Abstract
Mutations in Leucine-rich repeat kinase 2 (LRRK2) are linked to the most common familial forms and some sporadic forms of Parkinson’s disease (PD). The LRRK2 protein contains two well-known functional domains, MAPKKK-like kinase and Rab-like GTPase domains. Emerging evidence shows that LRRK2 contains kinase activity which is enhanced in several PD-associated mutants of LRRK2. However, the GTPase activity of LRRK2 has yet to be formally demonstrated. Here, we produced and purified the epitope-tagged LRRK2 protein from transgenic mouse brain, and showed that purified brain LRRK2 possesses both kinase and GTPase activity as assayed by GTP binding and hydrolysis. The brain LRRK2 is associated with elevated kinase activity in comparison to that from transgenic lung or transfected cultured cells. In transfected cell cultures, we detected GTP hydrolysis activity in full-length as well as in GTPase domain of LRRK2. This result indicates that LRRK2 GTPase can be active independent of LRRK2 kinase activity (while LRRK2 kinase activity requires the presence of LRRK2 GTPase as previously shown). We further found that PD mutation R1441C/G in the GTPase domain causes reduced GTP hydrolysis activity, consistent with the altered enzymatic activity in the mutant LRRK2 carrying PD familial mutations. Therefore, our study shows the biochemical characteristics of brain-specific LRRK2 which is associated with robust kinase and GTPase activity. The distinctive levels of kinase/GTPase activity in brain LRRK2 may help explain LRRK2-associated neuronal functions or dysfunctions in the pathogenesis of PD.
Keywords: bacterial artificial chromosome transgenics, GTPase, kinase, Leucine-rich repeat kinase 2, Parkinson’s disease
Parkinson’s disease (PD) is the second most common neurodegenerative disease. It is characterized clinically by a movement disorder that includes rigidity, resting tremor and bradykinesia, and pathologically by degeneration of dopamine neurons in the substantia nigra pars compacta as well as other selected brain regions. The etiology in most cases is unknown, but during the past several years, a number of gene mutations have been identified in some familial and sporadic forms of PD that provide an opportunity to elucidate the molecular mechanisms underlying the pathogenesis of cell death in PD. Recent studies suggest that missense mutations in Leucine-Rich Repeat Kinase 2 (LRRK2) or PARK8 are the most common cause of familial (autosomal dominant inherited) forms of PD and in addition have been identified in some sporadic cases (Paisan-Ruiz et al. 2004; Zimprich et al. 2004). Accordingly, it is particularly important to define the mechanism whereby these mutations lead to PD pathogenesis.
The LRRK2 gene product, also called dardarin, is a large and complex protein (285kD). LRRK2 contains multiple conserved domains including a MAPKKK kinase, leucine-rich repeat (LRR), GTPase, and WD40. These distinct structural moieties, some of which incorporate enzyme activities, could interact with or carry out biochemical reactions to modify a variety of proteins and participate in different cellular signaling pathways. Recently, much attention has been directed to the protein kinase and GTPase domains of LRRK2. The LRRK2 protein kinase domain shares sequence similarity to the Tyrosine-Kinase-Like subfamily of protein kinases (containing conserved sequences of both serine/threonine and tyrosine kinases). Several of pathogenic mutations of LRRK2 in PD have been found within the kinase domain (such as G2019S and I2020T) and the GTPase domain (such as R1441C/G), suggesting that these mutations may cause pathological conditions through altering the enzymatic activity of LRRK2 (Paisan-Ruiz et al. 2004; Zimprich et al. 2004). Recent biochemical studies have demonstrated that LRRK2 has kinase activity in vitro, and that the LRRK2 G2019S and I2020T mutations have elevated kinase activity as tested for autophosphorylation (West et al. 2005; Gloeckner et al. 2006; Greggio et al. 2006; Macleod et al. 2006; Smith et al. 2006). These results are consistent with the notion that LRRK2 mutations cause PD by way of a ‘gain-of-function’ or hyperactivity of LRRK2.
The putative GTPase domain of LRRK2 shows high sequence homology to the Rab small GTPase family. Most of the Rab family members are known to regulate vesicle transport and trafficking (Stenmark and Olkkonen 2001). Recent studies showed that LRRK2 binds to GTP and stimulates kinase activity (Smith et al. 2006; Ito et al. 2007). This result is consistent with the hypothesis that LRRK2 kinase activity is regulated by intra-molecular small GTPase function. Interestingly, despite the uncertain GTP hydrolysis activity of LRRK2, the R1441C/G mutation has been shown to affect the autophosphorylation level of LRRK2 (West et al. 2005), suggesting a possibility that R1441C/G may change GTPase activity of LRRK2 and thereby alter the kinase activity of LRRK2. However, a recent study suggests that LRRK2 may lack GTP hydrolysis activity (Ito et al. 2007). It is unclear whether the lack of GTPase activity could be an intrinsic feature of LRRK2 or due to tight regulation by co-factors.
Although LRRK2 (like all known PD-linked genes) are expressed in multiple tissues (Zimprich et al. 2004), the pathology caused by LRRK2 mutants seems to be restricted to the brains or regional brains, suggesting that the brain-specific context is critical for the onset of LRRK2-linked pathogenesis in PD. It remains to be determined whether LRRK2 produced in the brain has specific biochemical characteristics which may be associated with neuronal functions. In this study, we generated bacterial artificial chromosome (BAC)-mediated transgenic mice producing FLAG-tagged LRRK2 and purified FLAG-LRRK2 from the transgenic brains. We show that brain LRRK2 displays GTPase activity as assayed by GTP-binding and GTP-hydrolysis. We further show that LRRK2 purified from the brain has higher kinase activity than that from other mouse tissues or transfected HEK-293T cells. Our result suggests that LRRK2 GTPase can be active in the absence of LRRK2 kinase activity, whereas LRRK2 kinase activity requires GTPase of LRRK2 as previously shown (Smith et al. 2006; Ito et al. 2007). Interestingly, we find that GTPase domain containing PD mutation R1441C/G binds to GTP with similar efficiency to GTPase domain of wild-type LRRK2, but the ability of the mutants to hydrolyze GTP is significantly reduced. This result is consistent with the mechanism whereby LRRK2-linked PD mutations causes neuropathology via altered enzymatic activity. Therefore, our results suggest that LRRK2 produced in the brain is associated with distinctive levels of enzymatic activities, which may be related to specific neuronal functions of LRRK2 and may help explain a brain-specific pathology in mutant LRRK2-linked PD.
Materials and methods
Materials
Media, Calcium phosphate transfection kit were obtained from Invitrogen (Carlsbad, CA, USA). Anti-FLAG Affinity Gel Freezer-safe, GTP-agarose, GTP-γ-S, GTP, and ATP were purchased from Sigma (St Louis, MO, USA). Guanosine 5′-triphosphate [γ-32P], Guanosine 5′-triphosphate [α-32P] and Adenosine 5′-triphosphate [γ-32P] were purchased from PerkinElmer (Waltham, MA, USA). Purified Rac I protein was described previously (Guo et al. 2007). Quickchange Site-Directed Mutagensis kit was purchased from Strategene (Cedar Creek, TX, USA). PEI-F cellulose TLC plates were purchased from J.T. Baker Inc (Phillipsburg, NJ, USA).
Antibody
Anti-mouse LRRK2 polyclonal antibodies were raised in rabbit against a bacterially expressed glutathione S-transferase(GST)-fusion with LRR domain of mouse LRRK2 (GST-LRR) and were affinity purified by immobilized GST-LRR beads. Anti-FLAG antibody was purchased from Sigma. Anti-tyrosine hydroxylase was obtained from Chemicon (Billerica, MA, USA). Anti-synapsin Ia/b was obtained from BD (San Jose, CA, USA). Anti-synaptophysin, anti-CoxVI and anti-alph-synuclein antibodies were purchased from Abcam (Cambridge, MA, USA). All the FITC and Cy3-conjugated secondary antibodies were obtained from Molecular Probes (Carlsbad, CA, USA).
Generation of FLAG-LRRK2 BAC transgenic mice
BAC clone #RP23-312I9 (240 kb) containing entire mouse LRRK2 genomic sequence (promoter region with at least 35 kb and 3′ non-coding region with 60 kb) was purchased from BAC-PAC (Oakland, CA, USA) resource (CHORI). A nucleotide sequence containing FLAG epitope was inserted in-frame after start codon ‘ATG’ of LRRK2 by BAC modification previously developed in Heintz laboratory (Heintz 2001). The injection of purified BAC DNA and generation of transgenic mice was done in core facility of Mount Sinai School of Medicine. The BAC transgenic mice expressing FLAG-LRRK2 was genotyped by PCR with primers 5′GACTACAAAGACGATGACGACAAG3′ and 5′CAT-CCACCACCCAGATAATGTC3′.
LRRK2 wild type and mutant cDNA synthesis and HEK-293T cell transfection
Wild type mouse LRRK2 cDNA was synthesized by RT-PCR amplification of several partial sequences covering total LRRK2 proteins from mouse brain total RNA, followed by joining the DNA pieces through DNA restriction enzyme digestion and ligation with DNA ligase. GTPase domain (aa1321–aa1516) was obtained by RT-PCR from mouse brain total RNA. The cDNAs of LRRK2 were confirmed by repetitive sequence. Full length and GTPase domain of LRRK2 are subcloned into p3XFLAG-CMV-7.1 expression vector. The FLAG-LRRK2-full length or GTPase domains were produced in HEK 293T cells after transfection with the above expression vector and purified by using anti-FLAG affinity gel system according to the manual (Sigma). The singe site mutations (R1441C, R1441G and T1398N) within the GTPase coding sequence in p3XFLAG-CMV-ROC were generated by the Quick-change site-directed mutagenesis kit. HEK-293T transfection with above expressing plasmids was performed using standard calcium precipitation method (Invitrogen).
Immuno-purification of FLAG-LRRK2
FLAG-LRRK2-tranfected HEK-293T cells were lysed in lysis buffer (50 mmol/L Tris-HCl at pH7.5, 150 mmol/L NaCl, 1% Triton X100, 1 mmol/L EDTA, 1 mmol/L phenylmethanesulphonylfluoride, 10 μg/mL pepstain and mini complete protease inhibitor cocktail) for 30 min on ice. FLAG-LRRK2 transgenic mouse brain, lung or kidney or WT brain was homogenized in homogenization buffer (20 mmol/L HEPES at pH 7.4, 0.32 mol/L Sucrose, 1 mmol/L NaHCO3, 0.25 mmol/L CaCl2, 1 mmol/L MgCl2, 1 mmol/L PMSF, 10 μg/mL pepstain, and complete protease inhibitor cocktail), then 10% Triton X-100 was added to a final concentration 1% and incubated at 4°C on rotator for 30 min. The lysed cells and homogenized brain were clarified at 12 000 g for 10 min at 4°C, the FLAG-LRRK2 and FLAG-GTPase protein were purified using Anti-FLAG Affinity Gel according the manual (Sigma) with additional extensive wash before elusion. The protein was eluted by using 150 ng/μL FLAG-peptide and stored at −80°C until use.
Kinase assay
Kinase assay was carried out for 30 min at 25°C in 30 μL reaction mixture containing 20 mmol/L Tris-HCl at pH 7.5, 15 mmol/L MgCl2, 5 mmol/L EGTA, 20 mmol/L β-glycerol phosphate and 0.1 mg/mL bovine serum albumin (BSA) with [γ-32P]-ATP (6000 cpm/pmol), cold ATP was add to prevent the nonspecific binding. In some cases, assay was done with 3 μg of purified MBP (Myelin basic protein), or α-synuclein as substrate. The reaction was stopped by adding 6XSDS sample buffer, and subjected to SDS-PAGE, stained with Coomassie blue. 32P incorporation was measured by phosphor-imager system (GE Healthcare, Piscataway, NJ, USA).
GTP hydrolysis assay
GTP hydrolysis assay by TLC were performed according to previous report (Xia et al. 2003) with modification. Briefly, 100 nmol/L purified FLAG-LRRK2 protein and purified Rac1 protein were incubated at 30°C with 20 nmol/L [α-32P] GTP (3000 cpm/pmol) in reaction buffer (50 mmol/L Tris-HCl pH8.0, 100 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L dithiothreitol, and 0.1 mg/mL BSA) for 5 min to load the nucleotides, then Mg2+ was added to a final concentration of 5 mmol/L to stop the loading and start single turnover hydrolysis. The reaction was conducted at 30°C and 5 μL samples was removed at 0, 15, 30, 60, and 120 min and mixed with cooled 5 μL 0.5 mol/L EDTA to stop the hydrolysis reaction. 2 μL mixed samples were spotted and separated by PEI-cellulose TLC plate using 0.5 mol/L KH2PO4, pH3.4 buffer and quantified with a Storm860 phospho-image analyzer (GE Health-care). Hydrolysis rate (Kcat) was calculated by fitting the time course of GTP remaining to a single exponential by using Microcal Origin 6.0 (Northampton, MA, USA).
The steady-state GTPase activity was determined by the amounts of [32P]Pi released using the charcoal assay as previously described (Hart et al. 1990), 40 nmol/L of purified proteins were incubated for 5, 10, 20, 40, and 60 min at 25°C in a 30 μL of the reaction mixture containing 50 mmol/L Tris-HCl at pH = 8.0, 100 mmol/L NaCl, 1 mmol/L DTT, 1 mmol/L EDTA, 5 mmol/L MgCl2, 0.1% lubrol, 0.1 mg/mL BSA and 5 μmol/L [γ-32P]GTP (2000–3000 cpm/pmol). After the incubation, 0.3 mL of ice-cold 5% (w/v) charcoal in 20 mmol/L H3PO4 were added to stop the reaction. The mixture were centrifuged at 10 000 rpm for 10 min at 25°C, the amount of free 32P release from [γ-32P] GTP was then estimated by counting the radioactivity of 0.1 mL of the clear supernatant. In some cases, 200 μmol/L ATP was added to the GTP hydrolysis reaction system to prevent the nonspecific binding of GTP to the kinase domain, and the free 32P released from [γ-32P] GTP was counted.
GTP-binding assay
For GTP-agarose pull-down, transfected 293Tcells were lysed in lysis buffer (20 mmol/L HEPES at pH 7.4, 2 mmol/L EGTA, 20 mmol/L Glycerol phosphate, 1% Triton X100, 10% glycerol, 1 mmol/L DTT, 1 mmol/L PMSF, 10 μg/mL pepstain, 10 μg/mL aprotinin, 1 mmol/L Na3VO4, and 5 mmol/L NaF) for 30 min on ice, and centrifuged at 12 000 g for 10 min at 4°C. GTP-agarose beads were pre-treated with 1X TBS containing 100 μg/mL BSA for 1 h at 4°C on rotator. 100 μg of clarified cell lysate were incubated with the pre-treated GTP-agarose for 1 h at 4°C on rotator, then added 100 mmol/L GTP and ATP to a final concentration of 2 mmol/L, followed by continuous incubation for additional 2 h. After 2X washing using 500 μL cell lysis buffer, the binding protein was eluted by 20 μL 1X SDS sample buffer and subjected to western blot analysis. The GTPγS binding was determined by filter-binding assay. Briefly, affinity-purified FLAG-tagged proteins were incubated with 5 μmol/L [35S] GTPγS (3000 ~ 8000 cpm/pmol) in binding buffer (20 mmol/L Tris-HCl pH = 8.0, 100 mmol/L NaCl, 5 mmol/L MgCl2, 0.5 mg/mL BSA), at 25°C for 30 min. Reactions were terminated by adding 1 mL ice-cold wash buffer (20 mmol/L Tris-HCl pH = 8.0, 100 mmol/L NaCl, 10 mmol/L MgCl2). The mixtures were filtered through nitrocellulose filters. After washing four times with 1 mL ice-cold wash buffer, the filters were counted by a scintillation counter.
Immunofluorescent staining
Wild-type and FLAG-LRRK2 transgenic adult mice were perfused with 4% paraformaldehyde in phosphate buffered saline. Brains were dissected, post-fixed overnight in 4% paraformaldehyde and cryoprotected in 30% sucrose and coronal sections of 25 μm were cut on a sliding microtome. The slices were incubated overnight at 4°C with anti-FLAG-(M2) (1: 200, Sigma), affinity purified primary antibody (anti-LRRK2, 1: 1000) or anti-TH antibody (1: 500), followed by incubation with secondary antibodies conjugated to Alexa flourescence dyes (1: 500). The mounted sections were visualized on a Zeiss confocal microscope using fluorescent optics (Carl Zeiss Micro Imaging, Jena, Germany).
Results
Development of anti-LRRK2 antibody and BAC-transgenic mice expressing epitope-tagged (FLAG)-LRRK2
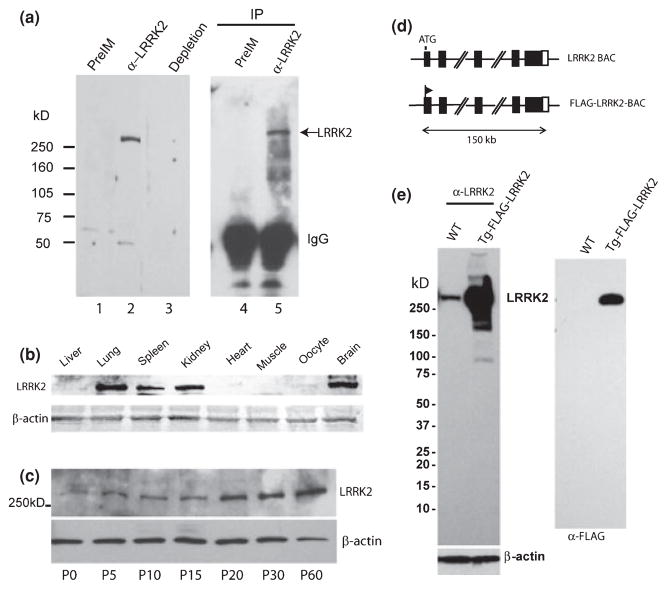
To detect endogenous LRRK2 protein expression, we raised a rabbit polyclonal antibody directed against the LRR region of LRRK2 protein. As shown by immunoblot analysis, the affinity purified anti-LRRK2 antibody, but not purified pre-immune rabbit IgG, detected endogenous LRRK2 protein (~280 kD) from mouse brain lysate (Fig. 1a). The detection of brain LRRK2 is eliminated by the pre-incubation with purified GST-LRR produced in the bacterial. Furthermore, endogenous LRRK2 protein from mouse brain was pulled down by immunoprecipitation with the same anti-LRRK2 antibody but not with pre-immune IgG (Fig. 1a). Using this antibody, we determined the total LRRK2 protein levels in different mouse tissues (Fig. 1b). Immunoblot analyses indicate that the highest expression levels of LRRK2 were in brain, lung, and kidney, followed by spleen. Liver, heart, oocytes, and muscle had significantly less LRRK2 expression than the brain, lung, or kidney (Fig. 1b). Total LRRK2 protein levels in the brain progressively increase through the period of postnatal development as shown in immunoblot analysis that LRRK2 protein levels are significantly higher in the adult brain (P60) than in the early postnatal brain (Fig. 1c). This result suggests that LRRK2 protein expression is regulated during postnatal development and that LRRK2 may be particularly important for the maintenance of certain functions of the mature neurons in the adult mice.
Fig. 1.
Tissue distribution and postnatal expression of LRRK2 protein in mouse brain and development of a bacterial artificial chromosome (BAC) transgenic line producing high levels of FLAG-LRRK2. (a) Endogenous brain LRRK2 protein is detected with purified anti-LRRK2 antibody (lane 2) but not with purified pre-immune (preIM) IgG (lane 1). The detection of brain LRRK2 with anti-LRRK2 antibody on the gel is depleted by the presence of competing purified GST-LRR protein (Lane 3). Brain LRRK2 is immmunoprecipitated (IP) by anti-LRRK2 antibody (lane 5) but not by pre-immune IgG (Lane 4). LRRK2 is detected by the same anti-LRRK2 antibody at size of about 280 kD (arrow). (b) Expression of LRRK2 protein in various mouse tissues as detected by anti-LRRK2 antibody. The protein amount used for immunoblot is controlled by β-actin levels. (c) Progressive increase in LRRK2 protein levels in postnatal mouse brain as shown at different ages. The protein amount used for analysis is controlled by β-actin levels. The brain LRRK2 levels are barely detected at P0 and reach the maximal at P60. (d) A diagram shows a mouse BAC containing complete LRRK2 gene and modified BAC with insertion of epitope tag FLAG after translation start codon ‘ATG’ of LRRK2 protein. (e) Expression of total LRRK2 in the brain of transgenic mice (Tg-FLAG-LRRK2) or wild type (WT) littermate is detected by anti-LRRK2 antibody (left panel). The protein amount used for analysis is controlled by β-actin levels. Specific expression of FLAG-LRRK2 in transgenic brain is confirmed by anti-FLAG antibody (right panel).
To better understand the biochemical activity and neuronal functions of LRRK2 in the brain, we generated transgenic mice expressing FLAG-tagged full-length LRRK2 using BAC transgenic approach (Yang et al. 1997; Heintz 2001), which ensures that FLAG-LRRK2 expression remains under endogenous regulation (Fig. 1d). We have identified a transgenic line producing high levels of FLAG-LRRK2 protein in the brain of transgenic mice as shown by immunoblot analysis with anti-FLAG antibody (M2) and anti-LRRK2 antibody developed in this study (Fig. 1e). Compared to wild-type mice, this transgenic line produced at least 20-fold increase in total brain LRRK2 over the endogenous LRRK2 protein as measured based on the intensity of LRRK2 detected by anti-LRRK2 antibody (Fig. 1e).
Distribution and subcellular localization of FLAG-LRRK2 in the transgenic brain
We investigated LRRK2 protein distribution in the transgenic brains with the anti-FLAG antibody and in wild type brains with anti-LRRK2 antibody. Overall, the staining pattern of LRRK2 in the CNS is very similar with either of the two antibodies. In the midbrain of transgenic mice, FLAG-LRRK2 staining was found particularly strong in cerebral cortex, ventral tegmental area (Fig. 2a), amygdale, and hippocampus (data not shown). FLAG-LRRK2 is also expressed in dopaminergic neurons of substantia nigra which degenerate in PD (data not shown). The distribution of FLAG-LRRK2 is consistent with previous in situ analysis of LRRK2 mRNA (Galter et al. 2006; Giasson et al. 2006; Melrose et al. 2006; Taymans et al. 2006) and the study using antibody staining (Biskup et al. 2006). Noticeably, LRRK2 was seen in the soma and the processes of pyramidal neurons (Fig. 2a). As a control, anti-FLAG antibody does not show any staining on non-transgenic brain slices. We further investigated the intracellular localization of FLAG-LRRK2 in CNS neurons of transgenic mouse brain using confocal laser scanning imaging. As shown by immunofluorescent labeling with anti-FLAG antibody, the staining reveals diffuse as well as vesicular localization of FLAG-LRRK2 protein in neurons (Fig. 2b). The similar intracellular distribution of LRRK2 in wild type mice is revealed by anti-LRRK2 antibody staining, which is largely depleted by the pre-incubation with the purified GST-LRR (Fig. 2b). Consistently, the subcellular localization of LRRK2 in vesicles was previously shown in rodent brains (Biskup et al. 2006).
Fig. 2.

Immunofluorescent analysis of Leucine-rich repeat kinase 2 (LRRK2) distribution and subcellular localization in neurons. (a) Co-immunofluorescent staining of brain slices from transgenic mice producing FLAG-LRRK2 with anti-FLAG (green) and anti-LRRK2 (red) antibodies in area of cortex (upper panels) and midbrain ventral tegmental area (VTA, bottom panels). Staining with two antibodies show largely co-localization of two different colors in ‘overlap’ panels. Scale bar, 100 μm. (b) Subcellular localization of LRRK2 in a neuron of VTA from wild type brain detected with anti-LRRK2 antibody (in green, two left panels). The pre-incubation of purified glutathione S-transferase-LRR deplete the staining with anti-LRRK2 antibody, suggesting the specificity of the LRRK2 staining in this assay. Subcellular localization of FLAG-LRRK2 in a neuron from FLAG-LRRK2 transgenic (Tg) brain detected with anti-FLAG antibody (in red, two right panels). As a control, non-transgenic (non-Tg) mice do not have staining with anti-FLAG antibody. Scale bar, 5 μm.
Purified brain LRRK2 contains robust kinase activity as compared to that from lung or HEK-293T cells
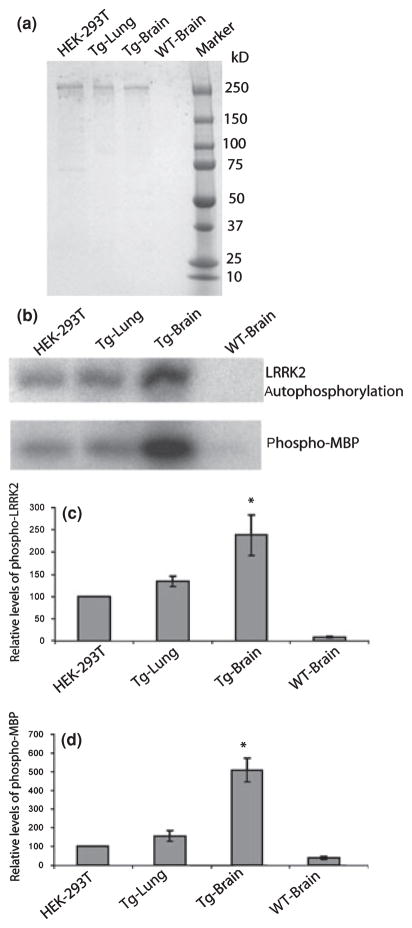
To characterize the enzymatic activities of brain-derived LRRK2, we took advantage of our transgenic mice with high yield of FLAG-LRRK2 and purified FLAG-LRRK2 protein from transgenic brain by immuno-affinity chromatography. Brain FLAG-LRRK2 specifically bound to M2 (anti-FLAG antibody)-immobilized sepharose was isolated, washed, eluted, and analyzed on SDS-PAGE. FLAG-LRRK2 was purified at high purity as assessed by Coomassie blue staining (Fig. 3a), silver staining and confirmed by western blot analysis using anti-LRRK2 antibody (data not shown). As the brain and lung produce comparable protein levels of LRRK2 (Fig. 1b), we also purified FLAG-LRRK2 from transgenic lung as well as from transfected cultured HEK-293T cells with FLAG-LRRK2 expressing plasmid (Fig. 3a). We performed kinase assays with the purified FLAG-LRRK2 from the brain, lung, and HEK-293T cells by analysis of autophosphorylation of LRRK2 and phosphorylation of a generic substrate of MBP (myelin-binding protein) (West et al. 2005). As shown in the analysis of the γ-32P labeling intensity with the equal amounts of purified FLAG-LRRK2, the brain FLAG-LRRK2 shows significantly higher auto-phosphorylation levels than that from lung (90% increased) or HEK-293T cells (130% increased) (*p < 0.05) (Fig. 3b and c). As tested for the levels of phosphor-MBP, the brain FLAG-LRRK2 displays even more robust kinase activity as compared with that from lung (300% increased), HEK-293T cells (400% increased) (*p < 0.05) (Fig. 3b and d), or kidney (data not shown). However, despite the robust kinase activity associated with brain LRRK2, it fails to phosphorylate α-synuclein (data not shown), a pre-synaptic protein also linked to autosomal dominant familial PD.
Fig. 3.
Brain Leucine-rich repeat kinase 2 (LRRK2) protein contains higher kinase activity than that from lung or transfected HEK-293T cells. (a) FLAG-LRRK2 proteins purified from transgenic brain (Tg-Brain), transgenic lung (Tg-Lung), and transfected HEK 293T cells, as well as the control immuno-isolated product from wild-type (WT) brain are examined on SDS-PAGE gel, followed by Coomassie blue staining. (b) kinase activity as tested for autophosphorylation (LRRK2) and MBP phosphorylation were performed with equal amounts of FLAG-LRRK2 proteins (or equal volume of WT-brain control) by incubation with [γ-32P]-ATP for 20 min at 25°C. The γ-32P labeling of each protein was separated on SDS-PAGE and visualized by exposure to X-ray film. Quantification of LRRK2 autophosphorylation (c) and phosphorylation of MBP (d) was done by measuring radioactivity of γ-32P labeled proteins through Phosphoimager. Phosphorylation of LRRK2 or MBP from brain or lung was normalized to that of LRRK2 from transfected HEK-293T cells. Data are presented as mean value (±SEM) from three independent experiments. *p < 0.05 comparing to data from transfected HEK-293T or transgenic lung (Student’s t-test).
Purified brain LRRK2 contains GTPase activity as assayed by GTP hydrolysis and binding
Despite the conserved GTPase domain in the LRRK2 protein, GTP hydrolysis of LRRK2 has yet to be directly shown. To determine the GTPase activity of LRRK2, we performed assays for GTP hydrolysis and GTP binding using purified brain FLAG-LRRK2. First, we performed thin layer chromatography (TLC) assay to test GTP hydrolysis. In the presence of purified FLAG-LRRK2, the radioactivity of α-32P-GTP is gradually reduced while the radioactivity of α-32P-GDP is increased as a function of time (Fig. 4a and b), indicating that FLAG-LRRK2 is capable of converting α-32P-GTP to α-32P-GDP in the assay. This GTP hydrolysis activity of LRRK2 is comparable to that of purified GTPase Rac1 (Guo et al. 2007), as Kcat calculated under this experiment condition for LRRK2 and Rac1 are 0.027 ± 0.005 per min and 0.027 ± 0.007 per min, separately. The Kcat of Rac1 in our assay is within the same range as previously reported (Zhang et al. 1998). In contrast, the immuno-isolated control product from wild-type brain is not able to carry out the change in α-32P-GTP radioactivity. The GTP hydrolysis activity of brain FLAG-LRRK2 was further tested by assay of radioactive phosphate (γ-32P) levels released from γ-32P-GTP. The result reveals that the incubation of brain FLAG-LRRK2 with γ-32P-GTP results in the linear increase in total free 32P in the solution during the incubation time, as compared to the little change in free 32P when incubation was done with controlled products (Fig. 4c). We also examined GTP hydrolysis activity in FLAG-LRRK2 purified from transgenic lung and HEK-293T cells (as shown in Fig. 3). While FLAG-LRRK2 from either transgenic lung or transfected HEK-293T cells displays GTP hydrolysis activity above background, both show lower GTP hydrolysis activity as compared with that from transgenic brain in TLC assay (Fig. 4a and b). Of note, the FLAG-LRRK2 purified from other cell lines, such as COS-7 and Neuro2A, also contains GTP hydrolysis activity above background levels (data not shown). These results clearly demonstrate for the first time that LRRK2 contains intrinsic GTP hydrolysis activity.
Fig. 4.
Analysis of GTPase activity in purified FLAG-Leucine-rich repeat kinase 2 (LRRK2) protein from transgenic brain, lung, and transfected cell lines. (a) Single turnover GTP hydrolysis assay was carried out by PEI-cellulose thin-layer chromatography (TLC). 100 nmol/L purified FLAG-LRRK2 protein from transfected HEK-293T cells, transgenic brain or lung was incubated with 20 nmol/L α-32P-GTP for indicated time at 30°C. The same purification procedure was performed at same time by using non-transfected 293T cells and wild type brain, lung and same volume of elution samples were used as negative control. 100 nmol/L purified Rac1 was used as positive control. The α-32P-GTP and α-32P-GDP were separated by TLC; (b) Quantification of remaining α-32P-GTP on the TLC was analyzed by Storm860 image analyzer. Data are presented as mean value from three independent experiments. Results are shown as percentages of radioactivity at time 0. (c) The steady-state GTP hydrolysis assay was carried out by charcoal assay. 40 nmol/L of purified FLAG-LRRK2 proteins from transgenic brain (Tg brain LRRK2) or the same volume of control products from WT brain (Wt brain control) was incubated at 25°C in a buffer with 5 μmol/L [γ-32P]-GTP, followed by measurement of the release of radioactive γ-32P at indicated time points. Data are presented as mean value (±SEM) from three independent experiments; (d) Filter binding assay to monitor the specific binding of [35S] GTPγS to purified FLAG-LRRK2 protein. 40 nmol/L of purified brain FLAG-LRRK2 protein was incubated with 5 μmol/L [35S] GTPγS with or without Mg2+ (5 mmol/L) for 30 min at 25°C. (e) The purified brain FLAG-LRRK2 was pulled down by GTP sepharose in the absence of GTP or ATP (lane 1), in the presence of 2 mmol/L ATP (lane 2) or in the presence of 2 mmol/L GTP (lane 3). Precipitates on GTP sepharose were resolved by SDS-PAGE, followed by detection with anti-FLAG antibody.
The GTP binding of brain FLAG-LRRK2 was examined by [35S] GTPγS filter-binding assay. 35S radioactivity in brain FLAG-LRRK2 was three times higher than that of control (Fig. 4d). In addition, the presence of Mg+2 (5 mmol/L) is able to stimulate the binding of GTP to FLAG-LRRK2 without changing the background level in the control (Fig. 4d). The GTP binding was further assessed by GTP sepharose precipitation analysis. The purified brain FLAG-LRRK2 was specifically pulled down by GTP sepharose as the presence of excess amount of free GTP, but not ATP, competes for FLAG-LRRK2 binding and eliminates the binding of FLAG-LRRK2 to GTP-sepharose (Fig. 4e).
Taken together, these results demonstrate LRRK2 as a complex protein containing both GTPase and kinase activity in the brain. The particularly robust kinase and GTPase activities of LRRK2 produced in the brain suggest a distinctive biochemical characteristic of LRRK2 in the brain.
GTPase domain of LRRK2 has GTP hydrolysis activity, which is reduced in mutant LRRK2 containing PD-linked mutation R1441C or R1441G
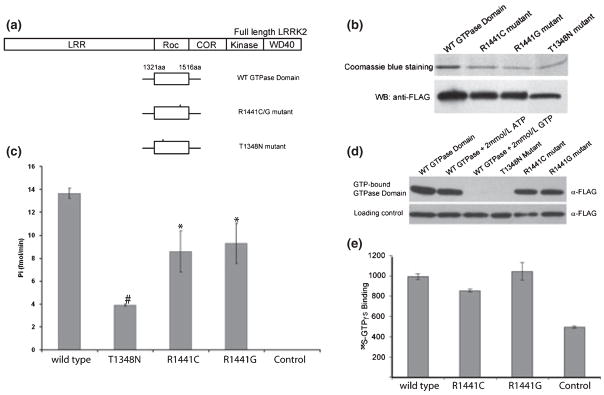
Two LRRK2 missense mutations (R1441C and R1441G) that are associated with PD occur in the GTPase domain, and are therefore considered to possibly affect the GTPase activity of the protein. To determine the potential effect of the R1441C/G mutations on GTPase activity, we generated several FLAG-tagged LRRK2 deletion constructs: wild type GTPase domain (aa1321–aa1516), mutant GTPase domain with an R1441C or R1441G mutation, and mutant GTPase domain with a T1348N mutation in which the predicted GTP binding activity is disrupted based on the bioinformatics analysis of conserved GTPase structures (Farnsworth et al. 1991; Bosgraaf and Van Haastert 2003) (Fig. 5a). These expressing plasmids containing variants of the LRRK2 GTPase domain were introduced into HEK-293T cells by transfection. Each protein was then isolated and purified from transfected cells by M2 sepharose immuno-affinity pull-down and confirmed by western blot analyses with anti-FLAG antibody (Fig. 5b). To test GTPase activity, we performed GTP hydrolysis with these purified GTPase domain variants. Wild-type GTPase domain displays GTP hydrolysis activity as indicated by the release of free γ-32P from pre-incubated γ-32P-GTP (Fig. 5c), suggesting that LRRK2 GTPase domain only contains GTPase activity independent of kinase activity. Interestingly, GTPase domain containing R1441C or R1441G mutation display significantly reduced GTP hydrolysis activity in comparison to the wild-type GTPase domain (Fig. 5c) (*p < 0.05). As expected, the T1348N mutation which is predicted to have impaired GTP binding, has markedly low GTP hydrolysis activity (#p < 0.0001) (Fig. 5c). To further investigate the GTPase activity of mutant R1441C or G, we examined the GTP binding of GTPase domain variants by GTP sepharose pull-down and GTPγS loading assays. The results show that the wild-type GTPase domain is specifically precipitated by GTP sepharose, as excess amounts of free GTP but not ATP compete for protein binding and eliminate precipitation of the GTPase domain (Fig. 5d). Under the same conditions, GTP sepharose precipitates mutant GTPase R1441C or R1441G with the similar efficiency to the wild-type. In contrast, the GTP binding mutant T1348N was not pulled down by GTP sepharose (Fig. 5d). Moreover, GTP binding of wild-type GTPase domain and mutant R1441C or G was further analyzed by GTPγS labeling assay. As shown by the measurement of γ35S radioactivity binding to each protein, no significant difference between wild-type and either of the R1441C/G mutants was detected (Fig. 5e). Therefore, we conclude that the R1441C or G mutation in GTPase domain of LRRK2 results in decreased GTP hydrolysis activity without reducing the binding to GTP as assayed by using non-hydrolysable GTP analog. Although further analysis is needed for understanding the effects of R1441C/G (or other PD mutations of LRRK2) on the GTPase activity in full-length LRRK2, our result implicates the PD mutants of LRRK2 R1441C/G as a relatively active form of GTPase by prolonging the binding to GTP, comparing to the wild-type GTPase of LRRK2.
Fig. 5.
GTPase activity in Leucine-rich repeat kinase 2 (LRRK2) GTPase domain and decreased GTP hydrolysis in GTPase domain containing pathogenic Parkinson’s disease (PD) mutation R1441C/G. (a) A diagram shows schematic of the full-length of LRRK2 protein containing functional domains, wild type GTPase domain (from aa1321 to aa1516), mutations in GTPase domain R1441C or G and T1348N. (b) FLAG-GTPase domain of wild type and mutant variants as indicated were purified from HEK-293T cells after transfection, followed by analysis on SDS-PAGE and visualization by Coomassie blue staining and western blot (WB) detection with anti-FLAG antibody. The protein levels were normalized for the GTP hydrolysis assay. (c) GTP hydrolysis of purified GTPase domain of wild type and mutant variants was assayed by measurement of the release of γ-32P from [γ-32P]-GTP at 40 min. The value of wild type GTPase domain is significantly higher than mutants R1441C (p < 0.05, student’s t test), R1441G (p < 0.05, student’s t test), and T1348N (p < 0.0001, student’s t test). The result is presented as mean values from three independent experiments (±SEM). (d) GTPase domain directly binds to GTP. The cell lysates (100 μg) from transfected cells with plasmids expressing GTPase domain of wild type, or individual mutant variant was incubated with GTP sepharose. The precipitated products by GTP sepharose pull-down were resolved by SDS-PAGE and detected by immunoblot analysis with anti-FLAG antibody. The presence of 2 mmol/L GTP but not ATP eliminates the binding of wild type GTPase domain to GTP sepharose. Wild type GTPase domain shares similar GTP binding efficiency to GTPase domain mutants R1441C or R1441G. (e) PD mutant R1441C/G show no significant difference (p = 0.2, student’s t test) in GTPγS binding as compared to wild type GTPase domain. The GTPγS loading of purified wild type GTPase domain and PD mutant R1441C/G was measured by filter-binding assay and result is presented as mean value (±SEM) from three independent experiments.
Discussion
Our study demonstrates a robust system which combines mouse transgenic approach and biochemical study to investigate LRRK2 structure/function in the CNS. Although LRRK2 contains a Rab-like GTPase domain and binds to GTP (Smith et al. 2006; Ito et al. 2007), GTP hydrolysis activity of LRRK2 has yet to be formally shown. In this study, we demonstrate for the first time that mammalian LRRK2 is an authentic GTPase as analyzed for both GTP hydrolysis and binding through multiple assays. By taking advantage of one of BAC-transgenic line which produce high levels of FLAG-LRRK2 in the brain and allow FLAG-LRRK2 for efficient purification, we performed TLC and radioactive γ-phosphate release assays separately in purified brain FLAG-LRRK2 protein. Each assay independently reveals the GTP hydrolysis activity of the brain-derived LRRK2. Furthermore, GTP binding of brain LRRK2 was shown in GTPγS labeling and GTP-beads pull-down assay separately, consistent with the result in previous study using LRRK2 obtained from transfected cells (Smith et al. 2006; Ito et al. 2007). In addition, we found that LRRK2 purified from other mouse tissues and several different transfected cell lines also contain GTP hydrolysis activity, although lower than that from the brain. However, a recent report by Ito et al. indicates that human LRRK2 may not have intrinsic GTP hydrolysis activity despite the results showing the GTP binding of LRRK2 and requirement of this binding for LRRK2 kinase activity (Ito et al. 2007). We speculate that different experimental conditions used by two groups are likely to contribute to the discrepancy in the result. In our study, the FLAG-LRRK2 protein was eluted and assayed for GTPase activity in the solution. The radioactive GTP and GDP in solution were separated on TLC. In contrast, Ito et al. used LRRK2 protein conjugated on the beads for the assay. The assay was followed by extensive wash and the radioactive GTP and GDP remained on the beads were eluded and separated on TLC. In their assay, the levels of radioactive GDP apparently depend on the GTP hydrolysis of LRRK2 as well as binding affinity of LRRK2 to GDP. Another possibility is that, although less likely, the mouse LRRK2 in our study has stronger GTPase activity than human LRRK2 used in their assay, as human LRRK2 shares ~86% homology in overall protein sequence and ~96% homology in GTPase domain (ROC) with the counterparts of mouse LRRK2.
Furthermore, we characterized the brain LRRK2 kinase activity which displays enhanced levels in comparison to that from other tissues and cultured cells, suggesting that specific cellular context in the brain is important for distinctive levels of LRRK2 enzymatic activities. The molecular mechanism for the increased enzymatic activities in brain LRRK2 is unknown. Multiple factors can contribute to the enhanced levels of enzymatic activities in brain LRRK2. For example, the involvement of tissue-dependent mRNA splicing, post-translational protein modification, brain-derived membrane/lipid binding or even small amount of proteins co-purified with brain FLAG-LRRK2. (Of note, we did not observe obvious difference in the protein migration rate on SDS-PAGE between FLAG-LRRK2 proteins derived from the brain, lung, and cultured cells.) It is also possible that intra-molecular interference between sub-domains of LRRK2 account for the relative low kinase or GTPase activity particularly from some cultured cell lines or other mouse tissues. Alternatively, brain LRRK2 protein may assume a specific structural conformation, thereby resulting in distinguished kinase and GTPase activities. It remains to be shown in the future the primary mechanism that causes specific biochemical characteristics of brain LRRK2 and whether this is linked to the pathogenic mechanism of PD mutant LRRK2.
Moreover, we found that LRRK2 GTPase domain only possesses GTPase activity independent of kinase activity. This is in contrast with the observation that LRRK2 kinase domain only does not have kinase activity and kinase activity requires GTPase activity (Ito et al. 2007). The relationship between these two enzymatic activities of LRRK2 remains to be further analyzed. Nonetheless, in analysis of the effect of PD-related R1441C/G mutation on the GTPase activity of LRRK2, we show that LRRK2 GTPase domain containing the R1441C or G mutation results in decreased GTP hydrolysis activity as compared to wild type control, whereas there is little change in GTP binding as assayed by binding to non-hydrolysable GTP analog. These results suggest that the PD pathogenic mutants R1441C/G may preferentially remain in GTP-bound state which behaves in constitutively active form. It is unclear whether R1441C/G-associated changes in LRRK2 GTPase activity is linked to the altered kinase activity which in turn causes PD pathogenesis, as LRRK2 mutant R1441G/C may have little change in kinase activity, if any, shown in several studies (West et al. 2005; Greggio et al. 2006; Smith et al. 2006). Alternatively, LRRK2 mutant R1441C/G may transduce signals continuously to the downstream independent of LRRK2 kinase by enhanced binding to GTP. A structural modeling of LRRK2 GTPase domain predicts that residue R1441 is distant from GTP hydrolysis site and could be exposed on the protein surface in a region that is implicated in effector or protein binding (Mata et al. 2006). Future study of LRRK2 interactors specifically bound to the GTPase domain will be important for testing this possibility.
Finally, the GTPase domain of LRRK2 shares a high sequence homology to the Rab small GTPase family. In line with the well-characterized role of Rab GTPase in vesicular trafficking and transport, a recent report (Biskup et al. 2006) and our study showed that LRRK2 is associated with vesicles in CNS neurons. Future study should also examine the relationship between GTPase activity of LRRK2 and vesicular membrane trafficking in the neurons. The identification of LRRK2 GTPase downstream effectors, and proteins that regulate GTPase activity (such as GEFs and GAPs), may prove important in better understanding the cellular pathways that are regulated by LRRK2 and how mutations might lead to the familial and sporadic forms of PD through interactions between genetic factors and environmental toxins (McNaught et al. 2001).
Acknowledgments
We thank Dr Q.-J. Wang for technique support. This study is supported by NIH (RNS055683A) and by Bachmann-Strauss Dystonia and Parkinson foundation to ZY.
Abbreviations used
- BAC
bacterial artificial chromosome
- GST
glutathione S-transferase
- LRR
leucine-rich repeat
- LRRK2
leucine-rich repeat kinase 2
- PD
Parkinson’s disease
- TLC
thin layer chromatography
References
- Biskup S, Moore DJ, Celsi F, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L, Van Haastert PJ. Roc, a Ras/GTPase domain in complex proteins. Biochim Biophys Acta. 2003;1643:5–10. doi: 10.1016/j.bbamcr.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Farnsworth CL, Marshall MS, Gibbs JB, Stacey DW, Feig LA. Preferential inhibition of the oncogenic form of RasH by mutations in the GAP binding/“effector” domain. Cell. 1991;64:625–633. doi: 10.1016/0092-8674(91)90246-u. [DOI] [PubMed] [Google Scholar]
- Galter D, Westerlund M, Carmine A, Lindqvist E, Sydow O, Olson L. LRRK2 expression linked to dopamine-innervated areas. Ann Neurol. 2006;59:714–719. doi: 10.1002/ana.20808. [DOI] [PubMed] [Google Scholar]
- Giasson BI, Covy JP, Bonini NM, Hurtig HI, Farrer MJ, Trojanowski JQ, Van Deerlin VM. Biochemical and pathological characterization of Lrrk2. Ann Neurol. 2006;59:315–322. doi: 10.1002/ana.20791. [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O’Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Guo D, Tan YC, Wang D, Madhusoodanan KS, Zheng Y, Maack T, Zhang JJ, Huang XY. A Rac-cGMP signaling pathway. Cell. 2007;128:341–355. doi: 10.1016/j.cell.2006.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MJ, Polakis PG, Evans T, Cerione RA. The identification and characterization of an epidermal growth factor-stimulated phosphorylation of a specific low molecular weight GTP-binding protein in a reconstituted phospholipid vesicle system. J Biol Chem. 1990;265:5990–6001. [PubMed] [Google Scholar]
- Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- Ito G, Okai T, Fujino G, Takeda K, Ichijo H, Katada T, Iwatsubo T. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson’s disease. Biochemistry. 2007;46:1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- Macleod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Olanow CW, Halliwell B, Isacson O, Jenner P. Failure of the ubiquitin-proteasome system in Parkinson’s disease. Nat Rev Neurosci. 2001;2:589–594. doi: 10.1038/35086067. [DOI] [PubMed] [Google Scholar]
- Melrose H, Lincoln S, Tyndall G, Dickson D, Farrer M. Anatomical localization of leucine-rich repeat kinase 2 in mouse brain. Neuroscience. 2006;139:791–794. doi: 10.1016/j.neuroscience.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taymans JM, Van den Haute C, Baekelandt V. Distribution of PINK1 and LRRK2 in rat and mouse brain. J Neurochem. 2006;98:951–961. doi: 10.1111/j.1471-4159.2006.03919.x. [DOI] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci USA. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Ma W, Stafford LJ, Liu C, Gong L, Martin JF, Liu M. GGAPs, a new family of bifunctional GTP-binding and GTPase-activating proteins. Mol Cell Biol. 2003;23:2476–2488. doi: 10.1128/MCB.23.7.2476-2488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Bio-technol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chernoff J, Zheng Y. Interaction of Rac1 with GTPase-activating proteins and putative effectors. A comparison with Cdc42 and RhoA. J Biol Chem. 1998;273:8776–8782. doi: 10.1074/jbc.273.15.8776. [DOI] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]