Abstract
An MR spectroscopic imaging (MRSI) pulse sequence based on Proton-Echo-Planar-Spectroscopic-Imaging (PEPSI) is introduced that measures 2-dimensional metabolite maps in a single excitation. Echo-planar spatial-spectral encoding was combined with interleaved phase encoding and parallel imaging using SENSE to reconstruct absorption mode spectra. The symmetrical k-space trajectory compensates phase errors due to convolution of spatial and spectral encoding. Single-shot MRSI at short TE was evaluated in phantoms and in vivo on a 3 T whole body scanner equipped with 12-channel array coil. Four-step interleaved phase encoding and 4-fold SENSE acceleration were used to encode a 16×16 spatial matrix with 390 Hz spectral width. Comparison with conventional PEPSI and PEPSI with 4-fold SENSE acceleration demonstrated comparable sensitivity per unit time when taking into account g-factor related noise increases and differences in sampling efficiency. LCModel fitting enabled quantification of Inositol, Choline, Creatine and NAA in vivo with concentration values in the ranges measured with conventional PEPSI and SENSE-accelerated PEPSI. Cramer-Rao lower bounds were comparable to those obtained with conventional SENSE-accelerated PEPSI at the same voxel size and measurement time. This single-shot MRSI method is therefore suitable for applications that require high temporal resolution to monitor temporal dynamics or to reduce sensitivity to tissue movement.
Keywords: Magnetic resonance spectroscopic imaging, single-shot encoding, Proton Echo Planar Spectroscopic Imaging, parallel imaging, SENSE, spectral quantification
Introduction
Ultra high-speed MR spectroscopic imaging (MRSI) with acquisition times in the range of a few seconds will enable novel research applications, such as the measurement of metabolic dynamics inhyperpolarized MRI for tumor diagnosis and treatment monitoring [1,2,3], as well as assessment of cardiac function [4]. Another possible application, which is complementary to diffusion sensitive MRI [5,6,7,8], is spatial mapping of metabolite diffusion to study intra-cellular mobility [9,10]. Ultra high-speed MRSI is also applicable to biochemical imaging in moving organs, such as the heart, liver and breast [11,12].
A number of laboratories have developed high-speed MRSI methods, mostly using echo-planar and spiral readout modules, that provide considerable acceleration as compared to conventional phase encoded MRSI [13,14,15,16,17,18,19,20,21,22]. Recently, Mayer et al. described ultra high-speed metabolic imaging of systems with sparse spectra for applications in hyperpolarized 13C imaging [23]. The imaging method in that study was based on spiral readout encoding and enabled single-shot mapping of 13C spectra in seconds, albeit at narrow spectral width, which required separate reconstruction of each spectral peak. In a follow-up publication, Levin et al. described a least squares reconstruction to optimize fast spiral spectroscopic imaging by taking into account chemical shift evolution during the readout period [24]. Both methods are most suitable for sparse spectra with well separated peaks.
Recent advances in parallel MRI employing information from different channels in an RF coil array enable considerable acceleration of MRSI. Dydak et al. demonstrated acceleration of conventional phase encoded MRSI with acquisition times of just a few minutes using sensitivity encoding (SENSE) [25,26]. Recently, we have developed even faster spatial-spectral encoding by combining Proton-Echo-Planar-Spectroscopic-Imaging (PEPSI) with parallel imaging to obtain minimum encoding times of 1 min for 3D encoding of a 32×32×8 matrix [27] and 2D mapping with encoding times as short as 16 s for a 32×32 matrix [28,29].
In the present study, we extend this approach by interleaving partial phase encoding into the PEPSI readout trajectory [30,31] and using SENSE reconstruction [27] to achieve 2-dimensional spatial and spectral encoding in a single-shot with spectral width large enough to encompass the major singlet resonances in 1H spectra in the human brain. The readout module was designed with symmetric forward and backward k-space trajectories to enable reconstruction of pure absorption mode spectra. We show quantitative mapping of major spectral peaks of Inositol, Choline, Creatine and NAA in vivo on a 3 T whole body MR scanner equipped with 12-channel receive-only coil array. We also compare single-shot PEPSI with our recently published PEPSI methodology using conventional phase encoding (conventional PEPSI) [22] and 4-fold SENSE acceleration (SENSE-PEPSI) [28], and with data from a pulse sequence that combines single-shot encoding and additional conventional phase encoding to characterize the point spread function (PSF-PEPSI). Conventional PEPSI data with spectral width identical to that of single-shot encoding (low bandwidth PEPSI – LBW-PEPSI) were extracted from PSF-PEPSI to characterize the effects of narrow spectral width on sensitivity and spectral quantification. Preliminary accounts of this work were presented at a recent meeting [32].
Methods
Theory
The single-shot method is based on the PEPSI encoding scheme described in previous publications [21,22] and requires a phased array radiofrequency receive coil that generates a sufficient number of spatially distinct sensitivity patterns to enable acceleration with parallel imaging. Spatial-spectral encoding is performed by interleaving phase encoding gradient blips into the alternating readout gradient train between pairs of positive and negative readout gradient pulses to form a series of 2-dimensional spatial encoding modules with duration Δt (Fig. 1). Repetition of this spatial encoding module encodes spectral information with reconstructed spectral width 1/Δt. The k-space trajectory is designed to (a) maximize signal-to noise ratio (SNR) by minimizing analogue-to-digital-converter (ADC) dead time using ramp sampling, (b) minimize eddy current effects using uniform readout gradient inversion periodicity, and (c) obtain symmetrical chemical shift related phase evolution in the even and odd echo data by positioning the center of the first readout module at the spin echo time TE. For each spatial encoding module a series of positive phase encoding gradients blips is followed by a series of negative phase encoding gradient blips, resulting in a zig-zag trajectory in kx-ky-t-space (Fig. 2). Each readout module in kx-ky space is traversed symmetrically forward and backward. The end of the forward k-space trajectory of the first readout module is positioned at the spin echo time TE to minimize first order phase error upon combination of even and odd echo data. Accelerated phase encoding steps through ky-space by skipping R ky-space lines.
Figure 1.
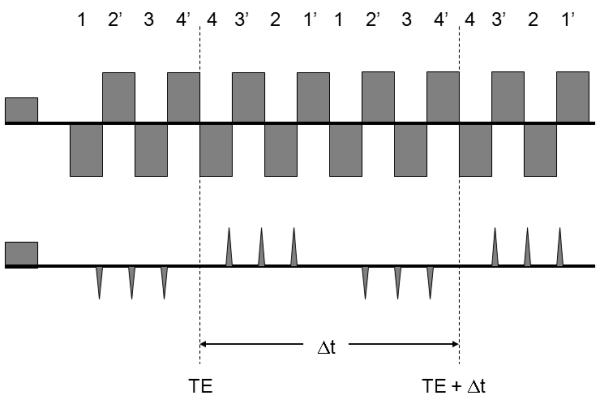
Gradient switching scheme for single-shot spatial-spectral encoding with negative and positive phase encoding gradient blips. A set of 8 readout gradients with duration Δt forms a readout module with negative readout gradients labeled 1,2,3 and 4, and positive readout gradients labeled 1, 2′, 3 and 4′.This module encodes an imaging matrix with 16 pixels in the phase encoding direction using 4-fold acceleration with partial parallel imaging. The readout module is repeated N-times to encode the spectral dimension.
Figure 2.
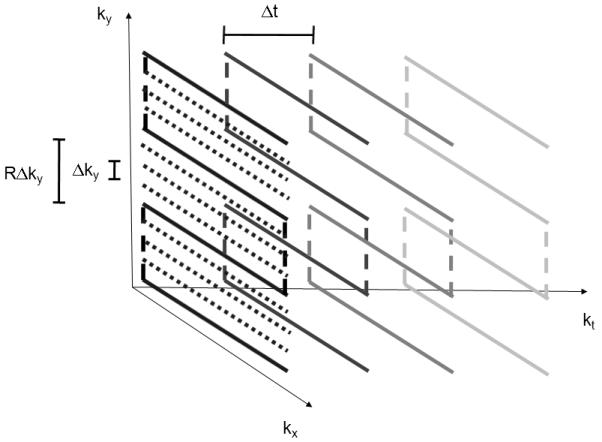
Kx-Ky-t-space trajectory for single-shot spatial-spectral encoding. Phase encoding lines are measured with ky-spacing RΔk, where R(=4) denotes the acceleration factor. Target ky-spacing after SENSE reconstruction is Δk. Each readout module, which consists of 4 forward and 4 reverse kx-ky-space encoding segments, is traversed forward and backward, and repeated N-times with spacing Δt to encode N spectral points. Data are only collected during the horizontal segments of the trajectory.
Single-shot encoding requires interleaving of phase encoding gradients, which reduces the sampling efficiency of the ADC as compared to conventional PEPSI encoding. With adaptation from the theory in Pohmann et al [33], the normalized SNR per unit time and unit volume is given by:
| (1) |
where SNRref is the reference SNR per unit time and unit volume [s−1/2cc−1] measured in data acquired with conventional PEPSI encoding, V: voxel size [cc], td: ADC dead time during readout module [s], tADC: ADC on time during readout module [s], TR: repetition time [s], N: number of scan repetitions number of experiment repetitions (taking into account R fold acceleration with parallel imaging), g = g-factor. The sampling efficiency σ of the ADC is defined as:
| (2) |
Pulse sequence implementation
Single-shot PEPSI was implemented on a 3T Siemens Tim Trio scanner equipped with 12-channel head array coils, respectively. Spatial-spectral encoding was performed with 2048 trapezoidal readout gradients (240 μs duration), 100 μs phase encoding blips, 4-fold uniform acceleration of phase encoding to encode a 16×16 spatial matrix with 192 mm minimum FOV. Four readout segments for the forward trajectory and four readout segments for the backward trajectory, respectively, resulted in a 2.56 ms readout module. Data were collected with 2-fold oversampling, regridded to correct the k-space trajectory distortion due to linear ramp sampling and decimated 2-fold to remove oversampling as described previously [21,22]. The reconstructed spectral width was 390 Hz and the digital spectral resolution was 1.5 Hz. The pulse sequence included 3-pulse water suppression and 8-slice outer volume suppression as described previously [21,22]. In addition, a modified pulse sequence was used to collect data to characterize the point spread function (PSF) of the single-shot encoding sequence (PSF-PEPSI) and to obtain conventionally phase encoded PEPSI data with spectral width identical to that of single-shot encoding (low bandwidth PEPSI – LBW-PEPSI). To collect these data the single-shot PEPSI sequence was repeated sixteen times with a stepwise incremented conventional phase-encoding gradient along the y-direction to fill an additional k-space dimension, referred to here as the point spread function (PSF) dimension.
Data acquisition, reconstruction and analysis
Measurements were performed on a spherical phantom filled with a metabolite solution with concentration values in the millimolar range and in healthy volunteers after obtaining institutionally approved consent. Data were acquired with single-shot PEPSI, PSF-PEPSI and with conventional PEPSI using TE: 11 ms, TR: 2 s and voxel size: 14×14×15 mm3. Single-shot PEPSI was performed with FOV: 230 mm and 16 averages resulting in 32 s scan time. PSF-PEPSI was performed with FOV: 230 mm and 1 average resulting in 32 s scan time. Conventional PEPSI was performed with 1087 Hz spectral width, 32×16 (readout × phase encode) image matrix, FOV: 460 mm × 230 mm and 4 averages resulting in 128 s scan time. Four-fold accelerated PEPSI data (SENSE-PEPSI) were obtained from data measured with conventional PEPSI using 4-fold undersampling along the ky dimension, resulting in an equivalent measurement time of 32 s.
Data obtained with PSF-PEPSI were edited to obtain conventional phase encoded low bandwidth PEPSI (LBW-PEPSI) data by extracting for each repetition of the readout module the 2 gradient echoes that correspond to the center of the ky-space encoded by the blipped phase encoding gradients (labeled 2 and 2′ in Fig.1).
Coil sensitivity maps were estimated using spectral water images from an extra fully phase encoded non-water-suppressed (NWS) acquisition as described previously [27,28]. Array geometry related noise amplification in the reconstruction was computed using the g-factor [27,28]. Even and odd-echo data were reconstructed separately using SENSE reconstruction with regularization as described previously [27]. Coil-by-coil SENSE reconstruction was generated by multiplying the combined SENSE reconstruction with the coil sensitivities in order to allow for application of spectral phase correction and frequency alignment to each channel separately. A sine bell filter was applied across the two k-space dimensions and Fourier transformation in the temporal domain were applied. The reconstructed NWS data were used to automatically correct zero-order phase error and frequency offset on a voxel-by-voxel basis. First order phase modulation in the even and the odd echo data sets canceled during combination of the two data sets, resulting in pure absorption mode spectra. Finally, the reconstructed multi-coil data were combined using sensitivity-weighted combination.
Spectra were quantified using LCModel fitting with analytically modeled basis sets as described previously [22]. Errors in metabolite quantification in LCModel (% SD) are expressed in Cramer-Rao lower bound (CRLB, the lowest bound of the standard deviation of the estimated metabolite concentration expressed as percentage of this concentration), which when multiplied by 2.0 represent 95% confidence intervals of the estimated concentration values [34]. SNR values were taken from LCModel output.
Metabolite concentration images at 3 T were created using the following thresholds to accept voxels: a) CRLB ≤ 30% for creatine (Cr) and N-acetyl-aspartate (NAA), CRLB ≤ 50% for choline (Cho), and inositol (Ins), and b) spectral line width (FWHM) ≤ 0.2 ppm. Larger CRLB thresholds (50 % for Cho, Cr and NAA) were used for the single-shot data at 1.5 T due to lower SNR. Finally, the metabolite concentration maps were interpolated to a 128×128 matrix using zero-filling of the k-space data to improve visualization.
Results
Data acquired with single-shot PEPSI encoding demonstrate adequate spectral width to encompass the major metabolites of interest in 1H brain spectroscopy. PSF-PEPSI k-space data obtained at 3 T in a spherical phantom confirmed that blipped interleaved phase encoding was consistent with conventional phase encoding: When plotting the k-space signal intensity as a function of PSF ky dimension versus the single-shot ky dimension, a nearly linear relationship was found.
Figure 3 shows metabolic images and water-suppressed spectra acquired on a phantom with conventional PEPSI, SENSE-PEPSI, LBW-PEPSI and single-shot PEPSI. The average g-factor of SENSE reconstruction was (2.1±0.4) (Fig.3b). The average SNR of the single-shot PEPSI data (7.8±6.8) was in a similar range as that in the SENSE-PEPSI data (7.5±2.3) and the LBW-PEPSI data (8.0±2.7). The average spectral line width measured with all four methods was similar (conventional PEPSI: 4.3±2.8 Hz, SENSE-PEPSI: 5.0±3.2 Hz, LBW-PEPSI: 4.0±2.4 Hz, single-shot PEPSI: 4.5±2.6 Hz) (Fig.3c’-f’). The baseline in the SENSE-PEPSI and single-shot PEPSI spectra was slightly distorted due to residual water aliased from distant regions with B0 shifts and spatial aliasing of peripheral lipid signals due to imperfections in SENSE reconstruction. In the LBW-PEPSI and single-shot PEPSI data the spectral aliasing of NAA onto the low-field side of the residual water peak resulted in increased baseline distortion, which increased fitting errors and as a result introduced inhomogeneity in the corresponding metabolite maps (Fig.3e,f).
Figure 3.
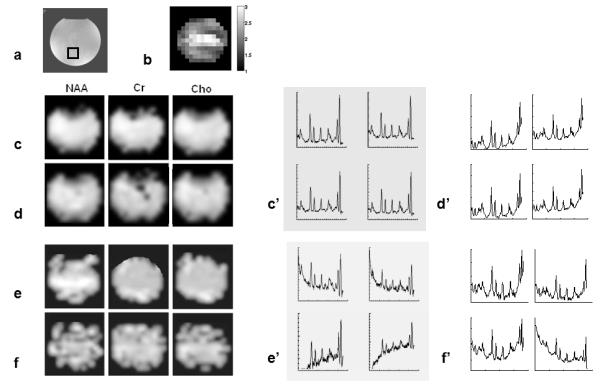
Comparison of MRSI methods on spherical phantom. (a) MRI of with volume location of spectral arrays shown below, (b) g-factor map for SENSE reconstruction, Spectroscopic images of choline (Cho), creatine (Cr) and N-acetyl-aspartate (NAA) (TR/TE: 2 s/ 11 ms, voxel size: 3 cc) using conventional PEPSI (c), SENSE-PEPSI (d), LBW-PEPSI (e) and single-shot PEPSI (f). The corresponding spectral arrays (c’-f’) display the spectral range from 1.8 to 4 ppm.
Figure 4 shows metabolic images and water-suppressed spectra acquired in an axial supra-ventricular slice in the human brain with PEPSI, SENSE-PEPSI, LBW-PEPSI and single-shot PEPSI. The placement of the outer volume suppression slices along the periphery of the brain is shown in Fig.4a. The mean g-factor (2.32±0.45) was comparable in magnitude to that in the phantom data (Fig.4b). The single-shot PEPSI metabolite maps (Fig.4f) show relatively uniform spatial distributions of Ins, Cho, Cr and NAA, consistent with the maps obtained with conventional PEPSI and LBW-PEPSI (Fig.4c,e). However, larger spatial non-uniformity was measured in the single-shot PEPSI data, which is in part due to g-factor related noise enhancement and spatial aliasing of residual water and peripheral lipid peaks as a result of SENSE reconstruction errors. The single-shot spectrum in Fig.4f shows clearly identifiable spectral peaks from Ins, Cho, Cr, NAA, as well as multiplet peaks between Ins and Cr at 3.9 ppm, consistent with the data obtained with conventional PEPSI, SENSE-PEPSI and LBW-PEPSI (Fig.4c–e). The LBW-PEPSI and single-shot PEPSI spectra show baseline distortion in the vicinity of NAA, which is aliased onto the low-field shoulder of the residual water peak. The slice averaged spectral line width of all 4 methods was in a similar range as well (conventional PEPSI: 8.7±3.8, SENSE-PEPSI: 8.1±3.8, LBW-PEPSI: 8.0±3.9, single-shot PEPSI: 10.2±5.3). Metabolite concentration values for the four methods (Table 1) were in the range reported in our previous study [22]. Concentration values measured with SENSE-PEPSI, LBW-PEPSI and single-shot PEPSI were slightly overestimated due to low signal-to-noise, consistent with our previous experience [22]. The CRLBs of SENSE-PEPSI, LBW-PEPSI and single-shot PEPSI were in similar ranges (Table 1).
Figure 4.
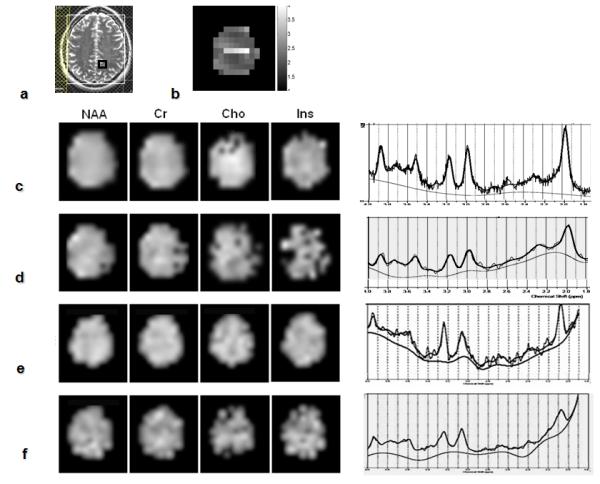
Comparison of MRSI methods in vivo. (a) MRI of an axial supra-ventricular slice with positioning of outer volume suppression slices and voxel location of spectra shown below, (b) g-factor map for SENSE reconstruction. Spectroscopic images of inositol (Ins), choline (Cho), creatine (Cr) and N-acetyl-aspartate (NAA) (TR/TE: 2 s/ 11 ms, voxel size: 3 cc) using conventional PEPSI (c), SENSE-PEPSI (d), LBW-PEPSI (e) and single-shot PEPSI (f). The selected spectra on the right display the superimposed LCModel fit and the estimated baseline in the spectral range from 1.8 to 4 ppm.
Table 1.
| Slice averaged metabolite concentration values [mmol] in human brain (+/−SD) | ||||
|---|---|---|---|---|
| Ins | Cho | Cr | NAA | |
| Convent. PEPSI | 5.5±2.2 | 2.0±0.7 | 8.6±3.7 | 11.7±3.8 |
| SENSE-PEPSI | 7.2±4.6 | 2.1±0.8 | 9.0±2.3 | 11.8±4.3 |
| LBW-PEPSI | 5.5±2.3 | 2.3±1.5 | 9.8±3.7 | 9.9±3.3 |
| Single-shot PEPSI | 6.8±4.6 | 2.4±1.7 | 14.1±10.9 | 13.6±8.2 |
| Slice averaged Cramer-Rao lower bound values [%] in human brain (+/−SD) | ||||
|---|---|---|---|---|
| Ins | Cho | Cr | NAA | |
| Convent. PEPSI | 20.4±9.4 | 11.1±7.2 | 8.1±4.8 | 6.0±4.8 |
| SENSE-PEPSI | 27.7±8.6 | 20.0±7.9 | 14.4±4.7 | 11.2±6.4 |
| LBW-PEPSI | 19.4±12.0 | 28.1±31.7 | 12.5±9.2 | 12.2±6.8 |
| Single-shot PEPSI | 22.2±19.2 | 25.6±28.2 | 14.4±17.3 | 14.3±18.6 |
Discussion
The results of this study demonstrate the feasibility of single-shot 2-dimensional MRSI with spectral width large enough to encompass the major singlet resonances in 1H spectra in the human brain. The methodology uses partial phase encoding interleaved into an echo-planar readout trajectory [30,31] and partial parallel imaging with SENSE reconstruction [25] to strongly accelerate spatial encoding. The achievable spectral width depends on gradient switching performance for interleaving phase encoding and on the spatial encoding capabilities of the RF coil array for acceleration with parallel imaging, which determines the number of required phase encoding blips. Large acceleration factors are required for achieving adequate spectral width and spatial resolution, which favors the use of large scale array coils and high magnetic field strength [35]. In this study we have used commercially available array coils, which exhibit relatively large g-factors at the 4-fold acceleration rate necessary for single-shot encoding. Alternate reconstruction schemes that avoid separation of even and odd echo data, such as the interlaced Fourier transform, could be used to double the spectral width [36,37]. Further acceleration of spectral encoding to increase spectral width can be achieved by generating multiple spin echoes that are individually phase encoded as originally described by Duyn et al. [38].
Equation 1 predicts that the sensitivity per unit time and unit volume of single-shot PEPSI is only slightly smaller than that of SENSE-PEPSI, which is consistent with our experimental data. Interleaving phase encoding gradient blips in single-shot PEPSI result in slightly lower ADC data sampling efficiency (σ = 77%) as compared to conventional PEPSI and SENSE-PEPSI (σ = 89 %). Theory also predicts that the sensitivity of single-shot PEPSI is similar to that of LBW-PEPSI when taking into account reduced sampling efficiency in the LBW-PEPSI data and g-factor related noise increases in the single-shot PEPSI data, which can be seen as follows: The extraction of the two echoes at ky=0 in the PSF-PEPSI data to generate the LBW-PEPSI data results in a 2.6-fold decrease in sensitivity per unit time as compared to single-shot PEPSI data, which is largely compensated by the g-factor related noise enhancement in the single-shot PEPSI data. This is confirmed by our data.
The narrow spectral width at 3 T leads to spectral aliasing of the NAA peak onto the low field shoulder of the residual water peak. This makes baseline estimation and spectral quantification, in particular for NAA, challenging. Overlapping multiplet resonances at the short TE used in this study further complicate spectral fitting. The simulated spectral basis sets in this study did not take into account spectral aliasing, which prevents accurate fitting of multiplet resonances and impairs estimation of the baseline. The LCModel fitting results in vivo in this study thus represent rough estimates of the actual metabolite concentration values. An approximately 10 % increases in spectral width could be achieved using triangular instead of rectangular readout gradients at the expense of increased ramp sampling. This would increase the separation of NAA and residual water and thus improve the quantification of NAA. In future studies we will use numerical optimization of the spectral width to minimize information loss due to overlapping spectral peaks and explicitly model basis sets for narrow spectral width to enable quantification of multiplet peaks. Increasing spectral width using array coils with larger number of coil elements and larger SENSE acceleration factors will further increase the robustness of spectral fitting.
Part of the baseline distortion in the single-shot PEPSI spectra arises from spatial aliasing of residual peripheral lipids signals as a result of imperfections in SENSE reconstruction due to errors in coil sensitivity estimation and intra-voxel coil sensitivity variation due to the low spatial resolution of the acquisition. The use of short TE, which we chose to maximize SNR and to compare with our previous short TE studies, increases this baseline distortion. The use the minimum-norm SENSE reconstruction technique would minimize spatial bleeding due to intra-voxel coil sensitivity variation [39].
Off-resonance effects in our 3T data blur the spatial response function of aliased signals originating from outside of the encoded spectral width as described by Mayer et al. [23]. Centering the spectrum at 2.6 ppm using transmitter and receiver offsets (not yet implemented in this study) will reduce spatial blurring for the major singlet resonances, but not for aliased macromolecular and lipid signals. Additional complexity arises from the reordering of the readout trajectories to form even and odd-echo data sets: spectral phase accumulation between individual readout segments, e.g. 1, 2′, 3, 4′, is slightly discontinuous leading to additional phase modulation in k-space resulting in broadening of the point spread function. Phase correction in k-space and the use of a first-order phase correction in the spectral domain as described by Mayer [23] to correct off-resonance effects for different resonances or least squares reconstruction [24] will result in narrower point spread function.
In conclusion, we have developed an MRSI method that allows collecting a complete spectroscopic image with two spatial dimensions and one spectral dimension in a single signal excitation with sensitivity per unit time and unit volume that is comparable to conventional SENSE accelerated MRSI. Combining echo-planar readout, interleaved phase encoding and partial parallel imaging using SENSE enables flexible tradeoff between gradient and RF encoding to maximize spectral width and spatial resolution. The method is suitable for applications that require high temporal resolution, such as hyperpolarized MRI, and to reduce motion sensitivity in vivo.
Acknowledgements
This work was supported by National Institutes of Health Grants R01 HD040712, R01 NS037462, R01 EB000790-04, P41 RR14075, R21 EB007298 and R01 DA14178-01 and by the Mental Illness and Neuroscience Discovery Institute (MIND). It was supported in part by Taiwan National Science Council under grants NSC-95-2221-E-002-179 and NSC 96-2320-B-002-085 and by the National Health Research Institute - Taiwan (E29C97N). We thank Dr. Lawrence Wald for his support and encouragement during the development of parallel high-speed spectroscopic imaging.
Footnotes
Presented in part at the 15th Annual Meeting of ISMRM, Berlin, Germany, 2007.
References
- 1.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc Natl Acad Sci USA. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golman K, in’t Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci USA. 2006;103:11270–11275. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66:10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 4.Golman K, Petersson JS. Metabolic imaging and other applications of hyperpolarized 13C1. Acad Radiol. 2006;13:932–942. doi: 10.1016/j.acra.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Posse S, Cuenod CA, Le Bihan D. Human brain: proton diffusion MR spectroscopy. Radiology. 1993;188:719–725. doi: 10.1148/radiology.188.3.8351339. [DOI] [PubMed] [Google Scholar]
- 6.Pfeuffer J, Tkac I, Gruetter R. Extracellular-intracellular distribution of glucose and lactate in the rat brain assessed noninvasively by diffusion-weighted 1H nuclear magnetic resonance spectroscopy in vivo. J Cereb Blood Flow Metab. 2000;20:736–46. doi: 10.1097/00004647-200004000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Valette J, Guillermier M, Besret L, Hantraye P, Bloch G, Lebon V. Isoflurane strongly affects the diffusion of intracellular metabolites, as shown by 1H nuclear magnetic resonance spectroscopy of the monkey brain. J Cereb Blood Flow Metab. 2007 Mar;27(3):588–96. doi: 10.1038/sj.jcbfm.9600353. Epub 2006 Jun 21. [DOI] [PubMed] [Google Scholar]
- 8.Le Bihan D, editor. Diffusion and perfusion: Magnetic resonance imaging. Raven Press; New York, NY: 1995. [Google Scholar]
- 9.Posse S, Cuenod CA, Le Bihan D. Motion artifact compensation in 1H spectroscopic imaging. (Series B).Journal of Magnetic Resonance. 1993;102:222–227. [Google Scholar]
- 10.Bito Y, Hirata S, Nabeshima T, Yamamoto E. Echo-planar diffusion spectroscopic imaging. Magn Reson Med. 1995 Jan;33(1):69–73. doi: 10.1002/mrm.1910330110. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs MA, Barker PB, Bottomley PA, Bhujwalla Z, Bluemke DA. Proton magnetic resonance spectroscopic imaging of human breast cancer: a preliminary study. J Magn Reson Imaging. 2004 Jan;19(1):68–75. doi: 10.1002/jmri.10427. [DOI] [PubMed] [Google Scholar]
- 12.Hu J, Vartanian SA, Xuan Y, Latif Z, Soulen RL. An improved 1H magnetic resonance spectroscopic imaging technique for the human breast: preliminary results. Magn Reson Imaging. 2005 May;23(4):571–6. doi: 10.1016/j.mri.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Mansfield P. Multi-planar image formation using NMR spin echos. J Physics. 1977;C10:L55–L58. [Google Scholar]
- 14.Norris DG, Dreher W. Fast proton spectroscopic imaging using the sliced k-space method. Magn Reson. Med. 1993;30:641–645. doi: 10.1002/mrm.1910300516. [DOI] [PubMed] [Google Scholar]
- 15.Posse S, DeCarli CS, Le Bihan D. Three-dimensional Echo-Planar MR Spectroscopic imaging at short echo times in human brain. Radiology. 1994;192:733–738. doi: 10.1148/radiology.192.3.8058941. [DOI] [PubMed] [Google Scholar]
- 16.Posse S, Tedeschi G, Risinger R, Ogg R, Bihan DL. High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn Reson Med. 1995;33:34–40. doi: 10.1002/mrm.1910330106. [DOI] [PubMed] [Google Scholar]
- 17.Adalsteinsson E, Irarrazabal P, Spielman DM, Macovski A. Three-dimensional spectroscopic imaging with time-varying gradients. Magn Reson Med. 1995;33(4):461–466. doi: 10.1002/mrm.1910330402. [DOI] [PubMed] [Google Scholar]
- 18.Guimaraes AR, Baker JR, Jenkins BG, Lee PL, Weisskoff RM, Rosen BR, González RG. Echoplanar chemical shift imaging. Magn Reson Med. 1999 May;41(5):877–82. doi: 10.1002/(sici)1522-2594(199905)41:5<877::aid-mrm4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Ebel A, Soher BJ, Maudsley AA. Assessment of 3D proton MR echo-planar spectroscopic imaging using automated spectral analysis. Magn Reson Med. 2001;46(6):1072–1078. doi: 10.1002/mrm.1301. [DOI] [PubMed] [Google Scholar]
- 20.Schuster C, Dreher W, Geppert C, Leibfritz D. Fast 3D 1H spectroscopic imaging at 3 Tesla using spectroscopic missing-pulse SSFP with 3D spatial preselection. Magn Reson Med. 2007 Jan;57(1):82–9. doi: 10.1002/mrm.21105. [DOI] [PubMed] [Google Scholar]
- 21.Otazo R, Mueller B, Ugurbil K, Wald LL, Posse S. Signal-to-Noise Ratio and Spectral Line Width Improvements between 1.5 and 7 Tesla in Proton-Echo-Planar-Spectroscopic-Imaging (PEPSI) Magn Reson Med. 2006;56(6):1200–1210. doi: 10.1002/mrm.21067. [DOI] [PubMed] [Google Scholar]
- 22.Posse S, Otazo R, Caprihan A, Bustillo J, Chen H, Henry P-G, Marjanska M, Gasparovic C, Zuo C, Magnotta V, Mueller B, Mullins P, Renshaw P, Ugurbil K, Lim KO, Alger JR. Proton Echo Planar Spectroscopic Imaging of J-Coupled Resonances in Human Brain at 3 and 4 Tesla. Magn. Reson. Med. 2007;58(2):236–244. doi: 10.1002/mrm.21287. [DOI] [PubMed] [Google Scholar]
- 23.Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM. Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized 13C imaging. Magn Reson Med. 2006 Oct;56(4):932–7. doi: 10.1002/mrm.21025. [DOI] [PubMed] [Google Scholar]
- 24.Levin YS, Mayer D, Yen YF, Hurd RE, Spielman DM. Optimization of fast spiral chemical shift imaging using least squares reconstruction: application for hyperpolarized (13)C metabolic imaging. Magn Reson Med. 2007 Aug;58(2):245–52. doi: 10.1002/mrm.21327. [DOI] [PubMed] [Google Scholar]
- 25.Dydak U, Weiger M, Pruessmann KP, Meier D, Boesiger P. Sensitivity-encoded spectroscopic imaging. Magn Reson Med. 2001;46(4):713–722. doi: 10.1002/mrm.1250. [DOI] [PubMed] [Google Scholar]
- 26.Dydak U, Pruessmann KP, Weiger M, Tsao J, Meier D, Boesiger P. Parallel spectroscopic imaging with spin-echo trains. Magn Reson Med. 2003;50(1):196–200. doi: 10.1002/mrm.10495. [DOI] [PubMed] [Google Scholar]
- 27.Otazo R, Tsai SY, Lin FH, Posse S. Accelerated Short-TE 3D Proton Echo-Planar Spectroscopic Imaging Using 2D-SENSE with a 32-Channel Array Coil. Magn Reson Med. 2007;58(6):1107–16. doi: 10.1002/mrm.21426. [DOI] [PubMed] [Google Scholar]
- 28.Lin F-H, Tsai S-Y, Otazo R, Caprihan A, Wald LL, Belliveau JW, Posse S. Sensitivity-encoded (SENSE) Proton-Echo-Planar- Spectroscopic-Imaging (PEPSI) in Human Brain. Mag. Reson. Med. 2007;57(2):249–257. doi: 10.1002/mrm.21119. [DOI] [PubMed] [Google Scholar]
- 29.Tsai S-Y, Posse S, Otazo R, Lin Y-R, Chung H-W, Lin F-H. Accelerated Proton-Echo-Planar-Spectroscopy-Imaging (PEPSI) using GRAPPA with Large-N phased-Array Coils. Magn. Reson. Med. 2008;59(5):989–98. doi: 10.1002/mrm.21545. [DOI] [PubMed] [Google Scholar]
- 30.Webb P, Spielman D, Macovski A. A fast spectroscopic imaging method using a blipped phase encode gradient. Magn. Reson. Med. 1989;12:306–315. doi: 10.1002/mrm.1910120303. [DOI] [PubMed] [Google Scholar]
- 31.Ebel A, Schuff N. Accelerated 3D Echo-Planar Spectroscopic Imaging at 4 Tesla Using Modified Blipped Phase-Encoding. Magn Reson Med. 2007;58(5):1061–1066. doi: 10.1002/mrm.21305. [DOI] [PubMed] [Google Scholar]
- 32.Posse S, Otazo R, Tsai S-Y, Lin F-H. Single-Shot MR Spectroscopic Imaging with Partial Parallel Imaging. Proc. International Society for Magnetic Resonance in Medicine (ISMRM) 2007 doi: 10.1002/mrm.21855. Abstract 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pohmann R, von Kienlin M, Haase A. Theoretical Evaluation and Comparison of Fast Chemical Shift Imaging Methods. J Magn Reson. 1997;129:145–160. doi: 10.1006/jmre.1997.1245. [DOI] [PubMed] [Google Scholar]
- 34.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 35.Wiesinger F, Van de Moortele PF, Adriany G, De Zanche N, Ugurbil K, Pruessmann KP. Parallel imaging performance as a function of field strength--an experimental investigation using electrodynamic scaling. Magn Reson Med. 2004 Nov;52(5):953–64. doi: 10.1002/mrm.20281. [DOI] [PubMed] [Google Scholar]
- 36.Metzger G, Hu X. Application of interlaced Fourier transform to echo-planar spectroscopic imaging. J Magn Reson. 1997;125:166–170. doi: 10.1006/jmre.1997.1114. [DOI] [PubMed] [Google Scholar]
- 37.Ebel A, Maudsley AA, Weiner MW, Schuff N. Achieving sufficient spectral bandwidth for volumetric 1H echo-planar spectroscopic imaging at 4 Tesla. Magn Reson Med. 2005;54(3):697–701. doi: 10.1002/mrm.20593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duyn JH, Moonen CT. Fast proton spectroscopic imaging of human brain using multiple spin-echoes. Magn Reson Med. 1993;30(4):409–414. doi: 10.1002/mrm.1910300403. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Gonzalez J, Tsao J, Dydak U, Desco M, Boesiger P, Pruessmann KP. Minimum-norm reconstruction for sensitivity-encoded magnetic resonance spectroscopic imaging. Magn Reson Med. 2006;55(2):287–295. doi: 10.1002/mrm.20758. [DOI] [PubMed] [Google Scholar]


