Abstract
Using two-colour imaging and high resolution TIRF microscopy, we investigated the assembly and maturation of nascent adhesions in migrating cells. We show that nascent adhesions assemble and are stable within the lamellipodium. The assembly is independent of myosin II but its rate is proportional to the protrusion rate and requires actin polymerization. At the lamellipodium back, the nascent adhesions either disassemble or mature through growth and elongation. Maturation occurs along an α-actinin–actin template that elongates centripetally from nascent adhesions. α-Actinin mediates the formation of the template and organization of adhesions associated with actin filaments, suggesting that actin crosslinking has a major role in this process. Adhesion maturation also requires myosin II. Rescue of a myosin IIA knockdown with an actin-bound but motor-inhibited mutant of myosin IIA shows that the actin crosslinking function of myosin II mediates initial adhesion maturation. From these studies, we have developed a model for adhesion assembly that clarifies the relative contributions of myosin II and actin polymerization and organization.
Adhesion assembly and turnover are highly dynamic and coordinated processes essential for cell migration1,2. Adhesions serve as traction points for cell translocation and mediate a network of signalling events that regulate protrusion, contractility and attachment1–7. Although much is known about the functions of adhesions in developing and responding to traction and the signalling networks they regulate, less is known about the mechanisms by which adhesions assemble and turnover.
In migrating cells, protrusions are generated by actin polymerization at the front1,8. Protrusions consist of two structurally and kinetically distinct actin networks9,10. The lamellipodium comprises a treadmilling network of Arp2/3-mediated, branched actin filaments, whereas the lamellum consists of actin filament bundles9,10. The location and mechanism of adhesion assembly are unclear. Recent evidence suggests that adhesions form at the base of the lamellipodium11,12 in response to waves of actomyosin-generated force and halted protrusive activity12. However, periodic contractions are not observed in all cells13,14, particularly highly protrusive or rapidly migrating cells, suggesting alternative mechanisms of adhesion assembly.
Adhesions are thought to mature by a sequential mechanism coupled to tension or myosin II activity12,15. Inhibition of several signalling components, including FAK, Src and ERK kinases, stops adhesion turnover and promotes maturation1, suggesting a role for phosphorylation-mediated affinity changes of adhesion components. However, contractile force has also emerged as a major regulator. Application of force induces adhesion growth, whereas inhibition of actomyosin contractility decreases adhesion size16–18. Myosin II is also an endpoint of the pathways regulated by Rho GTPases, which are downstream hubs for migration-related signalling pathways19,20.
Our goals in this study were to define the steps and mechanisms underlying the early events in adhesion formation during migration and determine the role of the actin cytoskeleton and myosin II contractility in these processes. These studies were facilitated by the identification of small adhesions near the leading edge of motile cells6,14,21,22 and rapid two-colour TIRF microscopy. We show that nascent adhesions assemble in the lamellipodium, are stable only within the lamellipodial dendritic actin network, and do not require myosin II activity. These adhesions grow and elongate at the lamellipodium–lamellum interface along an actin–α-actinin template. Finally, we show that the actin crosslinking property of myosin II contributes prominently to adhesion maturation. On the basis of these observations, we developed a model of adhesion assembly that provides a conceptual framework for the formation of adhesions in protrusions.
RESULTS
Nascent adhesions assemble and turnover in discrete phases
We and others have reported that small, dynamic adhesions are present near the leading edge of protrusions in motile cells6,14,21,22. These structures colocalize with integrins (Fig. 1a), contain molecules commonly associated with adhesions, contain phosphorylated FAK (Tyr 397) and paxillin (Tyr 31), and associate closely with the substratum6,14,21. They also follow the edge of the protrusion as it moves forward. To determine whether the adhesions were sliding outward during protrusion or undergoing rapid assembly and disassembly (turnover), we observed them at high temporal and spatial resolution using TIRF microscopy.
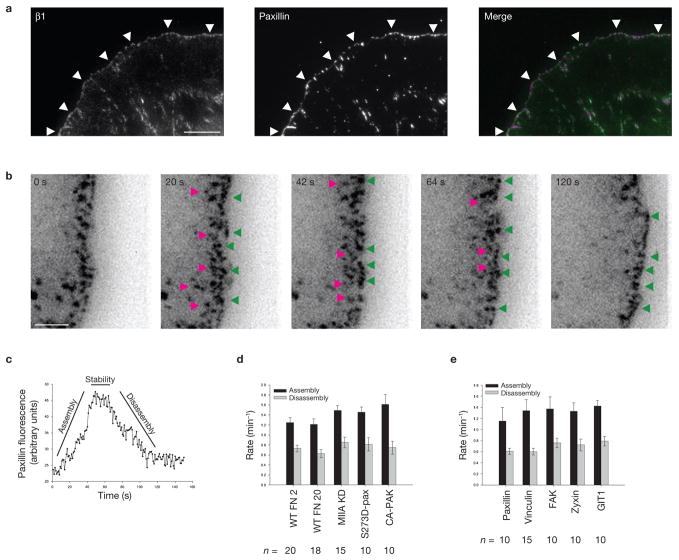
Figure 1.
Rapid turnover of nascent adhesions during protrusion. (a) Expressed human β1 integrin (green) and endogenous paxillin (magenta) imaged using TIRF optics in CHO.K1 cells. Both localize in nascent adhesions that form a thin outline near the leading edge (arrowheads). (b) The rapid assembly (green arrowheads) of new nascent adhesions near the leading edge as the pre-existing adhesions disassemble behind them (pink arrowheads), observed from TIRF time-lapse images of paxillin–GFP in CHO.K1 cells. These panels correspond to Supplementary Information, Movie 1. (c) Temporal fluorescence intensity profile of paxillin–GFP in a representative nascent adhesion. The lifetime of the nascent adhesion can be resolved into three discrete phases: assembly, stability and disassembly. (d) Rate constants for nascent adhesion assembly and disassembly. Nascent adhesions assemble and turnover at comparable rates independently of fibronectin (FN) concentration adsorbed at a concentration of 2 μg ml−1 and 20 μg ml−1) and protrusive phenotype (MIIA knockdown, expression of S273D–paxillin, expression of CA-PAK). Data are mean ± s.e.m., measured from 10–20 individual adhesions in 4–8 cells from independent experiments. (e) Rate constants for the assembly and disassembly of adhesion molecules in nascent adhesions. All molecules entered and exited the nascent adhesions simultaneously and at comparable rates. Data are mean ± s.e.m., measured from 10–15 individual adhesions in 4–6 cells from independent experiments. Scale bars are 10 μm (a) and 3 μm (b).
CHO.K1 cells expressing paxillin–mEGFP and plated onto fibronectin (2 μg ml−1) generated broad protrusions with an array of small, punctate adhesions near the leading edge (Fig. 1b; Supplementary Information, Movie 1). These adhesions were stationary relative to the substratum and disassembled as new, nascent adhesions formed in front of them. As a result, the periphery of active protrusions was always decorated with small adhesions undergoing continuous turnover.
The kinetics of formation and disassembly of these adhesions were quantified by measuring the fluorescence intensity for marker proteins, such as paxillin or vinculin1. Using a frame rate of 1 s and correcting for photobleaching, the kinetics revealed three distinct phases: assembly, stability and disassembly. The average life of these adhesions was 76.1 ± 22.0 s (Fig. 1b) and the rates of assembly and disassembly were 1.26 ± 0.10 min−1 and 0.73 ± 0.07 min−1, respectively (Fig. 1d). Higher fibronectin concentrations and highly protrusive phenotypes 6,14 had negligible effects on these rates.
The transient stability (11.8 ± 6.2 s) suggests that nascent adhesions do not grow indefinitely but instead stop at a target state. In support of this hypothesis, the relative intensity of the adhesions within a region peaked and remained at this plateau until disassembly. The nascent adhesions also reached a plateau at a common size of 0.19 ± 0.01 μm2, which is close to the diffraction limit, suggesting that their true size may be smaller than their observed size.
Finally, all components studied so far enter and leave nascent adhesions simultaneously. Paxillin, vinculin, FAK, G protein-coupled receptor kinase-interacting protein 1 (GIT1) and zyxin, for example, showed indistinguishable relative assembly and disassembly kinetics (Fig. 1e). This suggests that they enter and exit nascent adhesions either as preformed clusters or individually in response to a common or kinetically indistinguishable event(s).
Formation and turnover of nascent adhesions do not require myosin II
Recently, we reported that knockdown of myosin II by RNA interference (RNAi) produces protrusions that have only a rim of small adhesions near the leading edge15. This prompted us to investigate whether nascent adhesions can form in the absence of myosin II activity. Knockdown of myosin IIA (MIIA) resulted in adhesions near the leading edge that turned over continuously, showing the same three discrete phases described above for nascent adhesions in wild-type cells. The average life was 71.3 ± 21.5 s, with assembly and disassembly rates of 1.49 ± 0.10 min−1 and 0.85 ± 0.11 min−1, respectively, a stability time of 12.2 ± 6.4 s, and an average size of 0.20 ± 0.01 μm2. These properties are comparable to nascent adhesions from wild-type cells, indicating that they are similar, if not identical, adhesions.
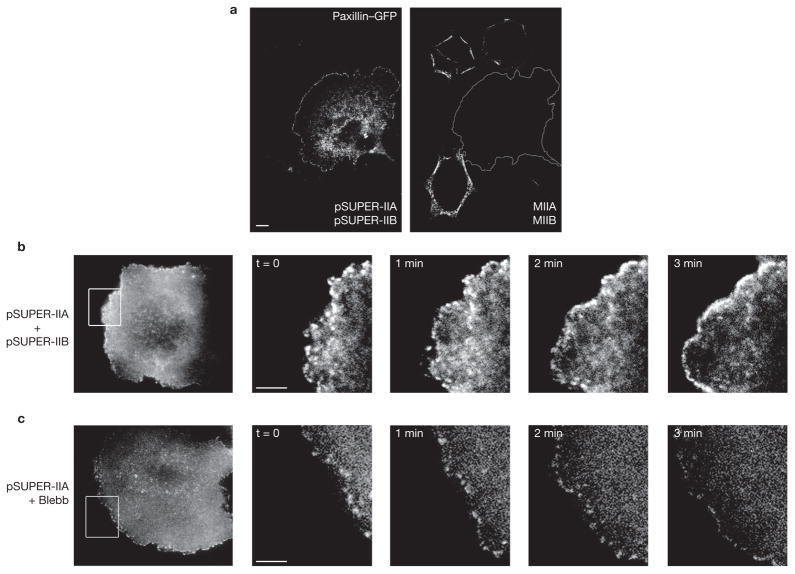
To demonstrate more rigorously that nascent adhesions can form without myosin II activity, we generated double RNAi knockdown of MIIA and MIIB. More than 90% of the total myosin II expression was inhibited, as assessed by residual fluorescence intensity in analysed cells (Fig. 2a). The cells no longer had large, stable adhesions in central regions, but the protrusions still contained small, dynamic adhesions at the periphery (Fig. 2b). We also treated MIIA knockdown cells with blebbistatin (an inhibitor of myosin II ATPase activity) to inhibit residual activity of MIIA and MIIB, the other isoform present in these cells. Blebbistatin-treated MIIA knockdown cells retained a rim of dynamic adhesions near the leading edge (Fig. 2c). Their life (74.1 ± 28.2 s) and size (0.22 ± 0.01 μm2) were comparable to those of both wild-type and myosin II knockdown cells. Finally, wild-type or MIIA knockdown cells plated in the presence of blebbistatin also had nascent adhesions near the leading edge of protrusions (data not shown). Taken together, these data show that the formation and turnover of the small, dynamic adhesions near the leading edge are independent of myosin II.
Figure 2.
Myosin II inhibition promotes nascent adhesion assembly and inhibits adhesion maturation. (a) CHO.K1 cells were transfected with pSUPER vector alone (control) or pSUPER-MIIA and pSUPER-MIIB. The cells were simultaneously stained for MIIA and MIIB. The reduced expression is evident in the transfected cell, which is silhouetted to show its outline. (b) Time-lapse TIRF images of a MIIA/MIIB-depleted cell expressing paxillin–mOrange. Images are representative of more than 10 cells from four independent experiments. (c) Time-lapse TIRF images of paxillin–mOrange in a MIIA-depleted cell treated with blebbistatin (20 μM, Blebb). Note the rim of nascent adhesions at the periphery. Scale bars are 10 μm (a) and 5 μm (b, c).
Finally, we compared the nascent adhesions with focal complexes, which are small adhesions at the cell periphery induced by constitutively activated (CA) Rac23. The focal complexes were about two times larger than nascent adhesions (data not shown), appeared mainly at the lamellipodium–lamellum interface (Supplementary Information, Fig. S1a) and did not turn over. Moreover, addition of blebbistatin induced disassembly of focal complexes into smaller adhesions that are indistinguishable from nascent adhesions (Supplementary Information, Fig. S1b). Thus, CA Rac-induced focal complexes are myosin II-dependent24 and distinct from the smaller, myosin II-independent nascent adhesions. This also shows that not all punctate, peripheral adhesions are nascent adhesions.
Nascent adhesions assemble and are stable in the lamellipodium
We next investigated where nascent adhesions form and what determines their lifespan. The lamellipodium was identified as the dense, 1–3-μm actin-rich band near the leading edge that contains barbed-end actin, Arp2/3 and cofilin10,11 (Supplementary Information, Fig. S2). When paxillin–mOrange (or vinculin–mOrange) and GFP–actin were co-expressed, time-lapse TIRF microscopy revealed that the nascent adhesions form and reside in the lamellipodium (Fig. 3a; Supplementary Information, Movie 2).
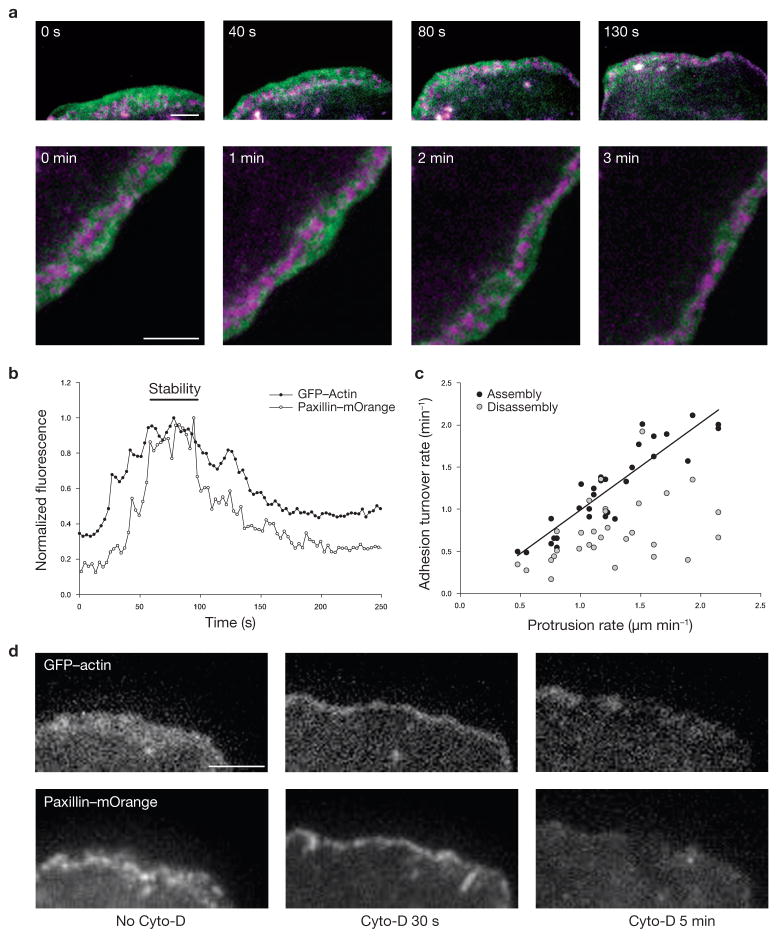
Figure 3.
Nascent adhesions reside in the lamellipodium. (a) Time-lapse TIRF images of GFP–actin (green) and paxillin–mOrange (magenta) show nascent adhesions forming and residing exclusively in the lamellipodium. These panels correspond to Supplementary Information, Movie 2. (b) Dual-colour temporal profiles of GFP–actin of the lamellipodium and paxillin–mOrange in a representative nascent adhesion. Relative changes in fluorescent intensities show that nascent adhesions begin to assemble after actin appears in the position of the adhesions (that is, assembly occurs within the protruding lamellipodium). Next, the stability phase correlates with residence time of the adhesion in the lamellipodium. Finally, the intensity of paxillin decreases simultaneously with that of actin, indicating that nascent adhesions turnover when the lamellipodium (dense actin band) moves by the adhesions. (c) Comparison of protrusion rate with assembly and disassembly rates of nascent adhesions from cells on fibronectin (2 μg ml−1). Each data point represents the kinetics of one adhesion and its adjacent cell periphery, which were quantified by measuring temporal fluorescent intensities of paxillin–GFP (nascent adhesion) and cytoplasmic mCherry (protrusion). The analyses were performed during the time of nascent adhesion assembly/disassembly. Regression line shows a linear correlation between the rates of assembly and protrusion. Rates were measured from 30 individual adhesions and protrusions in 11 cells from independent experiments. (d) Application of cytochalasin-D (1 μM) halts the protrusion and constricts the lamellipodium immediately (30 s). Minutes later (5 min), the thick band of GFP–actin characterizing the lamellipodium collapses, and the nascent adhesions residing in the band, indicated by paxillin–mOrange, disassemble. Scale bars are 3 μm (a, d)
Dual-colour imaging showed that the fluorescence intensity of paxillin (or vinculin) increased 10–35 s after the actin began accumulating in the lamellipodium (Fig. 3b) but not at its outer edge. After assembly, the nascent adhesions were transiently stable for 11.8 ± 6.2 s, which correlates with the time a fully assembled adhesion resides within the boundaries of the lamellipodium. Moreover, the adhesions began to disassemble at the rear of the lamellipodium as the dense actin band passed by them (Fig. 3a, b; Supplementary Information, Movie 2).
To determine whether the rates of nascent adhesion assembly, disassembly and protrusion were coupled, we compared the rate of increase or decrease in paxillin–mEGFP fluorescence in nascent adhesions with the rate of leading edge extension adjacent to the adhesions during phases of protrusion. The protrusion rates varied 3–5-fold within and among cells. The rate of adhesion assembly was linearly proportional to the local rate of leading edge extension (Fig. 3c). In contrast, the rate of adhesion disassembly was not affected by the speed of protrusion, indicating that the kinetics of adhesion disassembly and actin polymerization are not mechanistically linked. Interestingly, the relationship between the adhesion assembly and local protrusion rates was sustained under a variety of conditions, including plating in high fibronectin concentrations and myosin II inhibition, where retrograde flow is inhibited (data not shown). Taken together, these data show that the rate of nascent adhesion formation is directly coupled to the rate of protrusion and suggest that there is a link between nascent adhesion assembly and actin polymerization in the lamellipodium.
To ascertain whether these adhesions required dendritic actin for their stability, we disrupted the lamellipodium with cytochalasin-D, which caps barbed ends and inhibits actin polymerization. Addition of cytochalasin-D (1 μM) inhibited protrusion immediately; no new adhesions formed and those remaining in the lamellipodium were stabilized. However, as the dense actin band characterizing the lamellipodium narrowed and collapsed, the adhesions in the back disassembled as the band passed behind them (Fig. 3d). This shows that continued adhesion assembly requires actin polymerization, that these adhesions are only stable within the lamellipodium and that the adhesions disassemble when the dendritic actin of the lamellipodium moves past them.
Adhesions mature along an actin–α-actinin template
Whereas some nascent adhesions diassemble as the lamellipodium moves past them, others mature by growth and elongation at the lamellum–lamellipodium interface during pauses in protrusion (Fig. 4a; Supplementary Information, Movie 3). These maturing adhesions arise from the nascent adhesions (Supplementary Information, Movie 4). The oriented, centripetal nature of their growth suggests that the adhesions mature along a structural track or template. To investigate this, we imaged CHO.K1 cells expressing GFP–actin. We observed small, linear actin filaments emanating centripetally from the halted protrusion (Fig. 4a; Supplementary Information, Movie 5). This thin line of actin was most readily observed with a promoter-truncated GFP–actin vector25 that results in low expression levels, minimizing cytoplasmic background, suggesting that these structures consist of only a few actin filaments. Dual-colour imaging using GFP–actin and paxillin–mOrange revealed that short actin filaments seemed to grow from the nascent adhesions, which subsequently underwent elongation, raising the possibility that they arise from adhesion-associated actin polymerization (Fig. 4b; Supplementary Information, Movie 6). Dual-colour imaging of cells expressing GFP–α-tubulin and paxillin–mOrange did not show any colocalization of microtubules with the elongating adhesions (data not shown).
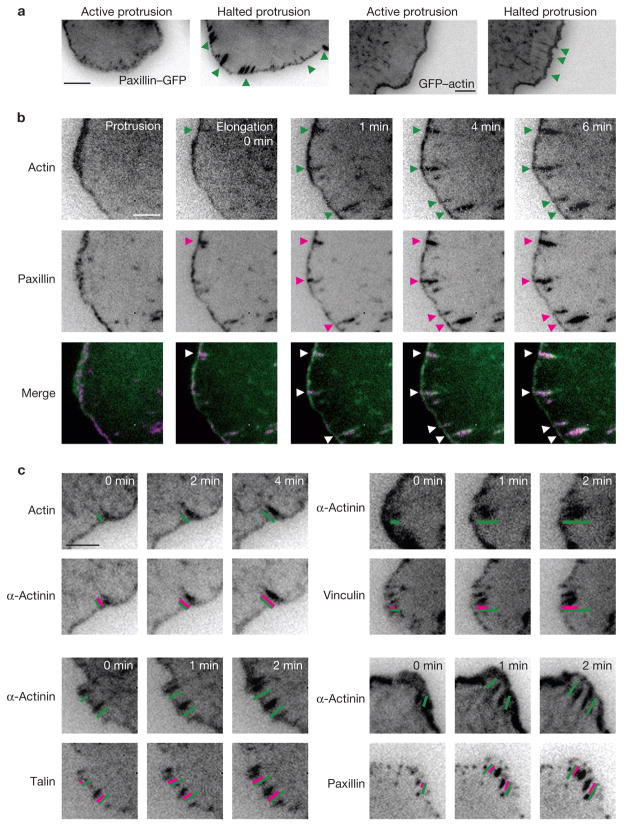
Figure 4.
An actin template organizes and promotes hierarchical adhesion maturation. (a) TIRF images of paxillin–GFP and GFP–actin exhibiting centripetal elongation when protrusion halts (green arrowheads). The linear structures emerged from the cell edge. These panels correspond to Supplementary Information, Movies 3 and 5. (b) Time-lapse TIRF images of GFP–actin (green) and paxillin–mKO (magenta) show elongating actin templates and elongating adhesions, respectively. During protrusion, nascent adhesions reside in the lamellipodium. Once the leading edge halts, a population of the adhesions matures along the growing actin filaments (arrowheads). These panels correspond to Supplementary Information, Movie 6. (c) Time-lapse TIRF images of actin and α-actinin shows simultaneous elongations from the cell periphery. Vinculin, talin and paxillin incorporation was delayed, compared with α-actinin. Inset bars indicate the degree of elongation (green, top panels; magenta, bottom panels). Scale bars are 5 μm (a) and 3 μm (b, c).
To determine the mechanism of adhesion maturation, we first compared the relative kinetics of elongation using GFP- and mOrange- or mCherry-labelled pairs of some core adhesion molecules. These studies revealed a hierarchy of entry into the elongating adhesion. α-Actinin and actin elongated simultaneously (Fig. 4c). Paxillin and talin entered after actin and α-actinin but slightly before vinculin, suggesting that vinculin entry into elongating adhesions requires the pre-assembly and/or activation of another adhesion component(s) (Fig. 4c). Tensin entered the adhesions after vinculin and its concentration increased as the adhesion matured further (data not shown). Finally, MIIA approached elongating actin filaments from the central part of the cell and linked up with the filaments as they elongated and thickened (Supplementary Information, Movie 7).
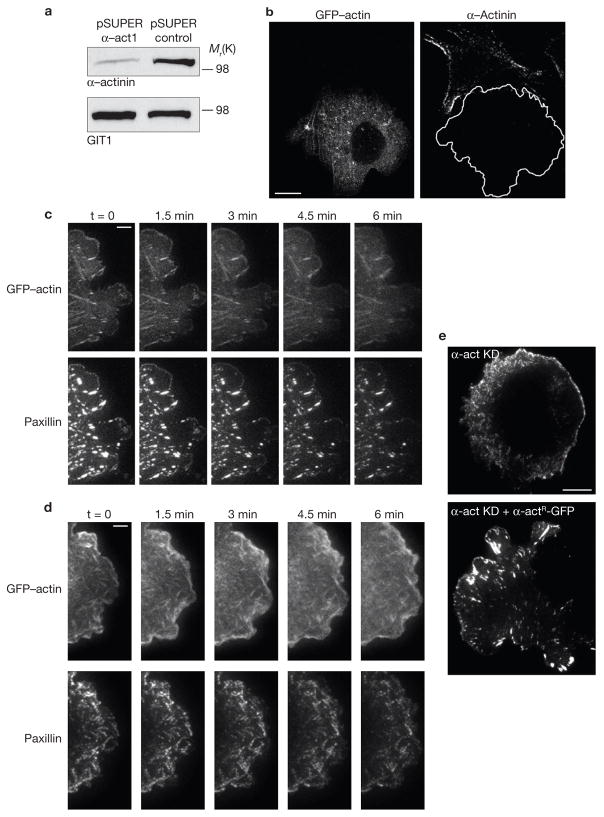
The simultaneous, initial accumulation of α-actinin and actin at sites of adhesion elongation points to a key role for α-actinin in the maturation process. To investigate this, we inhibited α-actinin expression in CHO.K1 cells using RNAi. Immunoblots revealed that the RNAi efficiency was close to 85% (Fig. 5a), similar to the transfection efficiency. Immunofluorescence microscopy showed that knockdown cells expressed undetectable amounts of α-actinin (Fig. 5b). The protrusion rate in α-actinin-deficient cells was reduced and discontinuous (Supplementary Information, Fig. 3). The actin filament bundles in protrusions no longer showed a centripetal orientation, and instead, were short and more randomly oriented (Fig. 5c, d, top panels). Small, punctate adhesions formed near the leading edge but they neither turned over nor matured into larger, elongated adhesions, even when the protrusion was halted (Fig. 5d, e; Supplementary Information, Movie 8). Interestingly, paxillin was distributed in punctate structures along the actin filaments throughout the cell rather than at the ends of large actin bundles (Fig. 5c). Expression of an RNAi-insensitive α-actinin–GFP rescued the phenotype produced by α-actinin knockdown (Fig. 5e and data not shown). These observations indicate that α-actinin has a crucial role in actin organization and adhesion maturation.
Figure 5.
α-actinin knockdown inhibits actin orientation and adhesion elongation in protrusions. (a) Immunoblot of α-actinin in CHO.K1 cells transfected with pSUPER–GFP vector (control) or pSUPER–GFP-RNAi against α-actinin (α-act1). The GIT1 immunoblot was used as a loading control. (b) Representative images of α-actinin-depleted cells stained for α-actinin. The transfected cell is silhouetted. (c, d) Time-lapse TIRF images of control (c) or α-actinin-depleted (d) cells expressing GFP–actin (top) or paxillin–mOrange (bottom). Panels and movies are representative of more than 25 cells in 6 independent experiments. Note the short, mis-oriented actin filaments and dot-like adhesions in α-actinin-depleted cells. Knockdown panels correspond to Supplementary Information, Movie 8. (e) TIRF images of α-actinin-depleted cells (α-act KD, top) and α-actinin-depleted cells rescued with an RNAi-insensitive α-actinin (α-act KD + α-actR–GFP, bottom) expressing paxillin–mOrange. Note the elongated adhesions in the rescued cells. Scale bars are 10 μm (b, e) and 5 μm (c, d).
Actin crosslinking by myosin II and α-actinin mediates adhesion maturation
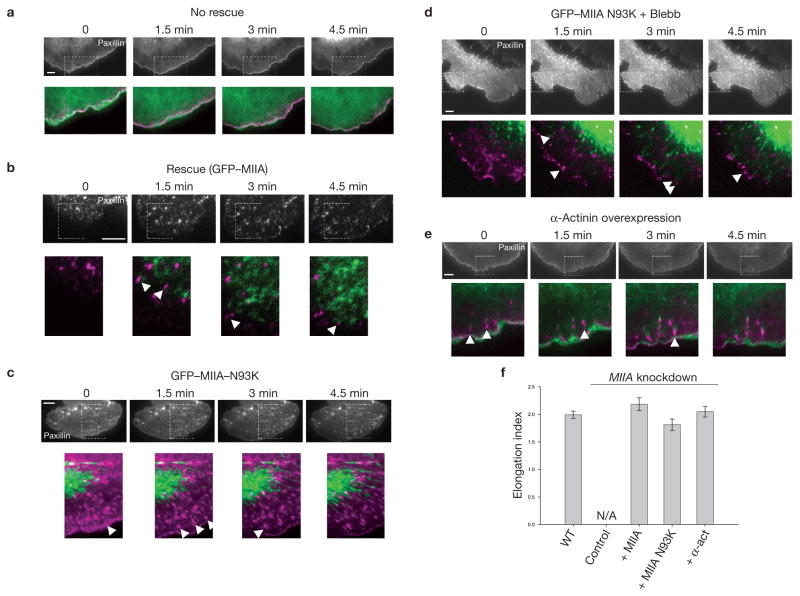
As α-actinin organizes F-actin through its crosslinking activity26, its marked effect on adhesion assembly prompted us to examine whether the crosslinking property of myosin II has a similar role. To separate the contribution of myosin II-dependent actin crosslinking from its contractile activity on adhesion maturation, we expressed paxillin–mOrange in MIIA knockdown CHO.K1 cells together with MIIAN93K27. In this myosin IIA mutant, ATPase activity is inhibited by 90%, and motor activity is inhibited completely27; but it constitutively binds to actin14,28. The adhesions elongated centripetally and were comparable in size and morphology to the control cells rescued with wild-type GFP–MIIA (Fig. 6b, c, f; Supplementary Information, Movie 9). This contrasts with the phenotype in MIIA knockdown cells in which adhesion maturation was inhibited14 (Fig. 6a; Supplementary Information, Movie 9). To inhibit residual MIIB contractility in MIIAN93K-rescued cells, we used blebbistatin, which inhibits the ATPase activity of myosin II but, unlike the mutant, does not sustain strong actin binding. We still observed elongated adhesions in the protrusions (Fig. 6d; Supplementary Information, Movie 10). This suggests that myosin II-mediated actin crosslinking has an important role in adhesion maturation.
Figure 6.
N93K–myosin IIA or overexpressed α-actinin restores adhesion maturation in myosin IIA-deficient cells. (a–e) Time-lapse TIRF images of MIIA-depleted CHO.K1 cells expressing GFP–actin (control, a), GFP–MIIA (rescue, b), GFP–MIIA–N93K (c), GFP–MIIA–N93K with blebbistatin (20 μM, d), and α-actinin–GFP (e). Black and white panels show adhesions as revealed by co-expression of paxillin–mOrange in each case. Colour inserts are magnifications of the indicated regions in the black and white panels. Paxillin–mOrange is depicted in magenta in all cases and green represents GFP–actin (a), GFP–MIIA (b), GFP–MIIA-N93K (c, d) or α-actinin–GFP (e). Arrowheads indicate representative maturing adhesions. Panels a–c correspond to Supplementary Information, Movie 9 and panels d to Supplementary Information, Movie 10. (f) Quantification of the elongation index of maturing adhesions under the different conditions. Data are mean ± s.e.m., measured from more than 25 adhesions from 5–6 cells per condition (n = 30 for each condition). N/A, not applicable (MIIA-deficient cells contain no elongating adhesions). Scale bars are 5 μm.
To further establish the importance of actin crosslinking, we investigated whether overexpressing one of the two crosslinking proteins, α-actinin and myosin II, can compensate for absence of the other. Overexpression of α-actinin–GFP in MIIA-deficient cells induced thick α-actinin bundles (Fig. 6e, green), which is consistent with its actin crosslinking activity29. This treatment also restored growth and centripetal elongation of the adhesions, although not to the levels seen in controls (Fig. 6e, f). In other respects, however, the cells still exhibited properties of the MIIA-deficient phenotype, for example, an inability to retract at the cell rear (data not shown).
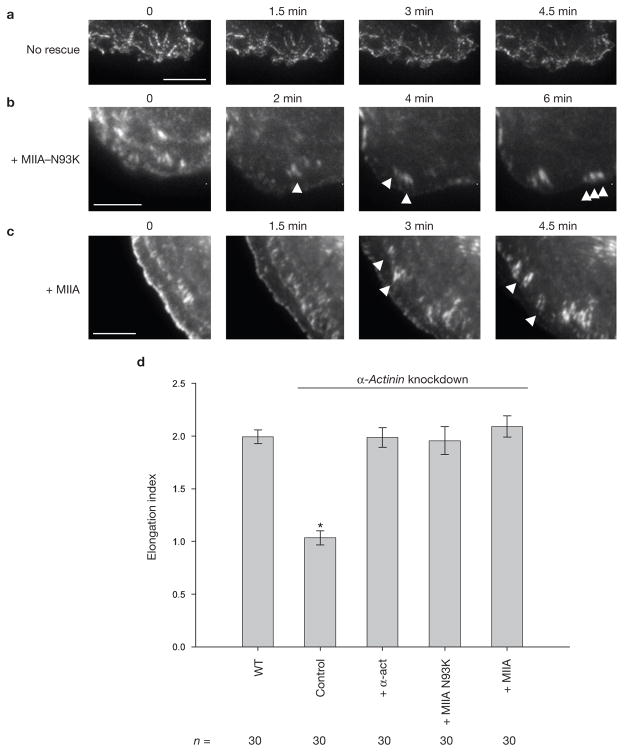
In the reciprocal experiment, overexpression of MIIAN93K, which bundles actin but is not contractile, restored the maturation of adhesions in α-actinin knockdown cells (Fig. 7a, b, d; Supplementary Information, Movie 11). Similar results were observed when wild-type MIIA was overexpressed in the α-actinin knockdown cells (Fig. 7c; Supplementary Information, Movie 11). Together, these observations show that actin crosslinking promotes adhesion maturation.
Figure 7.
Rescue of adhesion maturation in α-actinin-depleted cells by overexpression of MIIA. (a–c) Time-lapse TIRF images of α-actinin-depleted CHO.K1 cells expressing paxillin–mOrange and GFP–actin (a), GFP–MIIA–N93K (b) and GFP–MIIA (c). For convenience, only paxillin is shown. Arrowheads indicate representative maturing adhesions. Scale bars are 5 μm. These panels correspond to Supplementary Information, Movie 11. (d) Quantification of the elongation index of maturing adhesions in the different conditions. Data are mean ± s.e.m., measured from more than 25 adhesions from 5–6 cells per condition; *P = 6 × 10−9, Student’s two-tailed t-test.
DISCUSSION
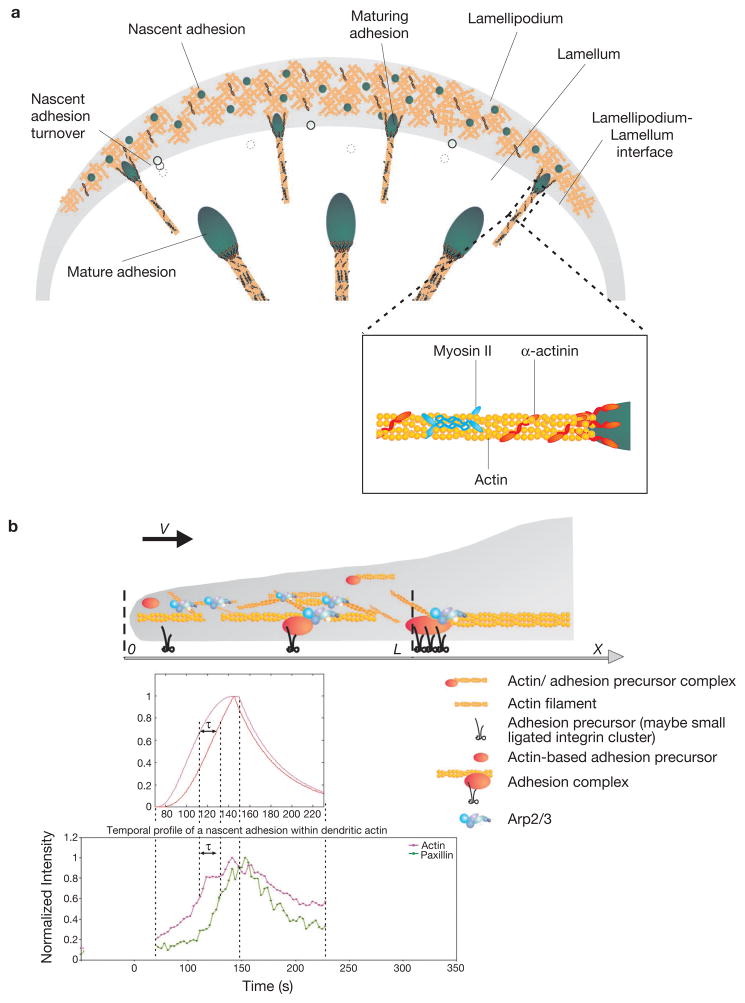
Our observations support a working model for adhesion assembly during cell migration (Fig. 8a). Nascent adhesions assemble in the lamellipodium in a single concerted step as the protrusion advances. The assembly rate is linked to the protrusion rate and probably to actin polymerization. The nascent adhesions are stable only within dendritic actin and disassemble as the wave of depolymerizing actin at the rear of the lamellipodium passes by them with the advancing protrusion. Both the assembly and stability of the nascent adhesions in the lamellipodium are myosin II-independent. When protrusion pauses, a subset of nascent adhesions grow and elongate centripetally from the base of the lamellipodium. The elongation is directed by actin filaments. α-Actinin associates with the emerging actin filaments and organizes and orients them centripetally. These α-actinin–actin filaments function as a template for the hierarchical addition of other adhesion components. The actin crosslinking properties of myosin II also have a major role in the formation of the template and initial stages of adhesion maturation.
Figure 8.
Working model for adhesion assembly, turnover and maturation (a) During protrusion, adhesions initially assemble as punta (blue circle) in the lamellipodium (gray band); their formation is driven by or linked to actin polymerization. Following assembly, these nascent adhesions remain small and stable within the lamellipodium. The nascent adhesions turnover (clear circle) when the depolymerizing dendritic at the rear of the lamellipodium passes by them; this links the stability of these adhesions to the integrity of the dendritic actin. The formation and turnover of nascent adhesions do not require myosin II activity. In addition, nascent adhesions can grow along an actin template (maturing adhesions), which elongates centripetally at the lamellipodium-lamellum interface. The cross-linking activities of both myosin II and α-actinin, possibly in conjunction with contraction, are critical for the initial elongation of adhesions, and α-actinin is also required for the proper positioning of adhesions on actin filaments. Working synergistically with contraction, the cross-linking of actin by myosin II and α-actinin mediate further development and maturation of the adhesions.(b) Quantitative analysis of the mathematical model of actin and adhesion assembly in the lamellipodium predicts accurately the dynamic and exclusive nature of nascent adhesions in the lamellipodium in migrating cells. Top plot, mathematical model; Bottom plot, representative experimental result. V = protrusion rate; L = width of the actin branching zone; X = distance from the front to the rear; τ = time lag. See the Supplementary Information, Materials section for a detailed description of the model, including its assumptions, governing equations, and solutions.
Our data show that nascent adhesions assemble in the lamellipodium as diffraction-limited puncta and then undergo myosin II-dependent maturation near the lamellipodium–lamellum interface. The prominent, readily visualized adhesions (for example, focal complexes reported to be present at the lamellipodium–lamellum interface11,12) differ from nascent adhesions as they are larger, more stable and myosin-II-dependent. The CHO cell is particularly useful for studies of adhesion assembly as the nascent adhesions have long lifespans and only a fraction of them mature. Presumably, this is due in part to lowered or localized myosin II activity. In contrast, nascent adhesions in MEFs or U2OS cells reside in the dendritic actin for only a few seconds (Supplementary Information, Fig. S4, Movie 12 and data not shown); most stabilize and mature at the lamellipodium–lamellum interface. Myosin II is a determining factor as blebbistatin inhibits the probability of adhesion maturation and promotes the assembly and turnover of the nascent adhesions in both cell types (Supplementary Information, Fig. S4 and data not shown). Conversely, MIIA overexpression in CHO.K1 cells inhibits protrusion and increases the probability that nascent adhesions grow and elongate, which occurs almost immediately after they form14 (Supplementary Information, Movie 13). In all of these cells, however, the elongating adhesions in protrusions arise from nascent adhesions. Previous observations also implicate myosin II in adhesion assembly12,20 and the periodic interruption and retraction of protrusions12,14,30.
The formation and stability of nascent adhesions within the lamellipodium, the correlation between the assembly and protrusion rates, and the inhibition of both protrusion and nascent adhesion assembly by cytochalasin-D suggest that adhesion assembly is mechanistically and kinetically linked to actin polymerization in the lamellipodium; this is also observed when net protrusion is driven without myosin II mediated retrograde flow. Two components of nascent adhesions, vinculin and FAK, interact with the Arp2/3 complex, which nucleates actin polymerization within dendritic actin and thereby provide a potential mechanism for the coupling31–33. A recent study showed actin-polymerization-based, protrusion-independent lateral movement of integrins to filopodia-like ripples in the lamellipodium34; however, they did not report a similar mechanism for adhesions outside of ripples.
The lamellipodium–lamellum interface emerges as a critical region where adhesion fate is determined. Nascent adhesions either disassemble or mature as the dendritic actin passes by them. It is also the region where the dendritic actin turns over35, suggesting that nascent adhesions are physically linked to dendritic actin and disassemble in response to its turnover.
We present a mathematical model, which assumes that an adhesion precursor binding to the dendritic actin is a limiting step for adhesion assembly, and that the adhesion disassembly is mechanically coupled to dendritic actin disassembly (Supplementary Information, Text). We also assumed that the Arp2/3 mediated branching takes place near the leading edge, either on adhesions, where the branching points are firmly anchored to the substratum, or in the immediate vicinity of adhesions (so that ‘daughter’ filaments branch off ‘mother’ filaments anchored to the substratum). The solutions of mathematical equations derived from these assumptions reproduced our quantitative observations. That is, as the branching rate is almost constant near the leading edge, total dendritic actin filament length builds up almost linearly from the leading edge towards the rear. Adhesion assembly lags by the characteristic time of the precursor binding to actin behind the front of the actin band. Actin–ATP hydrolysis, cofilin action and decrease of the branching activity behind the leading edge determine the rear of the dendritic actin band, where both actin and adhesions disassemble in synchrony (Fig. 8b). Consistent with our data, the model further predicts that the adhesion assembly rate is proportional to the leading edge extension rate, independent of the disassembly rate from the speed of protrusion, and correlates with the inverse duration of the adhesion stability phase with the speed of protrusion.
Nascent adhesions at the lamellipodium–lamellum interface can also grow and elongate, presumably in response to a changing or different organization of actin. Elongated, centripetal adhesions at the periphery are a hallmark of maturing adhesions22,36. Short, linear actin filaments emanate from this region and provide a template for the maturation of nascent adhesions. These filaments could arise from either the reorganization of existing filaments in the dendritic actin or from local polymerization.
α-Actinin and myosin II are essential for the formation and organization of the actin template. In the absence of α-actinin, actin filaments are abnormally short, discontinuous and misoriented. α-Actinin also positions adhesions along actin filaments as adhesion components in α-actinin knockdown cells are no longer restricted to the ends of actin filaments and appear as puncta spread along the entire filament. α-Actinin has been reported previously to participate in the later stages of adhesion maturation by forming large stress fibres and adhesions at their ends29,37,38. An earlier observation that α-actinin is a late entry into the larger adhesions was made with wide-field optics, which would not have seen the templates and early events described here38.
Myosin II is also required for the growth and elongation of nascent adhesions. Neither actin templates nor the elongation of nascent adhesions are observed in MIIA-deficient or inhibited cells14. Several reports suggest that myosin II-mediated contraction has a major role in the maturation of adhesions by tension-induced alterations in the conformation of adhesion-related proteins14,15,24,39,40. Our study shows that the actin crosslinking activity of myosin II is important in the initial stages of adhesion maturation, presumably by organizing and clustering actin and actin-associated adhesion components. Thus, although myosin II-mediated contractility seems to promote the formation of thick actin filament bundles and large adhesions at later stages of maturation, the contractile activity could function synergistically with myosin II-mediated actin crosslinking at early stages by organizing actin filaments. Myosin II-mediated actin crosslinking can also transmit distally generated actomyosin contractility to adhesions. Others have also ascribed roles for myosin II crosslinking: an ATPase-deficient myosin II restores cortical integrity in Dictyostelium discoideum41; a motor-impaired MIIB mutant rescues hydrocephalus in MIIB knockout mice42; the bundling function of myosin in adhesion assembly was proposed previously43.
In summary, the data presented here provide new insights into the mechanism of adhesion assembly. They identify and characterize a new class of adhesions, ‘nascent adhesions’, which reside in the lamellipodium and serve as precursors for other adhesions in the protrusion. Moreover, they clarify the role of myosin II in adhesion maturation, lead to a ‘template’ model for centripetal adhesion elongation along actin–α-actinin filaments, and demonstrate the importance of the actin-bundling activity of myosin II.
METHODS
Plasmids and antibodies
To generate α-actinin siRNA, the oligonucleotide GGAGATCAATGGCAAATGG, corresponding to nucleotides 2003–2021 of rat α-actinin1 (NM_031005) was inserted into the appropriate pSUPER cassette according to the vector manufacturer’s instructions (Oligoengine). pSUPER-MIIA and pSUPER-MIIB have been described previously14. siRNA-insensitive α-actinin was generated by site-directed mutagenesis (Quickchange kit, Stratagene) introducing two silent mutations (ATC to ATT: Ile to Ile; AAC to AAT: Asn to Asn) in the RNAi target region of human α-actinin1, which shares 100% homology with rat.
Promoter-truncated GFP–actin, GFP–MIIA, GFP–cofilin, GFP–tensin, GFP–zyxin and human β1 integrin cDNA were gifts from Tim Mitchison (Harvard Medical School, Boston, MA)25, Robert Adelstein (National Institutes of Health, Bethesda, MD)44, John Condeelis (Albert Einstein College of Medicine, New York, NY)45, David Brautigan (University of Virginia, Charlottesville, VA)46, Klemens Rottner (Ludwig-Maximilians-Universität München, Germany)47 and Martin Humphries (University of Manchester, UK), respectively. GFP–MIIA-N93K14, paxillin–GFP and α-actinin–GFP38, GFP–vinculin and GFP–FAK and GIT1 have been described previously1. Where indicated, GFP was replaced by mCherry from Roger Tsien48 or CoralHue monomeric Kusabira Orange (mOrange, MBL). Rabbit polyclonal antibodies against MIIA (1:1,000), MIIB (1:1,000) and GIT1 (1:1,000) were obtained from Covance; α-actinin (1:100, mouse, IgG1) from SantaCruz Biotechnology and TS2/16 (β1 integrin, 1mg ml−1) from Biolegend.
Cell culture and transfection
CHO.K1 cells, mouse embryonic fibroblasts and U2OS osteosarcoma cells were cultured under standard conditions and transfected using Lipofectamine (Invitrogen)14. For co-transfection experiments, plasmids containing the siRNA sequences were used in 10:1 excess to GFP or mCherry-containing plasmids to ensure knockdown in fluorescence-positive cells.
Immunofluorescence microscopy
Cells were plated onto fibronectin-coated cover-slips (2 μg ml−1) for 60 min, fixed using 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 for 5 min. Coverslips were incubated with primary antibodies and a species-appropriate secondary antibody coupled to either Alexa488 or Alexa568 (Invitrogen). Barbed-end staining was performed as described elsewhere49.
Microscopy and image processing
Cells were plated on 2 μg ml−1 fibronectin-coated glass-bottomed dishes (migration-promoting conditions) in CCM1 for 1 h and maintained at 37 °C at pH 7.4. Confocal images were collected on an Olympus Fluoview 300 microscope (1.45 NA (oil) PlanApo ×60 TIRFM objective (Olympus)). GFP and mCherry/mOrange were excited using the 488-nm laser line of an Ar ion laser and the 543-nm laser line of a He-Ne laser (Melles Griot), respectively. A Q500LP dichroic mirror (Chroma Technology) was used for GFP-labelled cells. For dual-colour imaging, a green-red cube (488/543/633) with a DM570 dichroic mirror (Chroma Technology) was used. Fluorescence images were acquired using Fluoview software (Olympus).
TIRF images were acquired using an Olympus IX70 inverted microscope (1.45 NA (oil) PlanApo ×60 TIRFM objective), fitted with a Ludl modular automation controller (Ludl Electronic Products) and controlled by Metamorph (Molecular Devices). The excitation laser lines used were as described for confocal microscopy. Mirrors and filters were supplied by Chroma Technology. A dichroic mirror (HQ485/30) was used for GFP-labelled cells. For dual GFP–mCherry/mOrange acquisition, a polychroic mirror (Z488/543rpc) and a dual emission filter (Z488/543) were used. Also, HQ525/50 and HQ620/60 emission filters were used for GFP and mCherry/mOrange, respectively. For simultaneous GFP–mCherry acquisition, Dual-View (MAG Biosystems) was utilized. All images were acquired with a charge-coupled device camera (Retiga Exi; Qimaging) and analysed using Metamorph or ImageJ (NIH).
Quantification of adhesion and protrusion dynamics
ImageJ was used to measure changes in fluorescent intensity of individual nascent adhesions over time in cells expressing fluorescent-tagged adhesion proteins1. Background and photobleaching corrections were applied to obtain true intensities of the adhesions. Assembly and disassembly rates were plotted and calculated using Microsoft Excel (Microsoft Corporation) or SigmaPlot (SPSS)1. Mean lifespan with standard deviation were measured from 30–50 individual adhesions in seven to fourteen cells. The elongation index of maturing adhesions was determined by measuring the long axis of the adhesions (that is, perpendicular to the membrane) and dividing it by the maximal perpendicular axis.
Protrusion was quantified using kymography14. Images were captured every second for 3 min. Kymographs were generated using Metamorph software along 1-pixel-wide regions oriented in the protrusion direction and perpendicular to the leading edge.
Supplementary Material
Figure S1 Activated Rac induced focal complexes differ from nascent adhesions. CHO.K1 cells expressing V12Rac, either without (A) or with 20 μM blebbistatin (B), were co-transfected with GFP-actin (left) and paxillin-mOrange (right). Scale bar = 10 μm. Color inserts show the relative positioning of paxillin-containing focal complexes (A) or nascent adhesions (B) with respect to the lamellipodium. Green, actin; magenta, paxillin. Note that the focal complexes reside the lamellipodium-lamellum interface and disassemble into smaller, lamellipodial localized nascent-like adhesions the presence of blebbistatin.
Figure S2 Molecular markers of the lamellipodium of migrating CHO.K1 cells. Characterization of dendritic actin of the lamellipodia. mRFP-actin (magenta) colocalize with GFP-cofilin and immunostaining of Arp3 (green). Barbed-end actin (green) outlines fluorescent phalloidin (magenta) at the leading edge.
Figure S3 Quantification of protrusion rates in α-actinin-knockdown cells. (A) Kymographs from control (pSUPER) and α-actinin-depleted cells. The knockdown cells were identified by coexpression of a GFP marker, imaged in brightfield, and then analyzed by kymography. (B) Quantification of protrusion rates from kymographs from (A). Average protrusion rates measured from >12 cells (three to five protrusions/cell) from four independent experiments. Error bars represent ±SEM.
Figure S4 The rapid assembly of nascent adhesions in MEFs. TIRF time-lapse images of paxillin-GFP in MEFs show the rapid assembly (green arrows) of new nascent adhesions near the leading edge. Most nascent adhesions undergo immediate maturation and do not turnover (red arrows). Scale bar = 3 μm. Elapsed time is in seconds. TIRF images of GFP-actin (green) and paxillin-mOrange (magenta) show nascent adhesions (green arrows) forming in MEFs and residing exclusively in the lamellipodium. Scale bar = 3μm.
Paxillin-containing nascent adhesions assemble and disassemble rapidly during protrusions. This movie corresponds to Fig. 1A. Images of paxillin-GFP collected every 1 sec using TIRF microscopy. Magenta and green arrows indicate representative assembling and disassembling adhesions, respectively. 10 frames/sec shown.
Rescue of the MIIA knockdown with the non-contractile MIIA mutant N93K and blebbistatin. This movie corresponds to Fig. 6D. Images of paxillin-mOrange collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Rescue of the α-actinin knockdown by the high actin affinity, non-contractile mutant N93K-MIIA or over expression of MIIA wild-type. This movie corresponds to Fig. 7A–C. Images of paxillin-mOrange collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Paxillin-containing nascent adhesions assemble and mature rapidly during protrusions in MEFs. This movie corresponds to Supplementary Fig. 40. Images of paxillin-GFP collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Myosin II regulates the probability and rate of nascent adhesion maturation. Left, MIIA-deficient CHO.K1cell. This movie is from Vicente-Manzanares et al., J. Cell Biol. 176(5)573–580, Suppl. Movie 7; center, wild-type; right, overexpression of MIIA in a CHO.K1 cell. Images of paxillin-GFP were collected every 5 s using TIRF microscopy. 24 frames/s are shown.
Nascent adhesions reside in the lamellipodium. This movie corresponds to Fig. 3A. Green indicates the lamellipodium marked by GFP-actin; magenta indicates nascent adhesions marked by paxillin-mOrange. Images collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Elongation of adhesions when a protrusion halts. This movie corresponds to Fig. 4A. Images of paxillin-GFP collected every 5 sec using TIRF microscopy. 24 frames/sec shown.
Long-term imaging of nascent adhesions showing formation, turnover and maturation. Images of paxillin-GFP collected every 5 sec using TIRF microscopy. 24 frames/sec shown.
Elongation of actin filaments when a protrusion halts. This movie corresponds to Fig. 4A. Images of GFP-actin collected every 1 sec using TIRF microscopy. 24 frames/sec shown.
Actin template elongates from maturing nascent adhesions. This movie corresponds to Fig. 4B. Merged images of GFP-actin (green) and paxillin-mOrange (magenta) collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
MIIA links to actin templates distal to the initial α-actinin. Merged images of GFP-actin (green) and mCherry-MIIA (magenta) collected every 5 s using TIRF microscopy. 24 frames/sec shown.
Adhesion turnover and maturation are inhibited by α-actinin knockdown. This movie corresponds to Fig. 5D. Images of GFP-actin and paxillin-mOrange collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Rescue of MIIA-knockdowns with wild-type-MIIA or the high actin affinity, non-contractile mutant, N93K-MIIA. This movie corresponds to Figs. 6A-C. Images of paxillin-mOrange collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Acknowledgments
We thank Laura E. Chopko for helping to characterize the α-actinin knockdown. Also, we thank Hannelore Asmussen for technical assistance with substrate preparation. This work was supported by NIH grants GM23244 (AFH), the Cell Migration Consortium (U54 GM064346) and NSF grant DMS-0715729 (AM).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
AUTHOR CONTRIBUTIONS
C.K.C and M.V.-M. designed and performed the experiments and wrote the paper; J.Z. and l.A.W. assisted with the research; A.M. developed the mathematical model and its presentation; A.R.H. designed the experiments and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Webb DJ, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nature Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nature Cell Biol. 2002;4:E65–68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 5.Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J Cell Biol. 2001;153:881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayal A, et al. Paxillin phosphorylation at Ser 273 localizes a GIT1–PIX–PAK complex and regulates adhesion and protrusion dynamics. J Cell Biol. 2006;173:587–589. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nature Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells — over and over and over again. Nature Cell Biol. 2002;4:E97–E100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- 9.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 10.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 12.Giannone G, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubin-Thaler BJ, Giannone G, Dobereiner HG, Sheetz MP. Nanometer analysis of cell spreading on matrix-coated surfaces reveals two distinct cell states and STEPs. Biophys J. 2004;86:1794–1806. doi: 10.1016/S0006-3495(04)74246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bershadsky A, Kozlov M, Geiger B. Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr Opin Cell Biol. 2006;18:472–481. doi: 10.1016/j.ceb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riveline D, et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katsumi A, et al. Effects of cell tension on the small GTPase Rac. J Cell Biol. 2002;158:153–164. doi: 10.1083/jcb.200201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amano M, et al. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 20.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell–matrix adhesions. J Cell Sci. 2007;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- 22.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 23.Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 24.Rottner K, Hall A, Small JV. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr Biol. 1999;9:640–649. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 26.Otey C, Carpen O. α-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton. 2004;58:104–111. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 27.Kim KY, Kovacs M, Kawamoto S, Sellers JR, Adelstein RS. Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II -C J Biol Chem. 2005;280:22769–22775. doi: 10.1074/jbc.M503488200. [DOI] [PubMed] [Google Scholar]
- 28.Hu A, Wang F, Sellers JR. Mutations in human nonmuscle myosin IIA found in patients with May-Hegglin anomaly and Fechtner syndrome result in impaired enzymatic function. J Biol Chem. 2002;277:46512–46517. doi: 10.1074/jbc.M208506200. [DOI] [PubMed] [Google Scholar]
- 29.Triplett JW, Pavalko FM. Disruption of α-actinin-integrin interactions at focal adhesions renders osteoblasts susceptible to apoptosis. Am J Physiol Cell Physiol. 2006;291:C909–C921. doi: 10.1152/ajpcell.00113.2006. [DOI] [PubMed] [Google Scholar]
- 30.Giannone G, et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 31.DeMali KA, Barlow CA, Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J Cell Biol. 2002;159:881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serrels B, et al. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nature Cell Biol. 2007;9:1046–1056. doi: 10.1038/ncb1626. [DOI] [PubMed] [Google Scholar]
- 33.Weed SA, et al. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J Cell Biol. 2000;151:29–40. doi: 10.1083/jcb.151.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galbraith CG, Yamada KM, Galbraith JA. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 2007;315:992–995. doi: 10.1126/science.1137904. [DOI] [PubMed] [Google Scholar]
- 35.Small JV, Resch GP. The comings and goings of actin: coupling protrusion and retraction in cell motility. Curr Opin Cell Biol. 2005;17:517–523. doi: 10.1016/j.ceb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Vasiliev JM. Spreading of non-transformed and transformed cells. Biochim Biophys Acta. 1985;780:21–65. doi: 10.1016/0304-419x(84)90006-4. [DOI] [PubMed] [Google Scholar]
- 37.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laukaitis CM, Webb DJ, Donais K, Horwitz AF. Differential dynamics of α5 integrin, paxillin, and α-actinin during formation and disassembly of adhesions in migrating cells. J Cell Biol. 2001;153:1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo CM, et al. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol Biol Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Y, et al. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91:3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu XS, et al. During multicellular migration, myosin II serves a structural role independent of its motor function. Dev Biol. 2001;232:255–264. doi: 10.1006/dbio.2000.0132. [DOI] [PubMed] [Google Scholar]
- 42.Ma X, Kawamoto S, Hara Y, Adelstein RS. A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Mol Biol Cell. 2004;15:2568–2579. doi: 10.1091/mbc.E03-11-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- 44.Wei Q, Adelstein RS. Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol Biol Cell. 2000;11:3617–3627. doi: 10.1091/mbc.11.10.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DesMarais V, Macaluso F, Condeelis J, Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eto M, Kirkbride J, Elliott E, Lo SH, Brautigan DL. Association of the tensin N-terminal protein-tyrosine phosphatase domain with the alpha isoform of protein phosphatase-1 in focal adhesions. J Biol Chem. 2007;282:17806–17815. doi: 10.1074/jbc.M700944200. [DOI] [PubMed] [Google Scholar]
- 47.Rottner K, Krause M, Gimona M, Small JV, Wehland J. Zyxin is not colocalized with vasodilator-stimulated phosphoprotein (VASP) at lamellipodial tips and exhibits different dynamics to vinculin, paxillin, and VASP in focal adhesions. Mol Biol Cell. 2001;12:3103–3113. doi: 10.1091/mbc.12.10.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nature Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 49.Schafer DA, et al. Visualization and molecular analysis of actin assembly in living cells. J Cell Biol. 1998;143:1919–1930. doi: 10.1083/jcb.143.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Activated Rac induced focal complexes differ from nascent adhesions. CHO.K1 cells expressing V12Rac, either without (A) or with 20 μM blebbistatin (B), were co-transfected with GFP-actin (left) and paxillin-mOrange (right). Scale bar = 10 μm. Color inserts show the relative positioning of paxillin-containing focal complexes (A) or nascent adhesions (B) with respect to the lamellipodium. Green, actin; magenta, paxillin. Note that the focal complexes reside the lamellipodium-lamellum interface and disassemble into smaller, lamellipodial localized nascent-like adhesions the presence of blebbistatin.
Figure S2 Molecular markers of the lamellipodium of migrating CHO.K1 cells. Characterization of dendritic actin of the lamellipodia. mRFP-actin (magenta) colocalize with GFP-cofilin and immunostaining of Arp3 (green). Barbed-end actin (green) outlines fluorescent phalloidin (magenta) at the leading edge.
Figure S3 Quantification of protrusion rates in α-actinin-knockdown cells. (A) Kymographs from control (pSUPER) and α-actinin-depleted cells. The knockdown cells were identified by coexpression of a GFP marker, imaged in brightfield, and then analyzed by kymography. (B) Quantification of protrusion rates from kymographs from (A). Average protrusion rates measured from >12 cells (three to five protrusions/cell) from four independent experiments. Error bars represent ±SEM.
Figure S4 The rapid assembly of nascent adhesions in MEFs. TIRF time-lapse images of paxillin-GFP in MEFs show the rapid assembly (green arrows) of new nascent adhesions near the leading edge. Most nascent adhesions undergo immediate maturation and do not turnover (red arrows). Scale bar = 3 μm. Elapsed time is in seconds. TIRF images of GFP-actin (green) and paxillin-mOrange (magenta) show nascent adhesions (green arrows) forming in MEFs and residing exclusively in the lamellipodium. Scale bar = 3μm.
Paxillin-containing nascent adhesions assemble and disassemble rapidly during protrusions. This movie corresponds to Fig. 1A. Images of paxillin-GFP collected every 1 sec using TIRF microscopy. Magenta and green arrows indicate representative assembling and disassembling adhesions, respectively. 10 frames/sec shown.
Rescue of the MIIA knockdown with the non-contractile MIIA mutant N93K and blebbistatin. This movie corresponds to Fig. 6D. Images of paxillin-mOrange collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Rescue of the α-actinin knockdown by the high actin affinity, non-contractile mutant N93K-MIIA or over expression of MIIA wild-type. This movie corresponds to Fig. 7A–C. Images of paxillin-mOrange collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Paxillin-containing nascent adhesions assemble and mature rapidly during protrusions in MEFs. This movie corresponds to Supplementary Fig. 40. Images of paxillin-GFP collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Myosin II regulates the probability and rate of nascent adhesion maturation. Left, MIIA-deficient CHO.K1cell. This movie is from Vicente-Manzanares et al., J. Cell Biol. 176(5)573–580, Suppl. Movie 7; center, wild-type; right, overexpression of MIIA in a CHO.K1 cell. Images of paxillin-GFP were collected every 5 s using TIRF microscopy. 24 frames/s are shown.
Nascent adhesions reside in the lamellipodium. This movie corresponds to Fig. 3A. Green indicates the lamellipodium marked by GFP-actin; magenta indicates nascent adhesions marked by paxillin-mOrange. Images collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Elongation of adhesions when a protrusion halts. This movie corresponds to Fig. 4A. Images of paxillin-GFP collected every 5 sec using TIRF microscopy. 24 frames/sec shown.
Long-term imaging of nascent adhesions showing formation, turnover and maturation. Images of paxillin-GFP collected every 5 sec using TIRF microscopy. 24 frames/sec shown.
Elongation of actin filaments when a protrusion halts. This movie corresponds to Fig. 4A. Images of GFP-actin collected every 1 sec using TIRF microscopy. 24 frames/sec shown.
Actin template elongates from maturing nascent adhesions. This movie corresponds to Fig. 4B. Merged images of GFP-actin (green) and paxillin-mOrange (magenta) collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
MIIA links to actin templates distal to the initial α-actinin. Merged images of GFP-actin (green) and mCherry-MIIA (magenta) collected every 5 s using TIRF microscopy. 24 frames/sec shown.
Adhesion turnover and maturation are inhibited by α-actinin knockdown. This movie corresponds to Fig. 5D. Images of GFP-actin and paxillin-mOrange collected every 3 sec using TIRF microscopy. 24 frames/sec shown.
Rescue of MIIA-knockdowns with wild-type-MIIA or the high actin affinity, non-contractile mutant, N93K-MIIA. This movie corresponds to Figs. 6A-C. Images of paxillin-mOrange collected every 3 sec using TIRF microscopy. 24 frames/sec shown.