrous DMF (5 mL); 60% NaH (120 mg) was added and the mixture stirred at r.t. for 10 min. Compound 7 (150 mg, 0.4 mmol) was added and the reaction mixture was stirred at r.t. overnight. The reaction was quenched by adding MeOH (0.5 mL), and diluted with ethyl acetate (30 mL) containing 5% MeOH, then washed with saturated aqueous NaHCO3. The organic phase was separated and concentrated to give crude product which was purified by column chromatography (CH2Cl2/MeOH 10:1) to give the product as yellow foam (55 mg, 25%). 1H-NMR (Acetone-d6, 400 MHz): δ 8.67(bs), 7.73 (d, 2H, J = 8.8Hz), 7.39-7.42 (m, 2H), 7.28 (d, 2H, J = 8.8Hz), 6.94 (dd, 1H, J = 2.4Hz, 8.8Hz), 6.89(d, 2H, J = 8.8Hz), 6.75(d, 2H, J = 8.4Hz), 4.13(t, 2H, J = 5.8Hz), 3.37(t, 2H, J = 7.2Hz), 2.93-2.96(m,2H), 2.71(t, 2H, J = 6.0Hz), 1.65-1.69(m, 2H), 1.49-1.54(m, 2H), 1.33-1.43(m,1H), 1.17-1.26(m, 2H); 13C-NMR (Acetone-d6, 100 MHz): δ 193.30, 164.04, 158.70, 156.34, 141.93, 140.81, 134.13, 132.77, 131.41, 131.30, 130.97, 125.84, 124.64, 116.43, 115.83, 115.12, 107.85, 67.27, 57.92, 54.89, 49.57, 35.96, 33.89, 32.86.
[4-[2-[4-(2-(Dansyl-epsilon-aminocapronamide) ethyl)-1-piperidinyl] ethoxy] phenyl] [6-hydroxy-2-(4-hyroxy phenyl)benzo[b]thien-3-yl] methanone (10)
Compound 9 (50 mg, 0.09 mmol) and Ph3P (42 mg, 0.15 mmol) were dissolved in the mixture of THF (1 mL) and H2O (20 μL) and stirred at r.t. overnight (solution A). Dansyl-epsilon-aminocaproic acid (93 mg, 0.17 mmol), N-hydroxyl succinimide (23 mg, 0.20 mmol), and DCC (46 mg, 0.22 mmol) were dissolved in anhydrous DMF (1 mL) and stirred at r.t. overnight (solution B). Solution A was concentrated to dryness, dissolved in DMF (300 μL) and added to solution B. The reaction mixture was stirred at r.t. overnight. After filtration, the solvent was removed under high vacuum. The residue was purified by column chromatography (CH2Cl2/MeOH 10:1 and 0.2% NH3•H2O) to give the product as a foam with slight green coloration (38 mg, 49%). 1H-NMR (Acetone-d6, 400 MHz): δ 8.54 (d, 1H, J = 8.5Hz), 8.40 (d, 1H, J = 8.7Hz), 8.20 (dd, 1H, J = 7.2Hz, 1.0Hz), 7.52-7.61 (m, 4H), 7.30 (d, 1H, J = 1.9Hz), 7.25 (d, 1H, J = 7.5Hz), 7.18 (d, 1H, J = 8.6Hz), 6.99 (t, 1H, SO2NH), 6.82-6.88 (m, 7H), 6.73(t, 1H, CONH), 4.00 (t, 2H, J = 5.8Hz), 3.16-3.21(m, 2H), 2.85-2.92(m, 10H), 2.65(t, 2H, J = 5.8Hz), 1.98-2.01(m, 2H), 1.61-1.64(m, 2H), 1.14-1.41(m, 13H); 13C-NMR (Acetone-d6, 100 MHz): δ172.94, 158.18, 156.66, 155.19, 152.71, 152.49, 140.38, 137.50, 137.36, 130.69, 130.58, 130.48, 129.61, 129.44, 128.59, 128.12, 126.61, 124.64, 124.15, 122.71, 120.41, 116.99, 116.55, 116.40, 115.99, 115.45, 108.80, 67.26, 58.33, 55.00, 45.59, 43.62, 37.26, 37.19, 36.46, 34.27, 33.95, 32.98, 26.73, 25.33.
5-(dimethylamino)-N- ((1-(2-(piperidin-4-yl) ethyl)-1H-1,2,3-triazol-4-yl)methyl)naphthalene-1-sulfonamide (12)
Azide 4 (250 μM), alkyne 11 (50 μM), CuSO4 (500 μM), Na ascorbate (1.0 M), and tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (100 μM) were stirred at room temperature for 1 h. The reaction product was identified by LC-MS and was quantified using as internal standard, 5-(dimethylamino) naphthalene-1-sulfonamide. The reaction yield in the presence of H2O2 (0, 100, 500, 1000 μM) was quantified in this way.
Rat liver microsomal incubation of azido raloxifene
A solution containing azido raloxifene (10 μM), rat liver microsomes (1.0 mg/mL), GSH (500 μM) and NADPH (1 mM) in 50 mM phosphate buffer (pH 7.4, 500 μL total volume) was incubated for 30 min at 37°C. For comparison, GSH was omitted and/or raloxifene was used as substrate. The reactions were terminated by chilling in an ice bath followed by the addition of a mixture of methanol/acetonitrile (100 μL, 1:1 v/v) and perchloric acid (50 μL/mL). The reaction mixtures were centrifuged at 13,000 rpm for 10 minutes; aliquots (80 μL) of supernatant were analyzed using LC-MS-MS. The HPLC analysis of the microsomal incubation was performed using an Agilent Eclipse XDB-C18 column (4.6 × 250 mm, 5 μm) with UV absorbance detection at 316 nm. The composition of HPLC mobile phase was a mixture of two solutions, solution A: water with 10% methanol and 0.1% formic acid; solution B: acetonitrile with 0.1% formic acid. The mobile phase consisted of a linear gradient of solution B from 10 - 30% over 20 min, 20 min gradient of solution B from 30 - 60%, and then 60 - 90% of solution B over 5 min at a flow rate of 1.0 mL/min.
ER competitive binding assays
Competitive displacement of [3H] estradiol (E2) radioligand (Amersham Biosciences, Piscataway, NJ) from full-length recombinant human hERα and hERβ (PanVera/Invitrogen, Carlsbad, CA) was assayed as described (33, 34). The reaction mixture consisted of 5 μL of compound in DMSO, 5 μL of hERα (0.5 pmol) in ER binding buffer (consisting of 10 mM Tris-HCl (pH 7.5), 10% glycerol, 2 mM dithiothrietol, 1 mg/mL BSA), 5 μL of “Hot Mix” [400 nM, prepared fresh using 95 Ci/mmol [3H] estradiol, diluted in 1:1 ethanol:ER binding buffer (NEN Life Science Products; Boston, MA)] and 85 μL of ER binding buffer. The incubation was carried out at room temperature for 2 h before 50% hydroxyapatite slurry (100 μL; HAPS) was added. The tubes were incubated on ice for 15 min with vortexing every 5 minutes. The appropriate ER wash buffer was added (1 mL), and the tubes were vortexed before centrifuging at 10,000g for 1 min. The supernatant was discarded, and this wash step was repeated three times. The HAPS pellet containing the ligand-receptor complex was resuspended in 200 μL of ethanol and transferred to scintillation vials. An additional 200 μL of ethanol was used to rinse the centrifuge tube. Cytoscint (4 mL/vial; ICN, Costa Mesa, CA) was added, and the radioactivity was counted using a Beckman LS 5801 liquid scintillation counter (Schaumburg, IL).
Induction of alkaline phosphatase in cultured Ishikawa cells
The Ishikawa cell line was provided by R. B. Hochberg (Yale University, New Haven, CT) and was maintained in Dulbecco's Modified Eagle medium (DMEM/F12) containing 1% sodium pyruvate, 1% non-essential amino acids (NEAA), 1% glutamax-1, 0.05% insulin, and 10% heat-inactivated FBS. The procedure of Liu et al. was used as described previously (33). Ishikawa cells (1.5 × 104 cells/190 μL/well) were preincubated in 96-well plates overnight in estrogen-free medium. Test samples (10 μL at varying concentrations in DMSO) were added to determine EC50 values, and the cells in a total volume of 200 μL media/well were incubated at 37 °C for 4 days. For the determination of antiestrogenic activity, 2 × 10−8 M estradiol was added to the 10 mL of media used to dilute the test samples. The induction plates were processed by washing the plates with PBS and adding 50 μL of 0.01 % Triton100 in 0.1 M Tris buffer (pH 9.8). Plates were subjected to a freeze thaw (−80 °C for at least 24 h before warming on a plate warmer to 37 °C). An aliquot (150 μL) of 1 mg/mL p-nitrophenylphosphate (phosphatase substrate) in 0.1 M Tris buffer (pH. 9.8) was added to each well. The enzyme activity was measured by reading the release of pnitrophenol at 405 nm every 15 s with a 10 s shake between readings for 16 readings using a Power Wave 200 microplate scanning spectrophotometer (Bio-Tek Instruments, Winooski, VT). The maximal slopes of the lines generated by the kinetic readings were calculated. For antiestrogenic determination, the percent induction as compared with the background induction control was calculated using: % antiestrogenic induction = 100× [1-((slopesample – slopecells)/(slopeestrogen – slopecells))].
Modification of model proteins
Model protein GST-P1 (0.3 mg/ml) or BSA (0.2 mg/ml) and tyrosinase (5,000 units/ml) were incubated with 50 μM tags (25 mM stock in DMSO) in PBS (pH 7.4), at 37 °C for 1 h. The azidoTag modified proteins were subsequently biotinylated via the modified Staudinger ligation by reaction with 500 μM phosphinyl biotin reagent (P-biotin; 50 mM stock in DMSO) at 47°C for 2 h or via click chemistry using click-it kit according to the manufacturer's instructions. Reaction mixtures were chilled on ice and 20 μL aliquots were separated in MOPS running buffer on 4-12% NuPAGE gels (Invitrogen), and run on 200 V, 1 h and detected with HRP-streptavidin (Pierce, 1:5,000) after transblotting onto PDVF membrane and blocking overnight (5% nonfat milk). For studying modified proteins in whole cell lysis, azidoTag was incubated with GST-P1 and tyrosinase as described above and unreacted tag was removed by 5,000 MWCM microcon to yield modified protein (“azido GST-P1”). The azido GST-P1 was then added to MDA-MB-231 breast cancer cell lysates (1 mg/ml) prepared with detergent or sonication, in a w/w ratio of GST-P1 to cell lysate proteins of 1:10. Ligation efficiency and background response were compared via chemiluminescence (for azidoTag and COATag) or in-gel fluorescence (for dansylTag) using 300 nm UV illumination.
Modification of proteins in rat primary hepatocytes
The cryo-preserved rat hepatocytes were thawed according to the protocol (Celsis Technologies). On the day of assay, the vials containing the rat cryopreserved hepatocytes were removed from liquid nitrogen storage. The cells were thawed by gently shaking the vials in a 37 °C water bath. The hepatocyte suspension was transferred into the prewarmed 50 mL thaw medium, the cells were centrifuged at 50 g for 3 min, and the resulting pellet was resuspended in Krebs-Hensleit buffer (KHB) at the appropriate cell density for further experiments. Viability was determined using the trypan blue exclusion assay. Tags dissolved in DMSO were added to the hepatocyte cultures to give a final concentration of 50 μM and the DMSO concentration was 0.1% (v/v). After incubation at 37 °C, 5% CO2 in the dark for 2 h, the hepatocytes were washed three times with cold PBS, collected by centrifuge, and immediately frozen at −75 °C and stored for future analysis. For the azidoTag treated hepatocytes, the whole cell proteins were harvested, lysed using 1% NP40/PBS and endogenous biotinylated proteins were pulled down using 1 mg/ml MagnaBind Streptavidin beads. The supernatants were collected and biotin conjugation was performed via Staudinger ligation or click chemistry as described above. Ligation efficiency and background response were compared via chemiluminescence (for azidoTag and COATag) or in-gel fluorescence (for dansylTag).
Avidin Affinity Concentration
The separation of biotinylated proteins from the unbiotinylated proteins was performed on an Ultralink Immobilized Monomeric Avidin column according to the manufacturer's instructions. Briefly, a monovalent avidin column was washed sequentially with 12 mL PBS (pH 7.2), 10 mL biotin buffer (2 mM D-biotin in PBS) to block any nonreversible biotin binding sites, 10 mL glycine buffer (0.1 M glycine, pH 2.8), and finally 12 mL PBS again. The protein mixture (1 mL) was incubated on the column at room temperature for 1 h and then washed extensively with 18 mL PBS to remove unbiotinylated proteins, followed by 12 mL biotin buffer to elute biotinylated proteins, and finally 10 mL glycine buffer to regenerate the column. The biotinylated protein eluate was concentrated to 250 uL by ultrafiltration through a 5,000 MWCM centricon at 4,000g for 1 h and aliquots were stored at −75 °C until analysis.
Confocal microscopy
ER positive MCF-7 cells were grown (1×106 cells/ml) on each of eight wells on a sterile Nunc™ chambered cover glass and incubated for 48 h at 37 °C with 5% CO2 in phenol red free RPMI medium supplemented with 10% stripped FBS. MCF-7 cells were treated with 1 μM dansyl DMA for 18 hours. Before imaging, cells were rinsed two times with PBS to remove the unincorporated dye and 1 μM of DRAQ5™ stain was added to the cells to see nuclear staining. In separate experiments, MCF-7 cells were treated with 1 μM dansyl DMA and confocal imaging was performed in 1 min intervals to see the time dependent localization of dansyl DMA. Imaging was performed with a Zeiss LSM510 laser-scanning confocal microscope with the detector gain adjusted to eliminate the background autofluorescence. The fluorescence signal from dansyl DMA was monitored with a 345-nm UV laser and a 420-nm band pass filter. DRAQ5™ nuclear staining signal was monitored with 568-nm argon/krypton laser and a 650-nm line pass filter. A x63 (1.2 numerical aperture) water immersion objective was used for all experiments. Images were analyzed using the analysis tool provided in Zeiss biophysical software package.
Two-dimensional PAGE
2DGE was performed at room temperature using ZOOM stripes and ZOOM IPG Runner System according to the manufacturer's instructions with minor modifications. Total 140 μL loading sample contains 10 μL cell lysate enriched using the Monomeric Avidin column (400 ng / μL), 128 μL ZOOM 2D protein solubilizer 1, 1.4 μL DTT (1M), 0.7 μL ZOOM Carrier ampholytes (pH 3-10), trace bromophenol blue and deionized water. The mixture was loaded into ZOOM IPGRunner cassettes and rehydrated in ZOOM pH 3-10 NL strips at room temperature. Isoelectric focusing was conducted using the following voltage gradient: 175 V for 15 mins, 175-2,000V ramp for 45 mins, and 2,000V for 30 mins. After focusing, the strips were equilibrated with 10 ml NuPAGE LDS sample buffer with sample reducing agent for 15 min, followed by incubation for 15 min with the same sample buffer supplemented with iodoacetamide (125 mM). In the sencond dimension, focused strips were then applied to the ZOOM 4-20% IPG gel, and the proteins were further separated by SDS-PAGE at 200 V for 60 mins. Proteins were transferred to PVDF, probed with HRP-streptavidin and visualized by chemiluminescence. In all separation experiments, broad range 2D protein markers (Pierce) and dual color precision plus protein markers (Bio-Rad) were used for calibration of the first and second dimension gels, respectively. The figures represented results from multiple experiments.
Mass Spectrometic Anaylsis of Tryptic Digests
The concentrated biotinylated proteins from rat hepatocytes were separated by 1D gel electrophoresis as described above. The gel was stained by Coomassie blue, excised, and digested with trypsin as described previously (23). The peptide mixture (30 μL, 10 μg enriched proteins) was injected onto a reversed phase column (75 μm × 150 mm Zorbax SB300 C-18, Agilent Technologies) connected to a Dionex Ultimate 3000 HPLC system and a Thermo Finnigan LTQ-FT mass spectrometer equipped with a nanospray interface. The samples were chromatographed using a binary solvent system consisting of A: 0.1% formic acid and 5% acetonitrile and B: 0.1% formic acid and 95% acetonitrile at a flow rate of 200 nL/min. A gradient was run from 15% B to 55%B over 60 minutes. The mass spectrometer was operated in positive ion mode with the trap set to data dependent MS/MS acquisition mode. The instrument was set to complete a mass scan from 400-1800 Daltons in one second. Peaks eluting from the LC column that had ions above 1,000 arbitrary intensity units triggered the ion trap to isolate the ion and perform an MS/MS experiment scan after the MS full scan. After every MS scan in the ion trap, ions were then transferred to the isolation cell of the 7T magnet in the FT where a high resolution (up to 500,000 at m/z 400) and high mass accuracy (1 ppm) mass spectrum of the precursor ions was obtained. Data analysis was carried out using the Mascot software platform (Matrix Science, UK) Bioworks 3.2 (Thermo Instruments, CA) and the X!tandem software platform. The library searching and de-novo interpretation identified the detected proteins from the individual peptides. The results for all proteins detected were collected and listed by protein name, detected peptide sequence(s), and search score. The reports were exported to an XML file for inclusion in a result database that was finally analyzed using Scaffold 2.1.0. Identifications were accepted only for proteins with 3 unique peptide identifications matching high quality MS/MS spectra.
RESULTS
Synthesis of SERM tags and labeling reagents
Azido raloxifene is an analog of raloxifene with close structural similarity. It was synthesized by a nucleophilic aromatic substitution reaction between intermediates 5 and 7 (Figure 2A). To prepare 5, compound 4 was obtained according to the literature with minor modifications (35); the subsequent N-alkylation of 4 gave the desired product. Intermediate 7 was conveniently synthesized by a one-pot, two-step procedure that has been used for the preparation of raloxifene (36). The synthesis of azido DMA has been reported previously (21); in this study, the azide group was reduced to an amino group prior to acylation to give dansyl DMA (Figure 2B).
Figure 2.
Synthesis of Ral AzidoTag (A), DMA DansylTag (B) and click ligation between model azide 4 and dansyl tagged alkyne 11 (C).
ER ligand binding and antiestrogenic activity
Radioligand displacement competitive binding to full length recombinant ERα and ERβ was assayed to determine the ability of tags to retain the molecular recognition characteristics of the parent molecule. For the DMA tags and parent drugs, nanomolar binding affinity was maintained despite the addition of substantial molecular baggage to the 3-position piperidine side chain (Table 1). Thus, the tags retain high affinity for binding to ERα relative to the parent molecule, DMA, and to the drug molecule, arzoxifene. Expectedly, given the greater steric requirement for binding to ERβ, the tags performed somewhat less favorably compared to DMA, but still retained nanomolar binding affinity.
Table 1.
ER binding data and antiestrogenic activity in cell culture for COATag, dansylTag, azidoTag compared to parent SERMsa
| Compounds | ER IC50 (nM)b |
Ishikawa cell IC50 (nM)c |
|
|---|---|---|---|
| ER-α | ER-β | ER-α | |
| DMA COATag | 9.37 ± 2.0 | 24.5 ± 4.7 | 6.7 ± 3.2 |
| Ral COATag | 37.7 ± 6.5 | n.s.d | 17.8 ± 2.5 |
| DMA DansylTag | 28.7± 4.0 | 49.2 ± 9.9 | 22.5 ± 1.9 |
| DMA AzidoTag | 14.8 ± 0.9 | 23.5 ± 4.4 | 6.5 ± 2.6 |
| Ral AzidoTag | 27.3 ± 7.6 | n.s. | 4.7 ± 0.5 |
| DMA | 16.5 ± 0.7 | 19.6± 3.6 | 0.4 ± 0.1 |
| Arzoxifene | 21.5 ± 6.5 | 66.3 ± 3.1 | 1.3 ± 0.3 |
| Raloxifene | 24.1 ± 1.9 | 399 ± 26 | 3.4 ± 0.4 |
Data show mean ± S.D. for at least triplicate measurements.
Competitive binding assay with recombinant ER.
Estrogen antagonist activity.
not studied.
The Ishikawa endometrial cancer cell line, used as a reporter for ERα-mediated classical estrogenic activity, also reveals antiestrogenic activity in intact cells in the presence of competing estradiol. All tags retained antiestrogenic activity in Ishikawa cells (Table 1) and were devoid of estrogenic activity (data not shown). Complete correspondence of isolated ER binding with cellular activity is not guaranteed, because classical antiestrogenic activity is thought to require ligand binding to the ER, receptor dimerization, and translocation to the nucleus to form a non-productive transcriptional complex at the ER response element (ERE). Therefore, in cell culture for an antiestrogen to retain the potency indicated from simple binding assays, it must freely enter the cytoplasm, and as part of a liganded ER complex translocate through the nuclear membrane, permitting binding at the ERE. The data suggest that all the SERM tags under study manifest this ability in Ishikawa cells.
Having determined that the synthetic probes retain ER binding capacity in cell culture, protein modification by these probes was examined.
Model protein modification by SERMs with COATag, azidoTag, and dansylTag
To compare modification by SERMs of isolated proteins, using different tag and conjugation strategies, GST-P1 and BSA were chosen as model proteins. GST-P1 is a key detoxification enzyme in phase II metabolism which has been reported previously to be a target for: the raloxifene COATag; SERM quinoid metabolites; and, o-quinones formed by oxidation of estrogens. GST-P1 contains four cysteine residues (Cys-14, Cys-47, Cys-101, and Cys-169); of these, Cys-47 and Cys-101 are highly susceptible to modification by quinoids derived from SERMs and estrogens (7, 9, 37). Alternately, BSA is a widely used model for study of protein modification by drugs (38). In model protein studies, tyrosinase was used to bioactivate the benzothiophene SERM moiety, since tyrosinase is not itself susceptible to covalent modification (7).
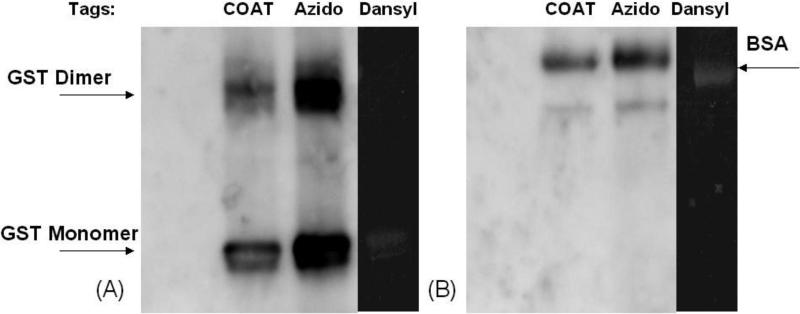
COATag, azidoTag, or dansylTag (50 μM) were incubated with either BSA or GST-P1 in the presence of tyrosinase, with solvent (DMSO) as vehicle control. The azidoTag modified proteins were subsequently labeled with biotin via the modified Staudinger ligation by reaction with phosphinyl biotin (P-biotin; 500 μM) at 37oC for 2 h. Protein modification was detected by western blots using HRP-streptavidin (for COATag and azidoTag) or in-gel fluoroscence (for dansylTag). Both GST-P1 and BSA were modified after incubation with tags for 1 h (Figure 3). GST-P1 readily oxidizes to form an intermolecular disulfide at Cys-47, leaving Cys-101 available for covalent modification (Figure 3A). Modification of GST-P1 dimer was not detected in the dansylTag incubation (Figure 3A). Densitometry indicated that the azidoTag appeared to give more protein modification than the COATag after incubation with GST-P1 for 1 h (Figure 3A). Explanations could rest on lower reactivity of the COATag or on decreased tyrosinase oxidation rates for the COATag. In PBS, BSA was modified by all three tags (Figure 3B).
Figure 3.
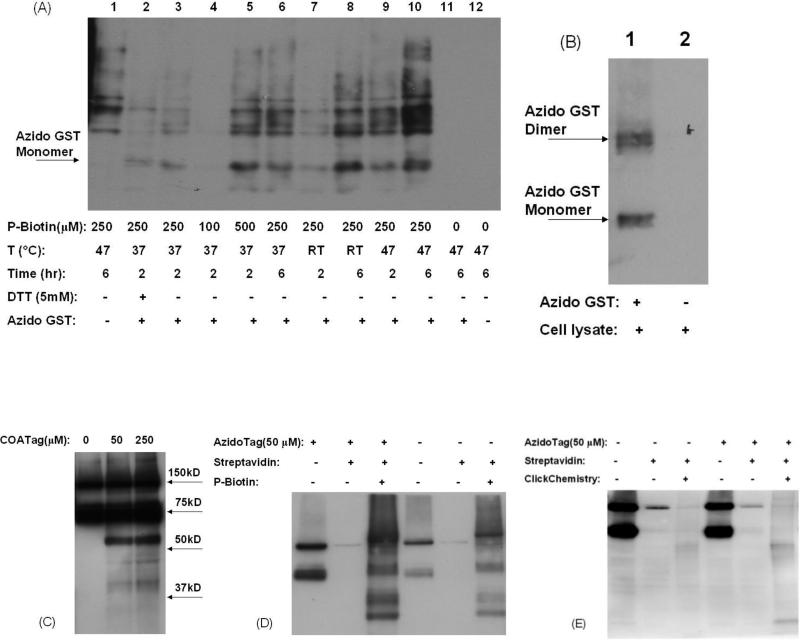
Modification of model proteins GST-P1 (A) and BSA (B) by COATag, azidoTag, and dansylTag. GST-P1 (0.3 mg/ml) and BSA (0.2 mg/ml) were incubated with tags (50 μM) and tyrosinase (5000 units/ml) in PBS (100 μL, pH 7.4) at 37 °C for 1 h. AzidoTag was further reacted with P-biotin (500 μM) at 47 °C for 2 h. Biotin tagged protein was visualized by western blots using HRP-streptavidin; dansylTag was visualized by ingel fluorescence (300 nm UV).
Sensitivity comparison using a model protein
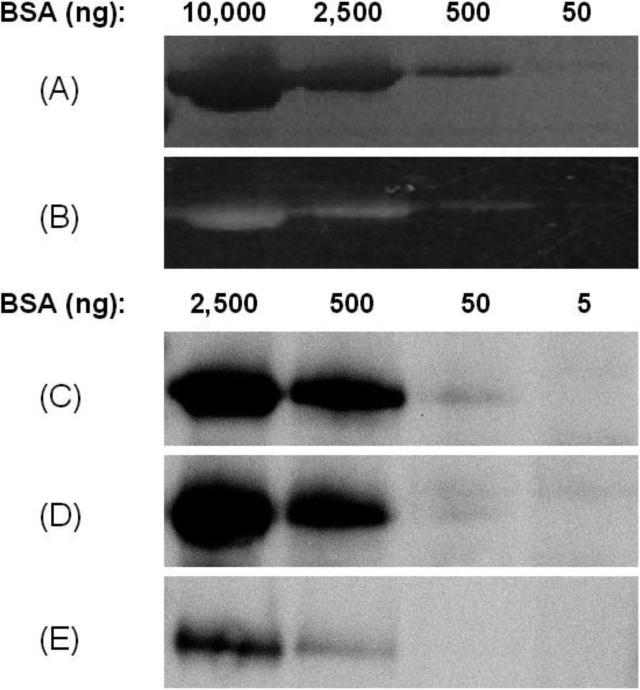
From the study of the two model proteins, using tyrosinase as an oxidizing system, it was seen that protein modification can be observed using all three methodologies. However, it is critical to be able to detect low abundance proteins both in whole cell lysates and in future experiments with tissues. Therefore, the sensitivity of the four conjugation strategies was tested using BSA as model protein (avoiding the ambiguity resulting from GST-P1 dimerization). Varied amounts of BSA (5 – 10,000 ng) were incubated with tags, followed by work-up. Electrophoresis on 1D gels visualized by immunoassay (for biotin-linked tags), in gel fluorosence (for dansylTag), or Coomassie blue staining (for dansylTag) was used to quantify the relative response and sensitivity to the four methodologies: (1) COATag; (2) azidoTag/Staudinger; (3) azidoTag/click; (4) dansylTag.
The results showed that both COATag and azidoTag/Staudinger ligation methodologies were able to detect BSA modified in incubations with only 5 ng protein and quantify protein modification in incubations with 50 ng protein (Figure 4C and 4D). AzidoTag/click chemistry and dansylTag methodologies were 10 fold less sensitive (Figure 4E and 4B), and were comparable in sensitivity to Coomasie blue staining (Figure 4A).
Figure 4.
Comparison of sensitivity of different tag methodologies towards BSA (5 -10,000 ng per lane) after bioactivation of tags (50 μM) by tyrosinase (5,000 units/ml) in PBS (pH 7.4) at 37 °C for 1 h. AzidoTag was further conjugated with biotin via modified Staudinger ligation or click chemistry. The sensitivity was measured by western blots using HRP-streptavidin (for COATag and azidoTag) or by in-gel fluorescence (for dansylTag). A) Coomassie blue staining of BSA in the presence of dansylTag showing comparative sensitivity of protein dye staining. B) In-gel fluorescence of BSA modified by dansylTag. C-E) western blots of BSA modified by COATag (C) and azidoTag/Staudinger (D) and azidoTag/click methodology (E).
The ability of oxidant systems to perturb the efficiency of conjugation by click chemistry was studied by the use of the click ligation between a model azide (4) and a dansyl tagged alkyne (11) (Figure 2B). As might be expected in the presence of H2O2 (0, 100, 500, 1000 μM), the efficiency of ligation was attenuated: 100%; 93%; 82%; and, 52% respectively. A solution of H2O2 does not fully mirror the oxidation potential of the catalytic oxidant systems under study, but the observations are in accord with the observed lower sensitivity of the azidoTag/click methodology. Although additional work-up procedures could be used to mitigate against the oxidation of cuprous ion in click ligation, the addition of an extra step in the conjugation procedure is a disadvantage.
Application of modified Staudinger ligation and click chemistry in cell lysates
The conditions used for modified Staudinger ligation of azides with phosphinyl biotin (P-biotin) after cellular incubation are widely variable in the literature (29, 31, 39, 40). Accordingly, time, temperature, and reactant concentrations were varied, in addition to variation of the common reagents used in cell lysis and SDS-PAGE. The model system selected to test the efficiency of each method in detecting modified protein in cell lysates was modified GST-P1 in MDA-MB-231 breast cancer cell lysates. AzidoTag was incubated with recombinant GST-P1 in the presence of tyrosinase and unreacted tag was removed by filtration to yield a protein mixture enriched in modified protein (“azido-GST”). Azido-GST was added to cell lysates, which contain no detectable endogenous biotinylated proteins (Figure 5A, lane 12), in a w/w ratio of GST-P1 to cell lysate proteins of 1:10. Modified Staudinger ligation and click chemistry were subsequently performed in the azido-GST cell lysate system and ligation efficiency and background response were compared using western blot.
Figure 5.
Comparison of protein tagging methodologies in rat primary hepatocytes. UPPER: Application of modified Staudinger Ligation and click chemistry for detecting modified protein targets in cell lysates. Biotin conjugation via modified Staudinger ligation or click chemistry is described in Materials and Methods. A). modified Staudinger ligation. Lane 1: cell lysate without azido GST-P1. Lane 2-11: cell lysate with azido GST-P1 under varied conditions. Lane 12: cell lysate only. B). Click chemistry. Lane 1: cell lysate + azido GST-P1. Lane 2: cell lysate only. LOWER: Comparison of protein tagging methodologies in rat primary hepatocytes. Cells were incubated with COATag (C) or azidoTag (D-E) (50 μM) in KHB buffer for 2 h and then labeled via modified Staudinger ligation (D) or click chemistry (E).
The results showed that many variables influenced the azidoTag/Staudinger methodology, but most prominent was the high background response in the control experiment in the absence of azidoTag and presence of the P-Biotin (Figure 5A; lane 1). The concentration of P-biotin was critical for Staudinger ligation, which poorly occurred at lower P-biotin concentrations (Figure 5A; lane 4). Thus, reducing P-biotin concentration was not useful in reducing the non-specific background. Increasing P-biotin concentration, incubation time and temperature all enhanced ligation, but concomitantly increased the background signal (Figure 5A, Lane 5-10). The reducing reagent DTT, commonly used to maintain reduced thiols, did not affect Staudinger ligation (Figure 5A, lane 2, 3), but detergents (i.e. NP40) interfered with the reaction (data not shown), consistent with previous reports (39). Even under optimal conditions, non-specific labeling was observed (Figure 5A, lane 5 and 8). Comparison reaction systems without azido-GST or P-biotin (Figure 5A, lane 1, and 11) indicated that these non-specific bands resulted from the reaction of proteins with P-biotin, which contains a benzoate ester moiety.
The azidoTag/click methodology was compared in the same model system consisting of cell lysate containing azido-GST, but in this case, using a commercial click labeling kit from Invitrogen requiring 1 h reaction at room temperature. The relative specificity of the azidoTag/click methodology was immediately apparent; both modified GST-P1 monomer and dimer were readily detected (Figure 5B, lane 1). Although modified GST-P1 dimer was also almost certainly present in the western blots resulting from use of the azidoTag/Staudinger methodology, this band was obfuscated by the high non-specific background of Staudinger ligation (Figure 5A, Lane 2-10). The azidoTag/click methodology was specific and sensitive in this model system; no biotinylated proteins were detectable in the control incubation of unmodified GST-P1 in cell lysates after reaction with the click chemistry reagents (Figure 5B, lane 2).
Application of COATag and azidoTag in hepatocytes
The next step from assay of model proteins in cell lysates was to compare methodologies in intact cells. Hepatocytes contain high levels of metabolic enzymes that can oxidatively bioactivate SERMs to electrophilic quinoids. Primary rat hepatocytes were treated with either solvent control, or the various tags (50 - 250 μM). The concentration and incubation time were optimized to give extensive protein modification in the absence of cell toxicity. The COATag was able to modify cellular proteins as shown by the concentration dependence of the immunoassay response, suggesting that the hepatocyte cell membrane is permeable to the COATag, and confirming the high metabolic capacity of hepatocytes (Figure 5C). Major modification bands were observed at about 60 kDa, 45 kDa, and 40 kDa, but low abundance proteins modified by SERM quinoids ranged from 15 kDa to 150 kDa (Figure 5C, lane 2 and 3). A drawback in the COATag methodology is seen in the obfuscation of modification of proteins at 75 kDa and 150 kDa because of large amounts of endogenous biotinylated proteins (Figure 5C).
In the azidoTag treated hepatocytes, proteins were harvested and endogenous biotinylated proteins were pulled down using MagnaBind Streptavidin beads (1 mg/ml). The supernatants were subsequently reacted with modified Staudinger ligation or click chemistry reagents. The capability to remove endogenous biotinylated proteins efficiently using streptavidin beads before conjugation of the tag using either Staudinger or click reactions represents an advantage of the azidoTag methodologies (Figures 5D and 5E). However, the azidoTag/Staudinger methodology again suffered from high non-specific background response (Figure 5D, lane 6). Varying of the concentration of P-biotin was not effective in attenuating background (Figure 5A and Supporting Information). Using the azidoTag/click methodology, very minimal non-specific background signal was observed, however, the ligation efficiency was lower than the COATag as reflected by the relatively weak visualization of only two bands of modified proteins (Figure 5E). The high non-specific labeling associated with modified Staudinger ligation and low ligation efficiency of the azidoTag/click chemistry in hepatocytes, led to abandonment of these methodologies.
Application of dansyl tag in breast cancer cells and hepatocytes
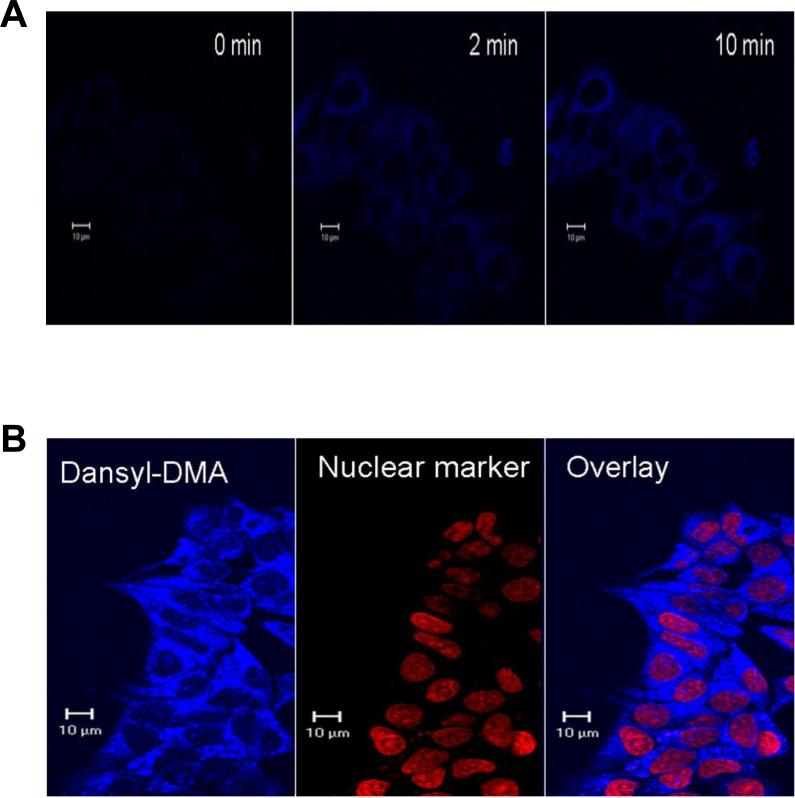
A major incentive for exploration beyond the COATag methodology was the anticipated poor cell membrane permeability of the large COATag, which must contain a relatively polar linker - biotin moiety to allow solubility at high micromolar concentrations. However, the hepatocyte experiments, where extensive protein modification was observed, suggest that the COATag is not inherently membrane impermeable (Figure 5C). A remote possibility was that the COATag was simply modifying extracellular proteins expressed by hepatocytes, but this was ruled out by study of cell culture supernatants prior to cell lysis (Supporting Information). The dansylTag (λex337nm, λem492nm), which may share similar size and hyphophilicity with biotin tags, was studied in both hepatocytes and in ERα positive MCF-7 breast cancer cells, in order to visualize cell permeability using confocal fluorescence microscopy. In MCF-7 cells, the dansylTag rapidly penetrated the outer cell membrane in a time dependent manner leading to relatively rapid accumulation in the cytoplasm (Figure 6A), but penetrated the nuclear membrane more slowly (after approximately 18 h incubation; Figure 6B). In hepatocytes, localization in the cytoplasm was also observed by fluorescence microscopy (data not shown), however, bands corresponding to modified proteins in hepatocyte lysates treated with dansylTag were not detectable by in-gel fluorescence (Supporting Information), indicating that the dansylTag methodology is not sufficiently sensitive for proteomic studies in intact cells.
Figure 6.
Localization of dansyl DMA in ERα positive MCF-7 cells. (A) Time dependent localization of dansyl DMA (1 μM) in MCF-7 cells. (B) Localization of dansyl DMA (1 μM) in MCF-7 cells after 18 h treatment. The nuclear marker staining defines the location of the nucleus and the overlay indicates the co-localization of dansyl DMA and the nuclear marker.
Two-dimensional resolution of COATagged proteins
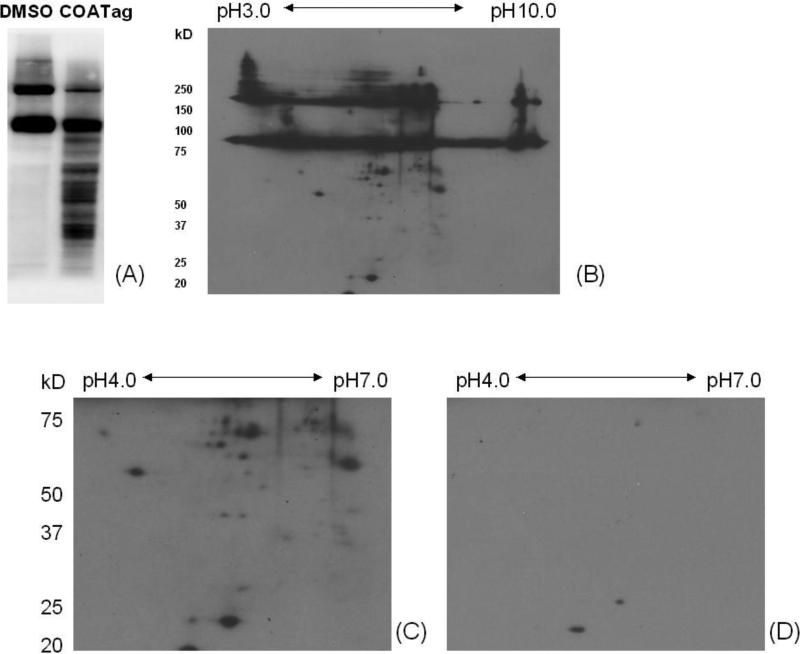
As seen above, a drawback to the use of the COATag methodology is obfuscation of modified proteins by endogenous biotinylated proteins. The use of 2D gels to overcome this obstacle was explored in hepatocytes treated with COATag (50 μM). After incubation, cells were lysed, enriched, desalted and concentrated; approximately 1 million hepatocytes yielded about 1 mg total protein; and, after enrichment and purification, the yield was approximately 40 μg. Enriched proteins (4 μg aliquots) were separated by electrophoresis on 1D and 2D gels, followed by transfer and western blot using HRP-streptavidin. 2D protein markers and solvent treated hepatocytes provided controls and calibration.
Endogenous biotinylated proteins were observed at high concentration in hepatocytes (Figure 7A; lane 1). The COATag clearly modified a large number of lower molecular weight proteins that could be readily isolated by in gel digest and subsequent analysis and identification by mass spectroscopy (Figure 7A; lane 2), but several spots in the 2D gel were localized close to the bands of endogenous biotinylated proteins (Figure 7B), suggesting that proteomic methods would be required for definitive classification of endogenous and COATagged proteins. A large fraction of the COATag modified proteins were detected at pI 4-7, MW 20-75 kDa; these proteins were studied further by 2D electrophoresis and the comparison between COATag and vehicle treated hepatocytes revealed COATagged and endogenous biotinylated proteins. Approximately 30 spots were observed reproducibly in the COATag treated hepatocytes, in contrast to vehicle treated cells (Figures 7C, D).
Figure 7.
Two-dimensional resolution of COATag modified cellular proteins. Hepatocytes were incubated with COATag (50 μM) or DMSO in KHB buffer at 37 °C for 2 h. Approximately 4 μg of enriched modified proteins were analyzed using 1D (A) and 2D (B) gel electrophoresis and visualized using HRP-streptavidin. An expanded section of the 2D analysis of COATag modified proteins (C) is compared with the similar vehicle control experiment (D).
Mass spectroscopic identification
Full proteomic analysis is not reported herein, however, in order to validate the use of the COATag methodology, several liver proteins were identified by MS analysis of in-gel digests. In addition to three modified proteins previously reported from rat liver microsomes, cytosolic glucose regulated protein, protein disulfide isomerase, and glutathione S-transferase, certain postulated target proteins such as peroxiredoxin-1 and heat shock proteins were also identified (Table 2).
Table 2.
Percent coverage of select proteins modified by Ral COATag in rat hepatocytesa
| |
|
Ral COATagb |
|||
|---|---|---|---|---|---|
| Protein | Accession# | Ctrl. | Rep. 1 | Rep. 2 | Rep. 3 |
| 78 kDa glucose-regulated protein precursor | IPI00206624 | - | 30.00 | 30.90 | 36.70 |
| Protein disulfide-isomerase precursor | IPI00198887 | - | 14.10 | 7.27 | 12.40 |
| Microsomal glutathione S-transferase 1 | IPI00230889 | - | - | 26.50 | |
| Glutathione S-transferase kappa 1 | IPI00327079 | - | - | - | 27.40 |
| Predicted Stress-70 protein, mitochondrial precursor | IPI00363265 | - | 9.72 | 25.90 | 11.60 |
| Heat shock cognate 71 kDa protein | IPI00208205 | - | 12.70 | 10.50 | - |
| Peroxiredoxin-1 | IPI00211779 | - | 29.60 | 24.60 | - |
This table presents select proteins identified from three independent experiments, in which rat primary hepatocytes were treated with raloxifene COATag (50 μM) or vehicle control.
The number value indicates the percent amino acid coverage of each protein identified by peptide fragmentation using LC-MS/MS analysis.
Metabolism of azidoTags
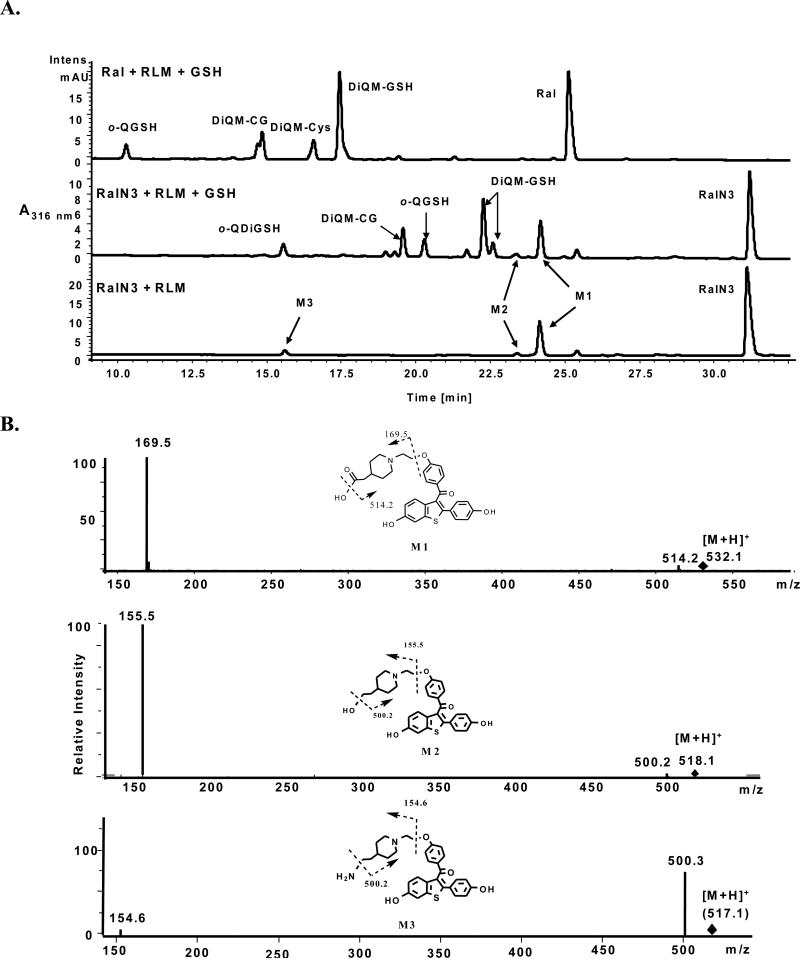
Several successful examples of the use of azidoTags have appeared in the literature, in particular using click chemistry for conjugation to various chemical ”handles” including biotin. The reactions of azide and alkynyl functionalities are generally viewed as bioorthogonal and therefore inert to reactions in biological systems, which might confound analysis. However, since the purpose of the COATag methodology, and by extension the azidoTag, is to study electrophilic metabolites generated by oxidative bioactivation, examination of the metabolic stability of the azidoTag was warranted. As has been previously reported, raloxifene in liver microsomes is oxidized substantially and predominantly to the diquinone methide that is efficiently trapped by GSH. In this oxidative environment dominated by P450 3A4, the o-quinone and its GSH conjugate are minor products. The raloxifene azidoTag was observed to yield metabolites that were observed in the presence or absence of GSH (Figure 8A). These metabolites were further interrogated by MS-MS and proved to be oxidative metabolites resulting from reaction and loss of the “bioorthoganol” azide moiety (Figure 8B), In addition to metabolism of the aliphatic azide, a spectrum of GSH conjugates of quinoid metabolites was observed qualitatively comparable to those observed for raloxifene itself (Figure 8A). Thus, the azidoTag was oxidized to quinoids, largely mirroring the parent drug, supporting its use as a probe for target proteins. However, the loss of the azido functionality in this same oxidative bioactivation process will inevitably decrease the sensitivity of the azidoTag methodology.
Figure 8.
Metabolism of raloxifene azidoTag compared to raloxifene in induced rat liver microsomes. A) Chromatograms of raloxifene and raloxifene azidoTag in microsomal incubations with and without GSH present to trap quinoid species. B) MS-MS spectra and identification of azido raloxifene metabolites M1, M2, and M3 corresponding to annotations in Figure 8A. MS-MS spectra of GSH conjugates are presented in supporting information.
Discussion
The COATag methodology was developed to study and identify the protein targets of electrophilic oxidative metabolites of small molecules (7). The proof-of-principle study focused on a clinical drug, the benzothiophene SERM raloxifene, which is oxidatively bioactivated to electrophilic quinoid metabolites (4, 6). The COATag technique was effective in identifying protein targets in microsomal incubations; including the abundant cytoplasmic disulfide isomerase proteins, chaperone proteins, and a detoxifying GST enzyme (7). In combination with protein-specific antibodies and mass spectroscopy, the COATag technique is transferable to other small molecules subject to bioactivation to electrophiles, including endogenous molecules such as estrogens and catecholamines. The technique has also been applied to assessment of the extent of protein modification and dependence on cofactors in microsomes (21). In order to proceed to identification of protein targets in intact cell incubations, the use of azidoTags was explored in anticipation of increased cell permeability relative to the larger more hydrophilic COATag.
The requirements for an effective tagging methodology are that the tagged small molecule must retain characteristics of the parent molecule including cell permeability and receptor binding characteristics. SERMs represent attractive subjects for study because side chain modification in COATags, azidoTags, and dansylTags does not significantly perturb binding to receptors that recognize the parent molecule; as demonstrated by the measured ER binding affinities and antiestrogenic activity in cell culture (Table 1). In contrast to tagging methodologies for inherently electrophilic biomolecules such as 9-HNE, the COATags and related tags studied herein require bioactivation to generate the electrophilic metabolites that target protein nucleophiles. Therefore, additionally, the tag must retain a similar metabolic profile to the parent molecule. Beyond these characteristics, tagging methodologies should provide sensitivity and specificity.
The use of biotin in affinity tags and the high affinity binding of biotin by immobilized avidin provides effective systems for protein purification and enrichment. Biotin affinity tags and comparable fluorophore affinity tags have both been subject to widespread study and usage (23), however, it was not certain that tagging with biotin or fluorophore plus the required linker moieties would limit cell permeability and thus limit use of these tagging approaches. An alternative approach is provided by the recent development of bioorthogonal reporter strategies in which azide-bearing molecules are labeled with biotin or a fluorophore via modified Staudinger ligation or click chemistry (26, 27, 30, 31). Azide is a small functional group assumed not to interfere with the properties of the parent molecule including receptor binding, metabolism, and cell permeability, and is assumed to be biologically inert. In addition, a two-step labeling strategy offers multiple methods for downstream protein detection and purification.
Four tagging methodologies were compared as depicted in Figure 9: two 1-step methods: Method 1) COATag; and, Method 2) dansylTag; and, two 2-step methods: Method 3) azidoTag/Staudinger; and, Method 4) azidoTag/click. The model systems employed were purified proteins (BSA and GST-P1), cell culture lysate, and primary cell cultures of rat liver hepatocytes. Both BSA, a highly abundant protein, and GST-P1, a known target of quinoid metabolites (7), contain thiol groups which are susceptible to modification by SERM quinoids formed from bioactivation by oxidase/oxygenase enzymes such as tyrosinase. Hepatocytes are major cell mediators of metabolism in vivo including SERM metabolism and have been implicated in liver carcinogenesis caused by the archetypal SERM, tamoxifen (41). Hence, hepatocytes represent both cells rich in metabolic enzymes and cellular targets for SERM quinoid electrophiles. Previously, protein modification by COATags was observed to be time and concentration dependent (7). In this study, concentrations of tags employed (50 μM) were comparable with similar studies using tagged electrophiles. This concentration is suprapharmacological, however, the purpose of these studies is to identify cellular target proteins. It should also be noted that exposure time (1 h for model proteins and 2 h for hepatocytes) is much shorter than the long-term clinical exposure typical for SERMs.
Figure 9.
Schematic depiction of the four methodologies explored in this work culminating in proteomic analysis.
In this study, the azidoTag/Staudinger methodology was found to offer high sensitivity but low specificity (Table 3). The modified Staudinger ligation reaction between triarylphosphine and azide functionalities produces an aza-ylide that is further stabilized by an electrophilic methyl ester via intramolecular cyclization (31). First introduced by Bertozzi, this ligation has been utilized for tagging and labeling surface proteins, expressed proteins, and engineered proteins in cells (29, 31, 42). The reaction is rapid in a physiological environment and is reported as compatible with living systems (25, 43). Correspondingly, in this study, the azidoTag/Staudinger methodology efficiently labeled protein targets and detected nanogram level of modified proteins in vitro (Figure 3 and 4). Compared to the alternative methodologies studied, this methodology was highly sensitive.
Table 3.
Comparison of tagging methodologiesa
| Tagging methodology | Tag is ER ligand & active in cells? | Sensitivity | Specificity |
|---|---|---|---|
| Method 1 (COATag) | Yes | higher | highb |
| Method 2 (dansylTag) | Yes | lower | low |
| Method 3 (azidoTag) | Yes | higher | N/Ac |
| Method 4 (azidoTag) | Yes | lower | high |
See figure 9 for depiction of details of each method.
With the exception of actually biotinylated proteins.
Modified proteins not detectable from hepatocyte incubations.
Using purified proteins, the azidoTag/Staudinger methodology appeared superior, however, when applied to the more complex cell lysates and whole cell treatments, a high background from modified Staudinger ligation was observed (Figure 5A). Attempts to attenuate the nonspecific background reactivity, including optimization of Staudinger reaction conditions and reagents, failed (Figure 5A). Control experiments in cell lysates showed that the non-specific background is linked to the second labeling reagent, P-biotin (Figure 5A), for which there is precedent in the literature (20). The non-specific labeling of abundant proteins is possibly associated with reaction of the methyl benzoate group of P-biotin with protein amino groups. The non-specific background obscures the detection of low-abundance protein targets of bioactivated metabolites limiting use of this method for analysis of protein modification by electrophilic metabolites in cells.
The azidoTag/click methodology was highly specific, but suffered from low sensitivity. Click chemistry was first reported for activity-based protein profiling by Cravatt, in which the [2+3] cycloaddition of an alkyne and azide functional group was catalyzed by cuprous ion (26, 30). In comparison to labeling via Staudinger ligation, click chemistry was considered to produce lower background (20). However, in the present study click chemistry both in model proteins and in cells was observed to have ligation efficiency much less than that of Staudinger ligation (Figure 4 and 5), and the methodology was observed to have lower detection sensitivity than either the COATag or azidoTag/Staudinger methodologies (Figure 4 and 5). Several studies have compared the efficiency of Staudinger ligation and click chemistry, describing varied time dependence for ligation efficiency, and shorter reaction times for click ligation, but these studies have not reported the click methodology as having low sensitivity (20, 43). A possible explanation for observed low sensitivity rests on the high oxidative capacity of the systems under study, containing either oxygenase (tyrosinase) or cytochrome P450 oxidase at high levels. Click ligation is catalyzed by Cu(I), which is attenuated by the oxidative environment that is essential for study of protein modification by the azidoTags. The attenuated efficiency of click ligation observed in the presence of H2O2 is compatible with the proposal. High ascorbate concentrations are used to maintain sufficient reduced copper ion for click ligation, but this is apparently not sufficient in highly oxidizing systems.
Metabolism studies in induced rat liver microsomes also reduced confidence in the use of the azidoTag methodologies because the azide group was observed to be susceptible to metabolism to an amino group and subsequent oxidative metabolism normal for a primary amine. Although the oxidative loss of the conjugating azide group may be at acceptable levels for many applications, the focus of these studies is oxidative bioactivation, therefore this is undesirable.
Fluorophore tagging is useful to provide an alternative or additional visualization method to biotin-avidin immunoassay, but the dansylTag methodology was the least sensitive of the methodologies compared herein. The most significant advantage of fluorophore tags is their use for direct imaging under fluorescence microscopy and by in-gel read-outs (24). Fluorophore tags may be of comparable size and hydrophilicity to biotin tags limiting cell impermeability (29), however, the dansylTag proved invaluable in visualization of the cell permeability of tagged SERMs, rapidly accumulating in the cytoplasm in intact cells (Figure 6). The use of dansylTags and related fluorophore tagged SERMs in this regard is clear, but no detectable protein modification was observed by dansylTag in hepatocytes (Supporting Information). The in-gel fluorescence read-out cannot be amplified in the same way as immunoassay signals. Important, cellular protein targets may be in low abundance; therefore dansylTag is insufficiently sensitive for detection of protein targets in cells.
Biotin is a commonly used affinity tag for affinity labeling; the high affinity of avidin/streptavidin makes it a powerful tool for both detection and enrichment of probe-labeled targets (23). However, cell impermeability caused by biotin and its linker was a major concern for the generalized application of COATags to cells (22-24). The COATag used herein was observed to modify a large number of cellular protein targets in hepatocytes suggesting that the COATag is cell permeable. Cell permeability for the dansylTag in MDA-MB-231 cells was readily visualized, showing rapid access to the cytoplasm. The COATag was also observed to be a potent antiestrogen in simile with the parent drug molecule in Ishikawa cells. Given the 4-day incubation with Ishikawa cells, it cannot be ascertained if the COATag manifests slow nuclear membrane penetration as was observed with the dansylTag in MBA-MB-231 cells. Nevertheless, the COATag methodology appears applicable for whole cell and tissue studies. Given that SERMs are good ER ligands, their binding with and transportation by ER may assist transport of these COATags into the cytoplasm and nucleus in ER positive cells. The COATag methodology also demonstrated high sensitivity and specificity, and furthermore, has the advantage of being a 1-step labeling method, preventing sample loss during multistep experimental processing.
One inevitable limitation of COATags was the presence of endogenous biotinylated proteins, observed in high abundance in hepatocytes at 75 kDa and 150 kDa (Figure 5C). Biotin-dependent carboxylases are especially abundant in liver tissues (44). In many other cells, such as MDA-MB-231 cells (Figure 5A), endogenous biotinylated proteins may not present a problem, however, for generalization of the use of COATags, further separation and discrimination is required. Exploration of the COATag in primary hepatocytes followed by further separation using 2D PAGE illustrated one such approach. Interestingly, COATag modified proteins clustered at low molecular weight (20-75 kDa) and acidic pI (pH 4-7) (Figure 7); thiol rich protein targets such as GST-P1 (MW: 23 kDa, PI: 6.9), Keap1 (MW: 69 kDa, pI: 6.0), and HSP70 (MW: 70 kDa, pI: 5.5) all occur within this range. The previous study of COATags in rat liver microsomes showed that the methodology is compatible with MS-MS analysis (7), therefore, antibodies specific for these potential targets and MS analysis can be used in combination, as a preferred methodology for future protein identification in cells treated with COATags. Preliminary MS-MS analysis was performed on gel digests using the COATag methodology, identifying modified cytosolic proteins previously described in liver microsomes (glucose regulated protein, protein disulfide isomerase, and glutathione S-transferase) (7) and certain proteins that were anticipated targets for quinoid electrophiles, such as peroxiredoxin-1 and heat shock proteins (Table 2), which were reported as targets for other electrophiles such as 4-HNE (20).
Conclusion
Endogenous small molecules, drugs, and xenobiotics are subject to oxidative bioactivation to electrophilic metabolites. SERMs are bioactivated to quinoids, implicated in carcinogenesis and chemoprevention, which illustrates the importance of understanding the cellular targets of such metabolites (14, 45, 46). Methodology development for detection and identification of cellular targets of electrophilic metabolites is therefore of importance in understanding and predicting the balance between cytotoxic versus chemopreventive effects. In this study, four methodologies were studied and compared using purified protein targets and rat primary hepatocytes (Figure 9 and Table 3). Importantly, the azidoTag/click methodology, using the currently preferred combination of azide-tagged molecules with click chemistry ligation, was limited in this application by the high oxidative capacity of the systems under study, leading to both metabolic loss of the azide group and reduced sensitivity. The COATag methodology, introduced in 2005, demonstrated superior characteristics in this application. The presence of endogenous biotinylated proteins in hepatocytes was overcome by application of 2D gel separation and proteomic analysis.
Supplementary Material
Abbreviations
- BHT
2,6-di-tert-butyl-4-methylphenol
- BSA
bovine serum albumin
- COATag
covert oxidatively activated tag
- 1DGE
1 dimensional gel electrophoresis
- 2DGE
2 dimensional gel electrophoresis
- DMA
desmethylarzoxifene
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- ER
estrogen receptor
- GST-P1
human glutathione S-transferase P1-1
- KHB
Krebs-Henseleit Buffer
- 4-HNE
4-hydroxynonenal
- LC-MS/MS
liquid chromatography with electrospray ionization tandem MS
- PBS
phosphate buffered saline
- Ral
raloxifene
- r.t.
room temperature
- SDS-PAGE
sodium dodecyl sufate polyacrylamide gel electrophoresis
- SERM
selective estrogen receptor modulator
- TFA
trifluoroacetic acid
- RalN3
azido raloxifene
- DiQM-CG
di-quinone methide CysGly conjugates
- DiQM-GSH
di-quinone methide GSH conjugates
- o-QGSH
o-quinone GSH conjugates
Footnotes
Supporting Information Available: Experiments of modified Staudinger ligation, COATag, and dansyl DMA in hepatocytes, and MS-MS spectra of GSH conjugates of azidoTag are described in supporting information. This material is available free of charge via the Internet at http://pubs.acs.org/BC.
Reference
- 1.McMeekin DS, Gordon A, Fowler J, Melemed A, Buller R, Burke T, Bloss J, Sabbatini P. A phase II trial of arzoxifene, a selective estrogen response modulator, in patients with recurrent or advanced endometrial cancer. Gynecol Oncol. 2003;90:64–69. doi: 10.1016/s0090-8258(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 2.Dowers TS, Qin ZH, Thatcher GR, Bolton JL. Bioactivation of Selective Estrogen Receptor Modulators (SERMs). Chem Res Toxicol. 2006;19:1125–1137. doi: 10.1021/tx060126v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariazi EA, Ariazi JL, Cordera F, Jordan VC. Estrogen receptors as therapeutic targets in breast cancer. Curr Top Med Chem. 2006;6:181–202. [PubMed] [Google Scholar]
- 4.Yu L, Liu H, Li W, Zhang F, Luckie C, van Breemen RB, Thatcher GR, Bolton JL. Oxidation of raloxifene to quinoids: potential toxic pathways via a diquinone methide and o-quinones. Chem Res Toxicol. 2004;17:879–888. doi: 10.1021/tx0342722. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Bolton JL, Thatcher GR. Chemical modification modulates estrogenic activity, oxidative reactivity, and metabolic stability in 4'F-DMA, a new benzothiophene selective estrogen receptor modulator. Chem Res Toxicol. 2006;19:779–787. doi: 10.1021/tx050326r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Liu J, van Breemen RB, Thatcher GR, Bolton JL. Bioactivation of the selective estrogen receptor modulator desmethylated arzoxifene to quinoids: 4'-fluoro substitution prevents quinoid formation. Chem Res Toxicol. 2005;18:162–173. doi: 10.1021/tx049776u. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Li Q, Yang X, van Breemen RB, Bolton JL, Thatcher GR. Analysis of protein covalent modification by xenobiotics using a covert oxidatively activated tag: raloxifene proof-of-principle study. Chem Res Toxicol. 2005;18:1485–1496. doi: 10.1021/tx0501738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Ngui JS, Doss GA, Wang RW, Cai X, DiNinno FP, Blizzard TA, Hammond ML, Stearns RA, Evans DC, Baillie TA, Tang W. Cytochrome P450 3A4-mediated bioactivation of raloxifene: irreversible enzyme inhibition and thiol adduct formation. Chem Res Toxicol. 2002;15:907–914. doi: 10.1021/tx0200109. [DOI] [PubMed] [Google Scholar]
- 9.Chang M, Zhang F, Shen L, Pauss N, Alam I, van Breemen RB, Blond SY, Bolton JL. Inhibition of glutathione S-transferase activity by the quinoid metabolites of equine estrogens. Chem Res Toxicol. 1998;11:758–765. doi: 10.1021/tx9702190. [DOI] [PubMed] [Google Scholar]
- 10.Aliau S, Delettre G, Mattras H, El Garrouj D, Nique F, Teutsch G, Borgna JL. Steroidal affinity labels of the estrogen receptor alpha. 4. Electrophilic 11beta-aryl derivatives of estradiol. J Med Chem. 2000;43:613–628. doi: 10.1021/jm990179s. [DOI] [PubMed] [Google Scholar]
- 11.Aliau S, El Garrouj D, Yasri A, Katzenellenbogen BS, Borgna JL. 17 alpha (haloacetamidoalkyl) estradiols alkylate the human estrogen receptor at cysteine residues 417 and 530. Biochemistry. 1997;36:5861–5867. doi: 10.1021/bi963111c. [DOI] [PubMed] [Google Scholar]
- 12.el Garrouj D, Aliau S, Aumelas A, Borgna JL. Steroidal affinity labels of the estrogen receptor. 2. 17 alpha-[(Haloacetamido)alkyl]estradiols. J Med Chem. 1995;38:2339–2348. doi: 10.1021/jm00013a011. [DOI] [PubMed] [Google Scholar]
- 13.Scott GK, Atsriku C, Kaminker P, Held J, Gibson B, Baldwin MA, Benz CC. Vitamin K3 (menadione)-induced oncosis associated with keratin 8 phosphorylation and histone H3 arylation. Mol Pharmacol. 2005;68:606–615. doi: 10.1124/mol.105.013474. [DOI] [PubMed] [Google Scholar]
- 14.Yu B, Dietz BM, Dunlap T, Kastrati I, Lantvit DD, Overk CR, Yao P, Qin Z, Bolton JL, Thatcher GR. Structural modulation of reactivity/activity in design of improved benzothiophene selective estrogen receptor modulators: induction of chemopreventive mechanisms. Mol Cancer Ther. 2007;6:2418–2428. doi: 10.1158/1535-7163.MCT-07-0268. [DOI] [PubMed] [Google Scholar]
- 15.Meier BW, Gomez JD, Kirichenko OV, Thompson JA. Mechanistic basis for inflammation and tumor promotion in lungs of 2,6-di-tert-butyl-4-methylphenol-treated mice: electrophilic metabolites alkylate and inactivate antioxidant enzymes. Chem Res Toxicol. 2007;20:199–207. doi: 10.1021/tx060214f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meier BW, Gomez JD, Zhou A, Thompson JA. Immunochemical and proteomic analysis of covalent adducts formed by quinone methide tumor promoters in mouse lung epithelial cell lines. Chem Res Toxicol. 2005;18:1575–1585. doi: 10.1021/tx050108y. [DOI] [PubMed] [Google Scholar]
- 17.Koen YM, Gogichaeva NV, Alterman MA, Hanzlik RP. A proteomic analysis of bromobenzene reactive metabolite targets in rat liver cytosol in vivo. Chem Res Toxicol. 2007;20:511–519. doi: 10.1021/tx6003166. [DOI] [PubMed] [Google Scholar]
- 18.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 19.Bogyo M, Verhelst S, Bellingard-Dubouchaud V, Toba S, Greenbaum D. Selective targeting of lysosomal cysteine proteases with radiolabeled electrophilic substrate analogs. Chem Biol. 2000;7:27–38. doi: 10.1016/s1074-5521(00)00061-2. [DOI] [PubMed] [Google Scholar]
- 20.Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21:432–444. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Qin Z, Thatcher GR, Bolton JL. Uterine peroxidase-catalyzed formation of diquinone methides from the selective estrogen receptor modulators raloxifene and desmethylated arzoxifene. Chem Res Toxicol. 2007;20:1676–1684. doi: 10.1021/tx7001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 23.Lennon-Dumenil AM, Bakker AH, Maehr R, Fiebiger E, Overkleeft HS, Rosemblatt M, Ploegh HL, Lagaudriere-Gesbert C. Analysis of protease activity in live antigen-presenting cells shows regulation of the phagosomal proteolytic contents during dendritic cell activation. J Exp Med. 2002;196:529–540. doi: 10.1084/jem.20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadaghiani AM, Verhelst SH, Bogyo M. Tagging and detection strategies for activity-based proteomics. Curr Opin Chem Biol. 2007;11:20–28. doi: 10.1016/j.cbpa.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 25.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 26.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 27.Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 28.Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 29.Hang HC, Loureiro J, Spooner E, van der Velden AW, Kim YM, Pollington AM, Maehr R, Starnbach MN, Ploegh HL. Mechanism-based probe for the analysis of cathepsin cysteine proteases in living cells. ACS Chem Biol. 2006;1:713–723. doi: 10.1021/cb600431a. [DOI] [PubMed] [Google Scholar]
- 30.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 32.Bolletta F, Fabbri D, Lombardo M, Prodi L, Trombini C, Zaccheroni N. Synthesis and Photophysical Properties of Fluorescent Derivatives of Methylmercury. Organometalllics. 1996;15:2415–2417. [Google Scholar]
- 33.Liu J, Burdette JE, Xu H, Gu C, van Breemen RB, Bhat KP, Booth N, Constantinou AI, Pezzuto JM, Fong HH, Farnsworth NR, Bolton JL. Evaluation of estrogenic activity of plant extracts for the potential treatment of menopausal symptoms. J Agric Food Chem. 2001;49:2472–2479. doi: 10.1021/jf0014157. [DOI] [PubMed] [Google Scholar]
- 34.Obourn JD, Koszewski NJ, Notides AC. Hormone- and DNA-binding mechanisms of the recombinant human estrogen receptor. Biochemistry. 1993;32:6229–6236. doi: 10.1021/bi00075a016. [DOI] [PubMed] [Google Scholar]
- 35.Ogino Y, Ohtake N, Kobayashi K, Kimura T, Fujikawa T, Hasegawa T, Noguchi K, Mase T. Muscarinic M(3) receptor antagonists with (2R)-2-[(1R)-3,3-difluorocyclopentyl]-2-hydroxyphenylacetamide Structures. Part 2. Bioorg Med Chem Lett. 2003;13:2167–2172. doi: 10.1016/s0960-894x(03)00350-0. [DOI] [PubMed] [Google Scholar]
- 36.Jones CD, Jevnikar MG, Pike AJ, Peters MK, Black LJ, Thompson AR, Falcone JF, Clemens JA. Antiestrogens. 2. Structure-activity studies in a series of 3-aroyl-2-arylbenzo[b]thiophene derivatives leading to [6-hydroxy-2-(4-hydroxyphenyl)benzo[b]thien-3-yl] [4-[2-(1-piperidinyl)ethoxy]-phenyl]methanone hydrochloride ( LY156758), a remarkably effective estrogen antagonist with only minimal intrinsic estrogenicity. J Med Chem. 1984;27:1057–1066. doi: 10.1021/jm00374a021. [DOI] [PubMed] [Google Scholar]
- 37.Chandrasena RE, Edirisinghe PD, Bolton JL, Thatcher GR. Problematic Detoxification of Estrogen Quinones by NAD(P)H-Dependent Quinone Oxidoreductase and Glutathione-S-transferase. Chem Res Toxicol. 2008;21:1324–1329. doi: 10.1021/tx8000797. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann KJ, Streeter AJ, Axworthy DB, Baillie TA. Structural characterization of the major covalent adduct formed in vitro between acetaminophen and bovine serum albumin. Chem Biol Interact. 1985;53:155–172. doi: 10.1016/s0009-2797(85)80093-4. [DOI] [PubMed] [Google Scholar]
- 39.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci U S A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Glycoproteomic probes for fluorescent imaging of fucosylated glycans in vivo. Proc Natl Acad Sci U S A. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tryndyak VP, Muskhelishvili L, Kovalchuk O, Rodriguez-Juarez R, Montgomery B, Churchwell MI, Ross SA, Beland FA, Pogribny IP. Effect of long-term tamoxifen exposure on genotoxic and epigenetic changes in rat liver: implications for tamoxifen-induced hepatocarcinogenesis. Carcinogenesis. 2006;27:1713–1720. doi: 10.1093/carcin/bgl050. [DOI] [PubMed] [Google Scholar]
- 42.Sprung R, Nandi A, Chen Y, Kim SC, Barma D, Falck JR, Zhao Y. Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J Proteome Res. 2005;4:950–957. doi: 10.1021/pr050033j. [DOI] [PubMed] [Google Scholar]
- 43.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A comparative study of bioorthogonal reactions with azides. ACS Chem Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 44.Lewis B, Rathman S, McMahon R. Dietary biotin intake modulates the pool of free and protein-bound biotin in rat liver. J Nutr. 2001;131:2310–2315. doi: 10.1093/jn/131.9.2310. [DOI] [PubMed] [Google Scholar]
- 45.Dunlap T, Chandrasena RE, Wang Z, Sinha V, Thatcher GR. Quinone formation as a chemoprevention strategy for hybrid drugs: balancing cytotoxicity and cytoprotection. Chem Res Toxicol. 2007;20:1903–1912. doi: 10.1021/tx7002257. [DOI] [PubMed] [Google Scholar]
- 46.Hagos GK, Carroll RE, Kouznetsova T, Li Q, Toader V, Fernandez PA, Swanson SM, Thatcher GR. Colon cancer chemoprevention by a novel NO chimera that shows anti-inflammatory and antiproliferative activity in vitro and in vivo. Mol Cancer Ther. 2007;6:2230–2239. doi: 10.1158/1535-7163.MCT-07-0069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.