Abstract
In many settings, molecular testing is needed but unavailable due to complexity and cost. Simple, rapid, and specific DNA detection technologies would provide important alternatives to existing detection methods. Here we report a novel, rapid nucleic acid detection method based on the accelerated photobleaching of the light-sensitive cyanine dye, 3,3′-diethylthiacarbocyanine iodide (DiSC2(3) I−), in the presence of a target genomic DNA and a complementary peptide nucleic acid (PNA) probe. On the basis of the UV–vis, circular dichroism, and fluorescence spectra of DiSC2(3) with PNA–DNA oligomer duplexes and on characterization of a product of photolysis of DiSC2(3) I−, a possible reaction mechanism is proposed. We propose that (1) a novel complex forms between dye, PNA, and DNA, (2) this complex functions as a photosensitizer producing 1O2, and (3) the 1O2 produced promotes photobleaching of dye molecules in the mixture. Similar cyanine dyes (DiSC3(3), DiSC4(3), DiSC5(3), and DiSCpy(3)) interact with preformed PNA–DNA oligomer duplexes but do not demonstrate an equivalent accelerated photobleaching effect in the presence of PNA and target genomic DNA. The feasibility of developing molecular diagnostic assays based on the accelerated photobleaching (the smartDNA assay) that results from the novel complex formed between DiSC2(3) and PNA–DNA is under way.
Photobleaching of cyanine dyes is usually seen as an undesirable characteristic which hampers their use in nucleic acid detection. However, we have discovered that in the presence of a target specific peptide nucleic acid (PNA) oligomer probe, the rate of 3,3′-diethylthiacarbocyanine iodide (DiSC2(3)) photobleaching is directly related to the amount of target DNA present. We call this type of reaction “smartDNA” and this rapid color loss can be used as a sensitive indicator for the presence of a specific DNA sequence.
PNAs, which are used in the current assay as hybridization probes, are oligonucleotide analogues where the negatively charged phosphoribose backbone has been replaced with a neutral N-(2-aminoethyl) glycine group.1,2 The absence of the negatively charged backbone gives PNAs unique physiochemical properties for binding to nucleic acid targets. PNAs rapidly hybridize to single-stranded DNA or RNA,3,4 and PNA–DNA or PNA–RNA duplexes have much higher thermal stabilities than the corresponding DNA–DNA or DNA–RNA duplexes. The melting temperature (Tm) of a PNA–DNA duplex is relatively insensitive to ionic strength and shows equal thermal stability under low (10 mM) and moderate (500 mM) salt concentrations.2,5 PNA–DNA hybridization is severely affected by base mismatches. A single base pair mismatch destabilizes a PNA–DNA duplex to a greater extent than a mismatch in a DNA–DNA duplex.2 These properties give PNA probes many advantages over conventional DNA probes. The exceptional characteristics of PNA probes are the basis for the development of several technologies such as PNA arrays,6,7 antisense DNA targeting,8,9 nucleic acid purification,10 and mutation analysis.11
Despite these examples, designing assays that require PNA invasion of duplex DNA remains difficult. Typically these assays benefit from PNA backbone modification,12 conjugation to “scanning” peptides,13 specific structural features of the target DNA, and overnight incubation with elevated temperature such as 55 °C.3,12,14,15 These requirements seriously impact the efficient application of PNAs to routine diagnostic product development.
The noncovalent interactions of small molecules, including sequence-specific DNA binding polyamides16 and cyanine dyes17 with DNA duplexes have been widely reported. Binding modes of cyanine dyes to DNA duplexes include electrostatic interactions of the cationic dye with the anionic phosphodiester groups of the nucleic acid,18,19 intercalation between base pairs,20 hydrophobic-associated interactions within the minor groove,21,22 and half-intercalation models.23 Cyanine dyes have been used in many different applications including detection of DNA and RNA in agarose gels,24 fluorescent labels in cellular imaging,25 sequence-specific DNA detection using cyanine dye-conjugated DNA oligonucleotide molecular beacons,26 and real-time PCR.27 The potential utility of cyanine dyes in nucleic acid detection has been improved by enhancing the fluorescence intensity of nucleic acid bound dye relative to unbound (free) dye28 or chemically modifying the light-sensitive cyanine dyes to be more resistant to photobleaching.29,30
While most traditional double-stranded DNA binding ligands do not bind PNA–DNA duplexes,31 the cyanine dye, 3,3′-diethylthiadicarbocyanineiodide(DiSC2(5)) does bind to PNA–DNA duplexes.32–35 DiSC2(5) and a closely related cyanine dye, DiSC3-(5), form aggregates in the minor groove of the PNA–DNA oligomer duplexes.35 This aggregate formation results in an approximate 114 nm absorbance shift (blue to violet) that can be observed by the eye.32 Furthermore, the UV–vis spectra of DiSC2(3) with the random PNA–DNA 10 base pairs duplex was scanned. An attenuated/broadened spectrum was observed and was regarded as indicating a lack of a well-defined complex of DiSC2(3) aggregation on the PNA–DNA duplex and was not further investigated.32
However, we have observed that under certain conditions, when DiSC2(3) is mixed with a PNA–DNA duplex, the mixture has a rapid color change (photobleaching of the original pink color of the dye), and we have pursued this phenomenon further. Here we report a rapid sequence-specific accelerated photobleaching reaction that occurs when a complementary PNA probe, a DNA target, and the cyanine dye DiSC2(3) are combined and exposed to 470 nm light. UV–vis, circular dichroism (CD), and fluorescence spectroscopy were used to examine the interaction of DiSC2(3) and related dyes with PNA–DNA oligomer duplexes. This accelerated photobleaching reaction is rapid, and the rate is directly related to the DNA target concentration in the sample. The extent of the color loss can be estimated by eye or measured with a simple photometer which allows sensitive DNA measurements to be performed on genomic DNA samples with minimal hardware. Thus, the utility of high affinity PNA–DNA binding is further improved by the discovery of the color loss reaction and a rapid room temperature binding process that occurs in the presence of DiSC2(3) and related dyes. We have named this PNA–dye–light mediated method of detecting DNA “smartDNA” and anticipate that assays developed with this rapid easy method will have wide applications including infectious disease diagnostics.
EXPERIMENTAL SECTION
Materials
The cyanine dyes with counterions 3,3′-diethylthiacarbocyanine (DiSC2(3)), 3,3′-dipropylthiacarbocyanine (DiSC3(3)), 3,3′-dibutylthiacarbocyanine (DiSC4(3)), and 3,3′-dipentylthiacarbocyanine (DiSC5(3)) (Chart 1A) were purchased from Sigma-Aldrich (St. Louis, MO) or FEW Chemicals (Wolfen, Germany) and were used without further purification. 3,3′-Di(3-propylpyridinium) thiacarbocyanine (DiSCpy(3)) (Chart 1B) was synthesized in the laboratory of Dr. Jerzy Paczkowski. Stock solutions of 7.5 mM dye in DMSO were prepared and stored at −20 °C.
Chart 1.
Chemical Structures of the 3,3′-Dialkylthiacarbocyanine Dyes Used in the Studya
a (A) n = 1 for DiSC2(3), 3,3′-diethylthiadicarbocyanine, n = 2 for DiSC3(3) 3,3′-dipropylthiadicarbocyanine, n = 3, for DiSC4(3) 3,3′-dibutylthiadicarbocyanine, n = 4 for DiSC5(3) 3,3′-dipentylthiadicarbocyanine; (B) tricationic thiacarbocyanine (3,3′-di(3-propylpyridinium)thiacarbocyanine) DiSCpy(3); and (C) structure of a photooxygenation product.
PNA oligomers were purchased from Panagene (Korea, ww-w.panagene.com), sequences shown in Supporting Information (S1), resuspended in molecular biology grade water (HyClone, Logan, UT) and stored at −20 °C. To increase solubility, all PNA oligomers have a N-terminal lysine. PNA oligomers with a C-terminal lysine had characteristics similar to the PNA oligomers with a N-terminal tag. DNA oligomers (Integrated DNA Technologies, Coralville, IA) were resuspended in molecular biology grade water and stored at −20 °C. Buffers were purchased from Research Organics (Cleveland, OH). Homopipes powder was dissolved in microbiology grade water to 20 mM, adjusted to pH 5.0 with 5 N NaOH, and filtered through 0.2 μm sterile filter (Nalgene PES filter, part no. 566-0020, Nalge Nunc International, Rochester, NY). EDTA was purchased as a 500 mM solution (Teknova, Hollister, CA) and diluted to the required concentration. Genomic DNA of Mycobacterium tuberculosis (MTB) strain CDC1551 was obtained from Colorado State University (http://www.cvmbs.colostate.edu/microbiology/tb/dna.htm). Genomic DNAs from other bacteria were purchased from ATCC (Manassas, VA) (Escherichia coli (700928D), Staphylococcus aureus (10832D), Streptococcus pneumoniae (BAA-334D), Haemophilus influenzae (51907D), Neisseria meningitidis (BAA-335D), Pseudomonas aeruginosa (17933D and 147085), and Klebsiella pneumoniae (707721D). Human placental DNAs (lots 14686, 02153787, and 123K3786) were purchased from Sigma-Aldrich (St. Louis, MO). All DNAs were resuspended in molecular biology grade water and stored at −20 °C.
Equipment
Reaction mixtures were prepared in 384-well white wall, optical bottom microplates (Nunc 242763, Nalge Nunc International, Rochester, NY). Measurement of UV–vis, fluorescence spectra, and time-course assays of the decrease in absorbance at 556 nm with exposure to 470 nm light were performed with a Tecan Safire2 microplate reader (Tecan US, Durham, NC). A solid state smartDNA activator was developed by Advanced Scientific Consulting (Toronto, Canada) and consists of 192 2000-mcd 470 nm light emitting diodes (LED), (Jameco Electronics part no. 183222, Belmont, CA) arranged in a 5 in. × 7 in. rectangular array placed 2 in. above the microplate surface. This configuration produced an irradiance of approximately 2 mW cm−2 at the microplate surface.
CD spectra were recorded on an Olis RSM 1000 circular dichroism spectrophotometer with a Quantum Northwest Peltier accessory located in the laboratory of Dr. Woolley at the Dept. of Chemistry, University of Toronto. Photolysis experiments were carried out in the laboratory of Dr. Jerzy Paczkowski.
Methods
For accelerated photobleaching assays performed in gels, the PNA–DNA oligomer duplexes were generated by mixing equal amounts of PNA (dissolved in water) and DNA complementary oligonucleotide (dissolved in 1X TE), heating the samples to 90 °C for 10 min, and allowing them to cool slowly to room temperature in a heat block. A volume of 4 μL (2 pmol) of DNA oligonucleotide or of the preannealed PNA–DNA duplex (2 pmol each) was mixed with 4 μL of a 25% glycerol solution and loaded into a 1X TBE, 10% (19:1) nondenaturing polyacrylamide gel. After electrophoresis for 60 min at 300 V, the gel was stained for 30 min in a 1X TBE solution containing 15 μM DiSC2(3). The gel was illuminated with a standard UV light box, and black and white gel photos were taken at time zero and after 10 min exposure of the gel to a 15 W white fluorescent light (Aurora 50/50, Fritz Industries, Mesquite, TX) at an irradiance of 2 mW cm−2. The photo shown was digitized and modified using PhotoShop CS 2.0 (Adobe Systems Inc., San Jose, CA) to simulate the colors that are observed by eye.
For the accelerated photobleaching assays performed in microtiter plates, a series of dilutions of M. tuberculosis DNA were prepared and aliquoted to the wells of the microtiter plate. Dye with or without PNA was added to DNA samples with final concentrations of 9 μM dye, 160 nM PNA probe, in 10 mM homopipes buffer, pH 5.0 (0.05% Tween-80). The reaction mixtures were incubated at room temperature for 10 min in the dark. After an initial reading of the absorbance at 556 nm, the reactions were recorded by alternating exposure to light from the smartDNA activator for 2 min and reading the absorbance at 556 nm, until the reactions were clear. Typically the absorbance change of DiSC2(3) in the reaction mixture was measured after the first 2 or 4 min of light exposure to determine the initial slope of the reaction (see Results for details).
For the spectra studies, 17mer PNA, 17mer single stranded DNA, or the 17 bp preannealed PNA–DNA duplex was titrated into a final dye solution of 9 μM and final buffer of 10 mM homopipes, pH 5.0. Spectra were scanned immediately.
For the photolysis studies, 500 mg of DiSC2(3) was dissolved in 4 L of KH2PO4/Na2HPO4 buffer (pH 7.0) and placed in a 5 L immersion well photoreactor equipped with a 400 W medium pressure mercury lamp (Photochemical Reactors Ltd., U.K.). The solution, in the presence of oxygen (bubbled using the gas inlet tube), was exposed to the filtered mercury lamp emission for 10–12 h until it became completely clear and the organic photobleached products were then extracted with chloroform. The UV portion of the mercury lamp’s emission was eliminated using a liquid K2CO3 filter. The solvent was evaporated, and the components of the residue were separated using column flash chromatography. This yielded two major products of which only one was stable. The structure of the stable product was analyzed using 1H NMR spectroscopy.
RESULTS
A previous report by Smith et al.32 focused on the interaction of DiSC2(5) with PNA–DNA oligomer duplexes. However, they also briefly looked at the interaction of other cyanine dyes with the PNA–DNA duplex, including DiSC2(3). In these spectral studies they observed that there was a broadened absorbance band with DiSC2(3) upon interaction with the short PNA–DNA duplex. No further observations were reported. We were interested in further characterizing the interaction of DiSC2(3) with the PNA–DNA duplex and focused on the development of the unique interaction into a practical DNA diagnostics method.
Spectra Studies
The interaction of carbocyanine dyes with PNA–DNA duplexes were studied through UV–vis, CD, and fluorescence spectra. Spectra were obtained for the dye only, the dye mixed with 17mer PNA, the dye mixed with the complementary 17mer DNA oligonucleotide, and dye mixed with a preannealed PNA–DNA 17mer duplex. The 17mer PNAs used in these studies were chosen based on a combinational approach of sequence analysis and practical PNA synthesis limits. For these experiments TB23, a 17mer PNA, with its complementary DNA strand was used. All spectra were obtained in the 10 mM, pH 5.0 homopipes buffer.
UV–vis Spectroscopy
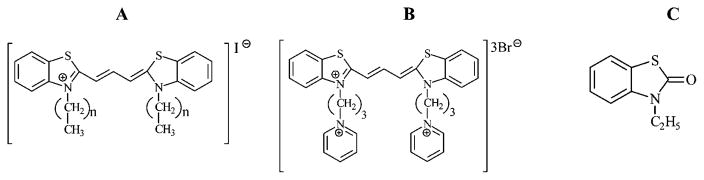
The UV–vis spectra of DiSC2(3) alone and of all five dyes (ethyl, propyl, butyl, pentyl, and propylpyridinium) combined with a preformed PNA–DNA 17mer duplex are shown in Figure 1A. All absorption spectra of dye alone in buffer are characterized by a major monomer absorbance band with a maxima at 556 nm and a shoulder at 520 nm. The size of this shoulder decreases in methanol solutions; thus, it may be due to the self-aggregation of dye dimer in aqueous solutions.36,37 Addition of PNA to solutions of any of the dyes did not result in a change in the spectra of the dye. Addition of single stranded oligomer DNA to solutions of the dyes resulted in a slight decrease in the monomer band. This indicates some interactions of the cationic dye with anionic DNA. However, the addition of PNA–DNA duplex to the dye solutions caused a dramatic decrease in the intensity of the monomer band and the appearance of a new band at approximately 470 nm, further blue-shifted from the 556 nm maxima. The observed blue shift in the spectra is similar to that previously reported by Armitage and co-workers32 upon the binding of DiSC2(5) to a 10 base pair PNA–DNA duplex. In this case, the formation of an H-aggregate of DiSC2(5) with the PNA–DNA duplex resulted in a blue-shifted band about ~100 nm shorter than the monomer band.
Figure 1.
(A) UV–vis spectra of N-3,3′-alkyl and tricationic dyes in the presence of 1 μM PNA–DNA (PD) oligomer duplex in pH 5 homopipes buffer, with dye final concentrations 9 μM. PNA used here is TB23, sequence shown in S1 in the Supporting Information. DiSC2(3) alone in the same buffer is also depicted as no PD. (B) CD spectra of DiSC2(3) in the presence of PNA, DNA, or PNA–DNA duplex, (left ordinate scale). The PNA used in the study is TB23, with its cDNA oligo. The mixture had final concentrations of 9 μM dye, 1 μM PNA, 1 μM DNA, or 1 μM PNA–DNA duplex. UV–vis spectra of dye with the same PNA–DNA duplex is included for comparison (right ordinate scale).
DiSCpy(3) did not show the new 470 nm band in the presence of the PNA–DNA duplex (Figure 1A), although there was a slight shift in the monomer band, probably due to other mechanisms of interaction (such as weakly binding in the groove of the duplex but failing to form an aggregate) between DiSCpy(3) and the PNA–DNA duplex. However, the propyl, butyl, and pentyl derivatives, (DiSC3(3), DiSC4(3), and DiSC5(3)), all show the appearance of a 470 nm band upon addition of the PNA–DNA oligomer duplex (Figure 1A). Bulkier substituents at 3,3′ positions, such as 3-propylpyridinium residues, might prohibit the formation of a dye–PNA–DNA complex due to unfavorable steric hindrances or better dye solubility in water caused by the presence of three positive charges in one dye molecule. Dyes with butyl and pentyl substituents still bind to the preannealed PNA–DNA duplex, although with decreasing affinity, as demonstrated by less decrease in the 556 nm absorbance as the bulk of the substituents increases. Furthermore, for DiSC5(3), the 470 nm absorbance was also decreased, compared to the 470 nm absorbance of DiSC2(3), DiSC3(3), and DiSC4(3). The approximate 86 nm blue-shift in absorbance maxima of the new band indicates that the propyl, butyl, and pentyl dyes also form higher aggregates in the presence of PNA–DNA complexes.
Circular Dichroism (CD)
DiSC2(3) is a symmetrical achiral molecule; thus, the dye alone has no CD spectra. Upon interaction of the dye with either the right-handed PNA–DNA duplex or DNA duplex, the dye adopts the chirality of the target duplex.32,38 If the dye interacts with the helical PNA–DNA duplex, an induced CD spectrum for the resulting dye–duplex complex is produced (Figure 1B). As expected, DiSC2(3) in the presence of PNA has no observable CD signal. DiSC2(3) combined with single-stranded DNA shows a small induced CD signal centered at about 525 nm due to the interaction of the dye with the naturally coiled DNA strand. The dimer band of DiSC2(3) has an absorbance maxima at 520 nm (Figure 1A). Thus, these 520 and 525 nm bands indicate that there is a weak interaction of DiSC2(3) dimer with single-stranded DNA. The dye with the PNA–DNA duplex has a much stronger multiphase CD signal centered at about 470 nm which corresponds to the proposed aggregate band observed in the UV–vis spectra. Upon electronic excitation of a pure dimer, only the transition to one of the two splitting energy levels is permitted, dependent upon the alignment of the two molecules and only one positive signal would be predicted.39 However, in a higher order dye aggregate, the close alignment of induced dipole moments (such as excited-state dye dimers or trimers) results in additional second order energy splitting of the excited state. Thus, transitions to both the second order upper and lower energy levels are possible, and a multiphase CD signal is produced. Therefore, the multiphase CD signal observed with DiSC2(3) and the PNA–DNA duplex indicates that DiSC2(3) interacts with the PNA–DNA duplex as a higher aggregate.
Fluorescence
There are three different interaction mechanisms of small molecules with duplexes: intercalation (and half-intercalation), groove binding, and nonspecific electrostatic interactions.19–23 For DNA–DNA duplexes, all of the above interaction modes have been reported, while for PNA–DNA or PNA–PNA duplexes, only groove binding and nonspecific electrostatic interactions have been reported. When a fluorescent molecule is intercalated into the base pairs of duplexes, internal rotation of the dye molecule is restricted and generally fluorescence enhancement is observed. If a single fluorescent molecule binds to the groove of the duplex, a similar fluorescent enhancement occurs. However, when the binding of an initial dye molecule facilitates the cooperative binding of a second or even third molecule and causes them to assemble in parallel, as observed in the H-aggregate, the fluorescence of the dye molecules is quenched.38
The fluorescence pattern of DiSC2(3) and DiSCpy(3) with PNA–DNA duplexes are very different (Supporting Information, S2). Upon binding to a PNA–DNA oligomer duplex, DiSCpy(3) exhibits enhanced fluorescence emission, indicating intercalation or a single molecule groove binding mode of interaction. DiSC2(3), on the other hand, shows quenching of fluorescence in the presence of the PNA–DNA oligomer duplex. This indicates that DiSC2(3) binds cooperatively as an aggregate. The data from the UV–vis and CD spectra further support the formation of a DiSC2(3) aggregate on a PNA–DNA oligomer duplex. Also, the DiSC2(3)–PNA–DNA complex exhibits a new fluorescence peak that is further red-shifted from the major fluorescence peak. This may be due to excimers (excited-state dimers) formed within the aggregates.
Dye–PNA–DNA Photobleaching Reaction on Gel
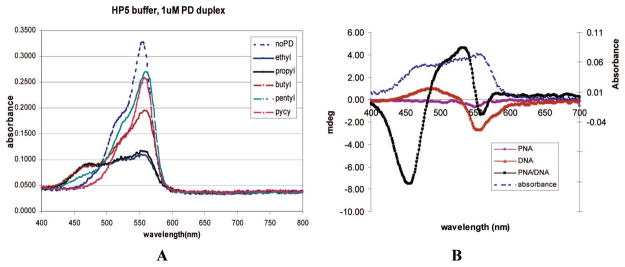
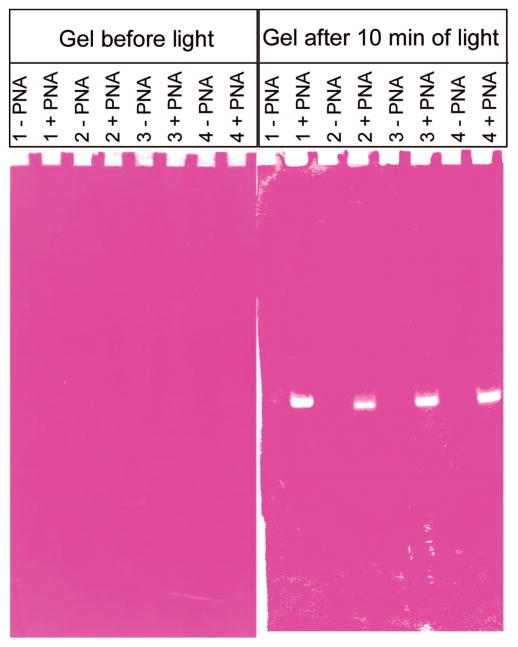
We wanted to determine if the decrease in the 556 nm dye monomer band and the appearance of the 470 nm band observed in solution with dye–PNA–DNA could be observed on a gel. The gel allows the separation of the free dye, the dye–PNA–DNA duplex, and the DNA (with or without dye). Thus, we could also determine the interaction of each complex with light. PNA has a neutral backbone; thus, in a PNA–DNA duplex, the lower molecular weight to charge ratio results in gel retardation. We used a modified nondenaturing polyacrylamide gel electrophoresis assay (PAGE) to observe the different effects of DNA vs PNA–DNA on dye photobleaching in a gel. Upon exposure of the DiSC2(3) stained gel to light, accelerated photobleaching of DiSC2(3) occurred at positions in the gel that correspond to the location of PNA–DNA duplexes (Figure 2, with four PNA and complementary DNA, sequences shown in Supporting Information, S1). Control lanes loaded with DNA alone showed none or a very weak photobleaching at a position on the gel that corresponds to the expected position of the DNA band. These results suggest that a direct association of DiSC2(3) with the PNA–DNA duplex results in accelerated photobleaching of DiSC2(3) upon exposure to light. As this accelerated photobleaching phenomenon occurs with PNA–DNA duplexes of four different sequences, it appears to be sequence independent.
Figure 2.
smartDNA assay run in a gel. See S1 (Supporting Information) for oligonucleotide and PNA sequences.
Aspects of the Dye–PNA–DNA Accelerated Photobleaching Reaction in Solution with Genomic DNA
The observed interaction and accelerated photobleaching of dye DiSC2(3) in the presence of PNA–DNA duplexes can be used in the detection of DNA. A novel phenomenon has been observed; when a PNA probe (TB14), genomic DNA, and DiSC2(3) were combined in buffer and exposed to light at the aggregate absorbance wavelength of 470 nm, accelerated photobleaching was observed only when the genomic DNA contained the PNA probe target. This entire process, which results in a rapid accelerated photobleaching reaction of the dye, is termed a smartDNA assay.
Estimation of Amount of DNA in a Sample Using the Accelerated Photobleaching Reaction
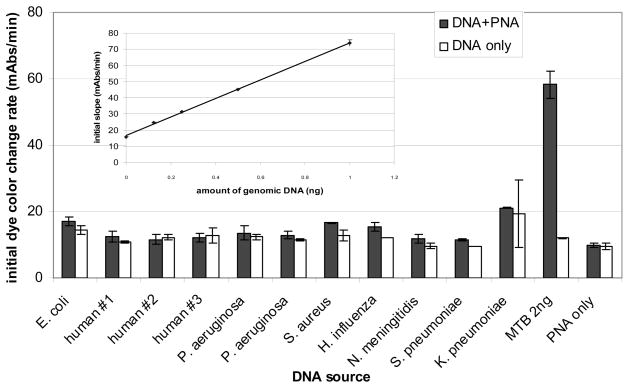
smartDNA reactions were set up with dye, PNA probe, and various amounts of genomic DNA isolated from M. tuberculosis. The photobleaching experiments were carried out in microplates with a final 50 μL reaction volume for each mixture. Generally 25 μL of samples containing different amounts of DNA in 1 mM EDTA were aliquotted into each well. Cyanine dyes were then mixed with the specific PNA probe in 20 mM homopipes, pH 5.0 buffer plus 0.1% Tween 80 and added to the DNA. After an incubation of 10 min in the dark, changes in absorbance at 556 nm as a function of light exposure time were recorded. The absorbance decreases at a greater rate in samples containing genomic M. tuberculosis DNA than in any of the controls. Initial rates of absorbance change (expressed in milliabsorbance units/min) are reasonably linear for the first 4 min and are proportional to the amount of target DNA present (Figure 3, inset).
Figure 3.
The initial (first 4 min) photobleaching rate of smartDNA reactions with 2 ng of DNA from M. tuberculosis (MTB) or from non-M. tuberculosis species in the presence and absence of M. tuberculosis specific PNA probe. The reaction was performed in 10 mM homopipes buffer, pH 5.0, with 0.05% Tween-80, and PNA probe used was TB19; the final concentration of PNA in the 50 μL reaction is 160 nM. The inlay shows the dose response curve of TB14 against M. tuberculosis genomic DNA. The photobleaching rate (milliabsorbance units/min) was calculated for each genomic DNA concentration and plotted.
Typically, mismatched probes are used as negative controls in DNA based assays. However, because of the lack of mature probe design rationale of PNA (which is currently under development), and also in our practice, we have found that some of the PNA probes we have designed do not work as expected in the smartDNA assay. Thus, using a mismatched PNA as a control yields no useful information unless it has first been shown to work in a separate genomic system. So far we have only developed a smartDNA assay for M. tuberculosis and have found that testing the PNAs that work in this system against other microorganisms that do not contain complementary binding sites is a more useful indicator of specificity.
Specificity of the Method for Detection of Genomic DNA Target Sequences
To further validate the potential utility of smartDNA for routine genomic DNA detection, the specificity of the system using DiSC2(3) and the specific M. tuberculosis PNA probe, TB14, was tested with DNA isolated from human, E. coli, S. aureus, S. pneumoniae, H. influenzae, N. meningitidis, P. aeruginosa, and K. pneumoniae. On the basis of genomic sequence data, in silico validation was performed to verify that the genomes of these organisms do not contain sites complementary to TB14. Figure 3 shows the photobleaching rate of DiSC2(3) with and without the PNA probe with nonspecific DNA only or with the specific M. tuberculosis DNA. Reactions that contained the specific (M. tuberculosis) DNA and the PNA probe had the accelerated photobleaching reaction. Reactions that contained nonspecific DNA and the PNA probe did not show the accelerated photobleaching reaction; rather the photobleaching rates were similar to the background photobleaching rate. Furthermore, mixtures of M. tuberculosis DNA with nonspecific DNA, (human, S. pneumoniae, or S. aureus) resulted in the accelerated photobleaching reaction in the presence of the specific PNA probe at levels similar to the reactions with M. tuberculosis DNA alone (Supporting Information, S3). Thus, the accelerated photobleaching reaction appears to be the result of a specific reaction occurring between DiSC2(3), the PNA probe, and its complementary DNA target. This suggests that the accelerated photobleaching can select against some single base mismatches present in potentially contaminating organisms. We have not explored this systematically with model systems.
Influence of Dye Structure on Accelerated Photobleaching Rate
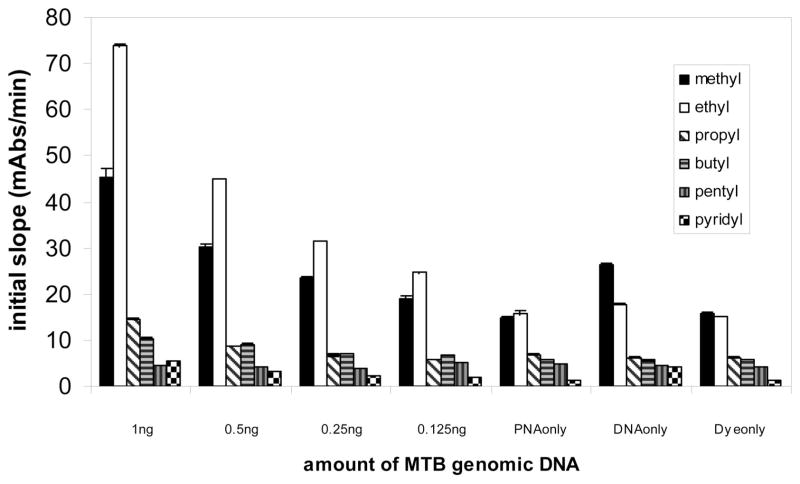
Among the five carbocyanine dye derivatives, DiSC2(3), the dye with ethyl substitutions, shows the most accelerated photobleaching reaction in the presence of PNA and genomic DNA. DiSC2(3) from two vendors shows similar accelerated photobleaching in the presence of PNA and genomic DNA. The other four dyes, DiSC3(3), DiSC4(3), DiSC5(3), and DiSCpy(3), exhibit substantially less photobleaching using the same conditions of PNA probe, buffer, genomic DNA, and light exposure (Figure 4). It is not surprising that DiSCpy(3), which exhibits no 470 nm band in the presence of preannealed PNA–DNA duplexes, does not participate in an accelerated photobleaching reaction (Figure 4). However, DiSC3(3), DiSC4(3), and DiSC5(3), which do interact with preannealed PNA–DNA duplexes (Figure 1A), do not exhibit the same accelerated photobleaching reaction with PNA–genomic DNA complexes. This suggests that the accelerated photobleaching reaction is dependent upon a unique complex structure formed between DiSC2(3), the PNA, and the DNA target. The effect of changes in the substituents at the N-3,3′ position on the rate of accelerated photobleaching indicates that the substituents play a central role in the formation of the dye–PNA–DNA complex.
Figure 4.
Comparison of the photobleaching rate of the thiacarbocyanine dyes with substituents of ethyl, propyl, butyl, pentyl, and pyridinium, (corresponding to DiSC2(3), DiSC3(3), DiSC4(3), DiSC5(3), and DiSCpy(3), respectively). The label on the X axis shows the amount of M. tuberculosis (MTB) genomic DNA in each reaction. The PNA probe used here is TB14, sequence shown in S1. The final concentration of PNA in the 50 μL reaction is 160 nM. All dyes had a final concentration of 9 μM. The buffer is 10 mM homopipes, pH 5.0, 0.05% Tween 80.
It is possible that the structure of the dye–PNA–DNA preannealed oligomer system might be different from that of the dye–PNA–DNA genomic DNA system. In the genomic DNA system, the PNA and dye are bound to high-molecular weight double stranded DNA. A significant, and as yet unexplained, sensitivity difference exists between smartDNA reactions run on oligomer DNA targets as compared to smartDNA reactions run on genomic DNA targets. The genomic DNA targets are detectable at approximately one million-fold lower concentration (femtomolar vs nanomolar) than the oligomer DNA targets. Investigations are ongoing to understand this important difference.
Photochemical Reaction Product
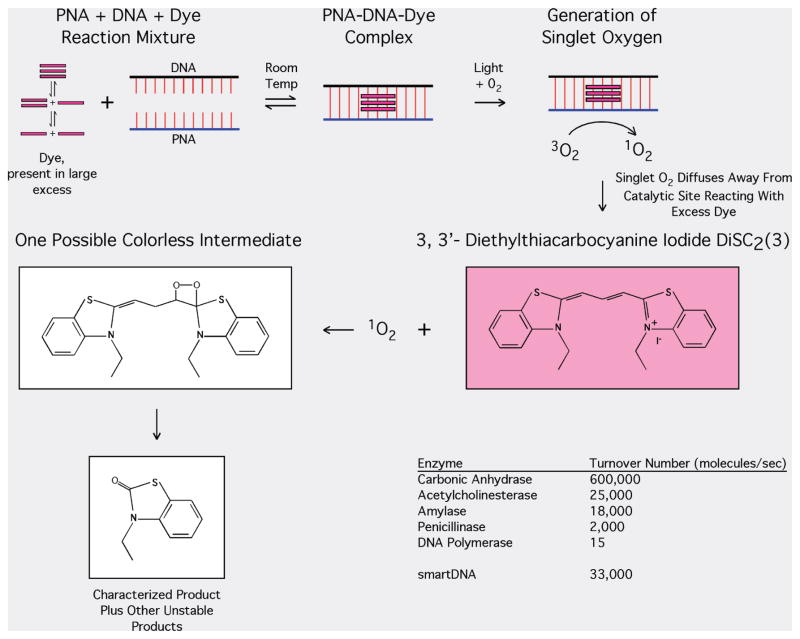
In order to obtain sufficient material for chemical characterization, steady-state photolysis of a large quantity, 500 mg, of DiSC2(3) was performed. Dye dissolved in aerated pH 7 phosphate buffer was exposed to the visible portion of light from a medium pressure mercury lamp for 10–12 h, until the pink color of the dye solution was completely cleared. Two major products of photolysis were separated using column chromatography; of which only one was stable. NMR spectra (Supporting Information, S4) indicate that during the photolysis of the dye in the presence of oxygen, 3-ethyl-2(3H)-benzothiazolone (shown in Figure 5) was the major stable product. Thus, when pure dye was photofragmented, the photofragmentation product is consistent with the involvement of singlet oxygen. The measuring of the bleaching kinetics of the dye in the presence and absence of oxygen in irradiated dye solution additionally supports this supposition. After removal of oxygen from the solution, there is a clear and distinct deviation from the zero reaction order that is typically observed for the bleaching in the presence of either air or oxygen (Supporting Information S5). It is believed that during the initial exposure of the argon saturated solution to light, the residual oxygen is consumed and this causes the decrease in the bleaching rate. After all the oxygen in solution is consumed, the bleaching reaction stops (Supporting Information, S5). We believe that the photochemical reaction which occurs between dye and the light activated PNA–DNA–dye complex is similar to that which occurs with pure dye alone. In reactions with dye–PNA–DNA, the rate of photobleaching is significantly decreased in the presence of azide (Supporting Information S6), a known singlet oxygen scavenger. The background photobleaching reaction on the other hand (PNA and dye only) remains similar with or without azide present. This suggests that the background photobleaching reactions occur via different mechanism. Characterization of the exact nature of this process is the subject of ongoing work.
Figure 5.
Proposed mechanism of accelerated photobleaching based on the generation of singlet oxygen.
DISCUSSION
Dye–PNA–DNA Complex As a Catalyst
Previous literature describes the interaction of cyanine dyes with PNA or DNA or PNA–DNA duplexes.32,35,38,40,41 Cyanine dyes are known to form dimer or higher aggregates in the minor groove of these duplexes and result in spectroscopic changes. In the present study, the UV–vis spectra (Figure 1A) of various dyes in the presence of PNA–DNA duplexes demonstrate the formation of a dye–PNA–DNA complex with DiSC2(3), DiSC3(3), DiSC4(3), and DiSC5(3) but not with DiSCpy(3). Fluorescent quenching of DiSC2(3) in the presence of PNA–DNA duplexes demonstrates the cooperative nature of the binding interaction of this dye with the minor groove of the duplex. CD spectra also show that DiSC2(3) interacts with PNA–DNA in an aggregate manner.
The present study demonstrates that in the presence of a PNA–DNA duplex, a novel DiSC2(3)–PNA–DNA complex can form. This complex is capable of rapidly catalyzing the accelerated photobleaching of a large excess of DiSC2(3), possibly through a singlet oxygen mediated mechanism (for cyanine dyes as 1O2 sensitizers, see ref 42). In a typical 50 μL smartDNA assay, DNA equivalent to 400 000 M. tuberculosis genomes causes the complete photobleaching of a large amount of dye, approximately 1014 molecules, in 4 min when the reaction mixture is exposed to 470 nm light. The calculated turnover rate of the dye molecules per PNA binding site on the target molecule at varying DNA target amounts remains surprisingly constant. Approximately 33 000 dye molecules are turned over per second. This suggests that the dye–PNA–DNA complex is acting in a catalytic manner to turnover the excess dye in the reaction. This fast turnover rate is not without precedent. The enzyme acetylcholinesterase turns over 25 000 molecules per second, and carbonic anhydrase turns over 600 000 molecules per second (Figure 5).
In the smartDNA system, the structure of the cyanine dye that can form a useful catalytic complex with PNA–DNA duplexes has low tolerance for changes. In addition to DiSC2(3), DiSC3(3), DiSC4(3), DiSC5(3), and DiSCpy(3), cyanine dyes with other modifications at the chromophore were screened. Oxazole substitution for the thiazole, or an increase in the length of the methane bridge between the two heterocycles, such as DiSC2(5) did not show accelerated photobleaching in the PNA–DNA oligomer duplex system. There is no observable difference between the photobleaching rate of the free dye in the presence or absence of a PNA–DNA oligomer duplex. Thus, we concluded in these studies that the benzothiazole heterocycle and the length of the bridge are all critical for the specific interactions that result in accelerated photobleaching in the genomic system.
With DiSC2(3), exposure of the dye–PNA–DNA complex (preannealed PNA–DNA oligomer duplex) to LED light sources near the dye’s absorption maxima or longer wavelengths (574 and 640 nm) or shorter wavelengths (390 and 410 nm) accelerated photobleaching was not observed. Exposure of the dye–PNA–DNA oligomer complexes to 470, 490, or 515 nm LED light sources did cause accelerated photobleaching. The 470 nm wavelength proved optimal. Compared to 490 and 515 nm, exposure to 470 nm light had a faster accelerated photobleaching rate and a slower rate of photobleaching of the free dye (data not shown). We suggest that the generation of singlet oxygen requires a stable excited triplet state, and dyes that form H aggregates which absorb at 470 nm were more effective than monomeric dyes in forming the required stable triplet state. The H aggregate formed should have a longer singlet lifetime, due to the forbidden radioactive transition to the ground state, thus is more efficient for the energy transfer of intersystem crossing than other configurations that resulted in absorbance at different wavelengths.43
Substituent Effects on Catalyst Formation
Even for dye derivatives with the same thiacarbocyanine chromophore, substituents at the N atom play a significant role in the interaction of the dye with the preannealed PNA–DNA duplex and/or on the rate of the accelerated photobleaching of the dye in the genomic DNA system. Detailed studies were performed with DiSC2(3), DiSC3(3), DiSC4(3), DiSC5(3), and DiSCpy(3), which have substituents of increasing bulkiness at the N atom of the benzothiazole (Figures 1 and 4). The spectra of DiSCpy(3) (3-alkylpyridinium substituents) with the PNA–DNA oligomer duplex showed no aggregate band at 470 nm. Thus, there is no or very low interaction affinity between DiSCpy(3) and the PNA–DNA oligomer duplexes. For DiSC5(3), the dye with the longest side chain in this group, the aggregate band at 470 nm is present but at low amounts and the 556 nm monomer band is present at high amounts. This result suggests a decreased interaction affinity of DiSC5(3) for the PNA–DNA duplex. DiSC4(3) does exhibit the 470 nm aggregate band and moderate amounts of the 556 nm monomer band. The decrease in the 556 nm band is directly related to the rate of the accelerated photobleaching in a smartDNA reaction with the dye–PNA–DNA oligomer duplex. Photobleaching experiments with M. tuberculosis genomic DNA and dyes other than DiSC2(3) did not exhibit the same accelerated photobleaching reaction. Among other things, these results suggest that the substituents on the nitrogen atom of the benzothiazole rings are important for the formation of a dye–PNA–DNA complex and are especially important for the formation of the complex that causes the accelerated photobleaching of the dye in the presence of genomic DNA and/or for the susceptibility of the “un-PNA–DNA-complexed”-dye to singlet oxygen oxidation.
Thus, DiSC3(3), DiSC4(3), and DiSC5(3) appear to bind to the PNA–DNA oligomer duplex as aggregates. However, under the conditions we tested, there is minimal accelerated photobleaching reaction with these dyes in the presence of PNA probe and M. tuberculosis genomic DNA. This might be due to several factors, including (1) the interaction affinity of dye to PNA–genomic DNA, (2) the structure of the dye–PNA–DNA complex and its efficiency at singlet oxygen generation, (3) the energy level of the excited states of dye molecules in the complex, and/or (4) the susceptibility of the dye to oxidation.
Rapid Dye Mediated PNA–DNA Binding
PNA invasion of duplex DNA, which requires breaking the Watson–Crick hydrogen bonding of duplex DNA, is a very slow and unfavorable process.44 Conjugation of acridine to a PNA,45 or bis-PNA in the presence of quinoxaline antibiotics46 has been reported to improve duplex DNA invasion. However, in the smartDNA system with PNA probes that have been empirically selected, a sequence specific PNA–genomic DNA complex is formed at room temperature in less than 10 min. DNA from other organisms has been tested with TB14, the M. tuberculosis specific PNA probe, and shows no accelerated photobleaching (Figure 3). Whether this complex is formed solely through a modified Watson–Crick hybridization process involving the presence of the cyanine dye or some completely different mechanism of interaction is under active investigation.
Proposed Mechanism
Figure 5 shows the proposed mechanism for the overall process of the singlet oxygen mediated accelerated photobleaching. The presented spectral evidence suggests that a complex containing an H aggregate of the dye forms when complementary PNA and DNA sequences bind to each other. PNA–DNA hybrids run out into a gel and stained with the dye exhibit photobleaching only where the hybrid migrates (Figure 2). This is consistent with the notion that the dye–PNA–DNA complex is required for photobleaching. Removing oxygen from a reaction solution of the dye slows its photobleaching. Azide also quenches the photobleaching reaction, apparently catalyzed by the complex. The structure of the characterized product produced from photobleaching free dye in solution is consistent with the involvement of singlet oxygen photooxidation of the bulk dye present in solution. Moreover, during this proposed 1O2 mediated process, the DNA might undergo oxidation, partially supported by the observation that readdition of PNA and dye after the photobleaching reaction did not resume the reaction.
The simplest mechanism is that the DiSC2(3)–PNA–DNA complex generates singlet oxygen when exposed to 470 nm light. Once formed, the singlet oxygen is free to diffuse into solution and causes the oxidation of the great excess (>99%) of free dye that is in solution and likely dye that is associated with DNA. Thus, the reaction rate should be directly dependent on the concentration of the dye–PNA–DNA catalyst present. Because the DNA target is the limiting reagent and all other reactants are present in about 105–106-fold excess, the predicted rate of photobleaching should be proportional to the DNA target concentration. This is observed experimentally.
CONCLUSION AND UTILITY OF SMARTDNA
The sequence specific accelerated photobleaching reaction of DiSC2(3) in the presence of the PNA probe and genomic DNA target forms the very interesting and previously unreported photochemical basis for a sensitive, rapid, simple, and potentially inexpensive method for DNA detection. The accelerated photobleaching is induced with an inexpensive LED and detected with a simple photodiode circuit. Measurement of the dye photobleaching rate in solution enables sensitive inexpensive DNA determinations to be performed. The method takes advantage of PNA’s natural high affinity and specificity for DNA. This is further improved by the discovery of a rapid room temperature binding process that occurs in the presence of the dye.
When combined with suitable sample processing and activating-reading hardware, the smartDNA technology could be used to produce sensitive, rapid, and potentially inexpensive diagnostic assays. Because of its inherent simplicity, the smartDNA detection is ideally suited for rapid diagnostics of infectious disease in resource constrained or point of care settings. To that end, assay development work is proceeding on applying the method to the detection of M. tuberculosis.
Supplementary Material
Acknowledgments
Dr. Bruce Armitage at Carnegie Mellon University for conversations regarding a proposed pathway, Dr. Andrew Woolley at University of Toronto for help obtaining the CD spectra, Dr. Peter E. Nielsen at Panum Institute, University of Copenhagen, Denmark, for helpful discussions, Dr. Maxim Frank-Kamenetskii at Boston University for helpful discussions, and Dr. Yuriy Fofanov and Dr. Catherine Putonti at University of Houston for the in silico validation of the TB14 sites are all acknowledged. The project described was supported by Grant Numbers R43AI069574 and R44AI069574 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nielsen PE, Egholm M, Berg RH, Buchardt O. Science. 1991;254(5037):1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 2.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. Nature. 1993;365(6446):566–8. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 3.Kuhn H, Demidov VV, Coull JM, Fiandaca MJ, Gildea BD, Frank-Kamenetskii MD. J Am Chem Soc. 2002;124(6):1097–103. doi: 10.1021/ja0041324. [DOI] [PubMed] [Google Scholar]
- 4.Jensen KK, Orum H, Nielsen PE, Norden B. Biochemistry. 1997;36(16):5072–7. doi: 10.1021/bi9627525. [DOI] [PubMed] [Google Scholar]
- 5.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen PE, Norden B, Gralslund A. J Am Chem Soc. 1996;118:5544–52. [Google Scholar]
- 6.Weiler J, Gausepohl1 H, Hauser N, Jensen ON, Hoheisel JD. Nucleic Acids Res. 1997;25:2792–9. doi: 10.1093/nar/25.14.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukeman PS, Mittal AC, Seeman NC. Chem Commun (Cambridge, UK) Vol. 15. 2004. pp. 1694–5. [DOI] [PubMed] [Google Scholar]
- 8.Larsen HJ, Bentin T, Nielsen PE. Biochim Biophys Acta. 1999;1489:159–66. doi: 10.1016/s0167-4781(99)00145-1. [DOI] [PubMed] [Google Scholar]
- 9.Nulf CJ, Corey D. Nucleic Acids Res. 2004;32:3792–8. doi: 10.1093/nar/gkh706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demidov VV, Bukanov NO, Frank-Kamenetskii MD. Curr Issues Mol Biol. 2000;2(1):31–5. [PubMed] [Google Scholar]
- 11.Orum H, Nielsen PE, Egholm M, Berg RH, Buchardt O, Stanley C. Nucleic Acids Res. 1993;21(23):5332–6. doi: 10.1093/nar/21.23.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohse J, Dahl O, Nielsen PE. Proc Natl Acad Sci USA. 1999;96:11804–8. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaihatsu K, Braasch DA, Cansizoglu A, Corey DR. Biochemistry. 2002;41(37):11118–25. doi: 10.1021/bi0263659. [DOI] [PubMed] [Google Scholar]
- 14.Demidov VV, Protozanova E, Izvolsky KI, Price C, Nielsen PE, Frank-Kamenetskii MD. Proc Natl Acad Sci USA. 2002;99:5953–5958. doi: 10.1073/pnas.092127999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smolina IV, Demidov VV, Soldatenkov VA, Chasovskikh SG, Frank-Kamenetskii MD. Nucleic Acids Res. 2005;33:e146. doi: 10.1093/nar/gni151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kielkopf CL, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, Rees DC. Science. 1998;282:111–5. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- 17.Wemmer DE. Ann Rev Biophys Biomol Struct. 2000;29:439–61. doi: 10.1146/annurev.biophys.29.1.439. [DOI] [PubMed] [Google Scholar]
- 18.Glazer AN, Rye HS. Nature. 1992;359(6398):859–61. doi: 10.1038/359859a0. [DOI] [PubMed] [Google Scholar]
- 19.Rye HS, Glazer AN. Nucleic Acids Res. 1995;23(7):1215–22. doi: 10.1093/nar/23.7.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biver T, De Biasi A, Secco F, Venturini M, Yarmoluk S. Biophys J. 2005;89(1):374–83. doi: 10.1529/biophysj.105.059790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannah KC, Gil RR, Armitage BA. Biochemistry. 2005;44(48):15924–9. doi: 10.1021/bi051298e. [DOI] [PubMed] [Google Scholar]
- 22.Mikheikin AL, Zhuze AL, Zasedatelev AS. J Biomol Struct Dyn. 2000;18(1):59–72. doi: 10.1080/07391102.2000.10506647. [DOI] [PubMed] [Google Scholar]
- 23.Yarmoluk SM, Lukashov SS, Ogul’Chansky TY, Losytskyy MY, Kornyushyna OS. Biopolymers. 2001;62(4):219–27. doi: 10.1002/bip.1016. [DOI] [PubMed] [Google Scholar]
- 24.Zipper H, Brunner H, Bernhagen J, Vitzthum F. Nucleic Acids Res. 2004;32(12):e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waggoner A. Curr Opin Chem Biol. 2006;10(1):62–6. doi: 10.1016/j.cbpa.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Antony T, Subramaniam VJ. Biomol Struct Dyn. 2001;19(3):497–504. doi: 10.1080/07391102.2001.10506757. [DOI] [PubMed] [Google Scholar]
- 27.Yin JL, Shackel NA, Zekry A, McGuinness PH, Richards C, Putten KV, McCaughan GW, Eris JM, Bishop GA. Immunol Cell Biol. 2001;79(3):213–21. doi: 10.1046/j.1440-1711.2001.01002.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee LG, Chen C, Liu LA. Cytometry. 1986;7:508–17. doi: 10.1002/cyto.990070603. [DOI] [PubMed] [Google Scholar]
- 29.Renikuntla BR, Rose HC, Eldo J, Waggoner AS, Armitage BA. Org Lett. 2004;6(6):909–12. doi: 10.1021/ol036081w. [DOI] [PubMed] [Google Scholar]
- 30.Matsui M, Kawamura S, Shibata K, Muramatsu H. Bull Chem Soc Jpn. 1992;65(1):71–4. [Google Scholar]
- 31.Wittung P, Kim SK, Buchardt O, Nielsen PE, Norden B. Nucleic Acids Res. 1994;22(24):5371–7. doi: 10.1093/nar/22.24.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith JO, Olson DA, Armitage BA. J Am Chem Soc. 1999;121:2686–95. [Google Scholar]
- 33.Wilhelmsson LM, Norden B, Mukherjee K, Dulay MT, Zare RN. Nucleic Acids Res. 2002;30(2):e3. doi: 10.1093/nar/30.2.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M, Armitage BA. Methods Mol Biol. 2002;208:131–42. doi: 10.1385/1-59259-290-2:131. [DOI] [PubMed] [Google Scholar]
- 35.Wang M, Dilek I, Armitage BA. Langmuir. 2003;19:6449–55. [Google Scholar]
- 36.West W, Pearce S. J Phys Chem. 1965;69:1894–903. [Google Scholar]
- 37.Chibisov AK, Zakharova GV, Görner H. Phys Chem Chem Phys. 1999;1:1455–60. [Google Scholar]
- 38.Seifert JL, Connor RE, Kushon SA, Wang M, Armitage BA. J Am Chem Soc. 1999;121(13):2987–95. [Google Scholar]
- 39.McRae EG, Kasha M. J Chem Phys. 1958;28:721. [Google Scholar]
- 40.Armitage B. Top Curr Chem. 2005;253:55–76. [Google Scholar]
- 41.Dilek I, Madrid M, Singh R, Urrea CP, Armitage BA. J Am Chem Soc. 2005;127:3339–45. doi: 10.1021/ja045145a. [DOI] [PubMed] [Google Scholar]
- 42.Santos PF, Reis LV, Almeida P, Serrano JP, Oliveira AS, Vieira Ferreira LF. J Photochem Photobiol, A: Chem. 2004;163:267–9. [Google Scholar]
- 43.Hoebeke M, Seret A, Piette J, van de Vorst A. J Photochem Photobiol, B. 1988;1(4):437–46. doi: 10.1016/1011-1344(88)85004-8. [DOI] [PubMed] [Google Scholar]
- 44.Demidov VV, Yavnilovich MV, Belotserkovskii BP, Frank-Kamenetskii MD, Nielsen PE. Proc Natl Acad Sci USA. 1995;92:2637–41. doi: 10.1073/pnas.92.7.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bentin T, Nielsen PE. J Am Chem Soc. 2003;125(21):6378–9. doi: 10.1021/ja029936t. [DOI] [PubMed] [Google Scholar]
- 46.Møllegaard NE, Bailly C, Waring MJ, Nielsen PE. Biochemistry. 2000;39(31):9502–7. doi: 10.1021/bi000254x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.