Abstract
Genetic association studies in systemic lupus erythematosus (SLE) have been extremely successful in recent years, identifying a number of loci associated with disease susceptibility. Much work remains to integrate these loci into the functional pathogenic pathways which characterize the disease. Our working hypothesis is that many of the genetic variations linked to SLE and autoimmunity mediate risk of disease by altering cytokine profiles or responses to cytokine signaling. Genetic polymorphisms affecting cytokine signaling could alter thresholds for immune responses, resulting in pro-inflammatory presentation of self antigens and the subsequent misdirection of adaptive immunity against self which is observed in autoimmune disease. SLE is clinically heterogeneous and genetically complex, and we expect that individual genes and cytokine patterns will be more or less important to different disease manifestations and subgroups of patients. Defining these genotype-cytokine-phenotype relationships will increase our understanding of both initial disease pathogenesis as well as subsequent response/non-response to various therapies. In this review we summarize some recent work in the area of SLE cytokine genetics, and describe the implications for SLE, autoimmunity, and immune system homeostasis which are revealed by these investigations.
Introduction
Systemic lupus erythematosus (SLE) is a systemic autoimmune disorder characterized by involvement of multiple organs including the skin, musculoskeletal, renal, and hematologic systems. The clinical presentation of SLE is highly heterogeneous, and it is not possible to predict the spectrum of organ system involvement in an individual patient with the disease. SLE is caused by a combination of genetic risk factors and environmental events which lead to an irreversible break in immunologic self-tolerance. Humoral autoimmunity is a hallmark of SLE, and circulating autoantibodies directed against double stranded DNA, as well as small nuclear RNA-binding proteins (such as anti-Ro, anti-La, anti-Sm, and anti-RNP) are found in many patients. A number of circulating cytokine abnormalities have been reported in SLE, including decreased interleukin-2 (1) and interleukin-12 (2), as well as increased interleukin-10 (3, 4) and interleukin-6 (1, 3). Serum tumor necrosis factor alpha (TNF-α) (5, 6) and osteopontin (OPN) (7) levels are elevated in many patients with SLE, and high circulating levels of interferon alpha (IFN-α) have been recognized as an important primary cytokine abnormality in SLE patients (8-11). Many of these cytokine abnormalities are shared with various murine models of SLE, although differences exist between mouse strains in the importance of particular cytokines to the observed SLE-like pathology. Cytokine abnormalities in murine SLE models have recently been reviewed extensively (12).
Given the importance of cytokines in immune system regulation, these molecules are of high interest not only in the “effector phase” of autoimmune disease in which self-tolerance has already been broken, but also in the “initiation phase” of autoimmunity, in which a lasting immune response against self antigens is first generated. We have been working under the hypothesis that initial susceptibility to autoimmune disease lies at least partly in the genetics of cytokine regulation, and that many genes will influence cytokine patterns to set a threshold for autoimmunity which varies greatly between different people. This idea is not completely new, however our current understanding of disease biology coupled with modern techniques in human genetics, cytokine measurement, and population science has enabled work in this area to proceed rapidly and we are now providing experimental support for this hypothesis.
Cytokine measurement has been classically performed using ELISA techniques, although microarray studies examining peripheral blood gene expression can frequently provide insights into circulating cytokine levels as well. In SLE, peripheral blood microarray studies demonstrated a dominant IFN-α-induced gene expression “signature” (11, 13), generating a renewed interest in this cytokine which is difficult to measure using commercial ELISA. Gene expression array data is often reported as a “signature”, and this term is used to indicate over-expression of numerous transcripts which are classically and coordinately up-regulated downstream of a particular signaling molecule, suggesting exposure of the blood cells to this particular molecule. In our work we have used a functional reporter cell assay to measure IFN-α in stored serum samples from large populations of SLE patients, enabling sufficient statistical power to establish genetic correlations (14, 15).
In this review, we will summarize recent work characterizing the genetics of serum cytokine traits in the prototype autoimmune disease, SLE. While temporal changes in serum cytokine levels related to the current state of immune system activation likely exist for many cytokines, thus far these potential fluctuations have not been sufficient to obscure genetic associations. If serum cytokine levels showed large variations over time with no stable background component, then genetic associations with cytokine levels would likely be undetectable. The studies discussed in this review support the opposite scenario, in which genetic risk factors are associated with stable differences in serum cytokine levels within the population. Longitudinal studies of IFN-α in SLE support this concept of stable differences in background cytokine levels between individuals, upon which some temporal variations also occur (16).
Genetic Loci Associated with SLE Susceptibility
Recent genetic studies in SLE have been extremely successful, and currently more than 20 loci are definitively linked to SLE susceptibility in case-control genetic studies (17, 18). Many of these successes are recent, and the first high density genome-wide scans in 2008 provided a detailed and unbiased look at genetic differences present in SLE patients. As expected, the HLA locus consistently provides the strongest evidence for association among the common genetic variants linked to SLE. The HLA locus is linked to many other autoimmune diseases and contains over 100 genes. Most of the genes in the HLA region have functions in the immune system, including the major histocompatibility (MHC) molecules which are involved directly in antigen presentation. The HLA locus is characterized by extensive and strong linkage disequilibrium, and in current studies it is difficult to separate individual gene effects upon SLE susceptibility. It is likely that a number of independent SLE risk factors may exist within this locus. Many non-HLA loci have been linked to SLE, and these represent a wide range of genes, largely with known or presumed functions in both the innate and adaptive immune systems. Case-control genetic studies in SLE have been reviewed recently (17, 18), and a summary of SLE-associated loci and potential function of these genes is provided in Table 1. Notably, a number of these genes are involved in responses to cytokines or cytokine production.
Table 1.
| Locus/Gene Name | Odds Ratio | Comments/Potential Function |
|---|---|---|
| HLA* | 2.4 | Multiple immune system genes contained within this locus |
| TNFAIP3* | 2.3 | Termination of immune responses by protein ubiquitination |
| IRF5 / TNPO3* | 1.7 | Transcription of IFN-α and IFN-α-induced genes |
| ITGAM* | 1.6 | Facilitates leukocyte adhesion to endothelial cells |
| FcGR3A+ | 1.6 | Clearance of immune complexes |
| STAT4* | 1.5 | Downstream responses to IFN-α and other cytokine signals |
| BLK / FAM167A / XKR6* |
1.5 | Modulation of B-cell activation |
| BANK1* | 1.4 | Modulation of B-cell activation |
| FcGR2A* | 1.4 | Clearance of immune complexes |
| PTPN22* | 1.3 | Modulation of lymphocyte activation and cytokine profile |
| CRP+ | 1.3 | Clearance of immune complexes |
| TNFSF4^ | 1.3 | Involved in T-cell/antigen presenting cell interactions |
| PHRF1 (KIAA1542) / IRF7* |
1.3 | Associated variant in linkage with IRF7, which is involved in production of IFN-α |
| PXK* | 1.3 | Unknown function currently |
| SPP1 (OPN)+ | 1.3 | Gene encoding the cytokine osteopontin, interacts with IFN-α pathway |
| MECP2 / IRAK1^ | 1.2 | Found on X-chromosome, involved in DNA methylation |
| PDCD1+ | 1.2 | Involved in lymphocyte signaling |
| TREX1 and early complement proteins (C1Q, C2, C4A, C4B)+ |
All >2 | Rare variants of these genes show high penetrance for SLE |
= confirmed association with p value <10−7,
= confirmed association with p value <10−5,
= probable association observed in multiple studies
Familial Aggregation of Serum IFN-α
IFN-α is a type I interferon which plays an important role in viral defense. Some of the physiologic functions of IFN-α include activation of dendritic cells and other antigen presenting cells, as well as increased expression of MHC class I and II molecules, leading to increased antigen presentation (19). In this way IFN-α can bridge the innate and adaptive immune systems, and may lower the threshold for productive pro-inflammatory antigen presentation. Serum IFN-α is elevated in many SLE patients, and elevations correlate with disease activity (8, 11, 20-22). In addition, a number of patients treated with recombinant human IFN-α for malignancy and chronic viral hepatitis have developed de novo SLE, which typically resolves after the IFN-α is discontinued (23, 24). These data suggest a primary role for IFN-α in the initiation of SLE.
SLE family members are at higher risk of developing not only SLE, but also other forms of autoimmunity, such as autoantibodies, autoimmune thyroid disease, autoimmune cytopenias, and others (25, 26). A similar spectrum of autoimmunity can be induced by recombinant IFN-α used as a therapy for malignancy or chronic viral hepatitis (27). These findings suggest that a genetic predisposition to increased IFN-α production in SLE families could explain some of the burden of autoimmunity in this population. We measured serum IFN-α in both healthy and affected members of SLE families, including 266 SLE patients and 405 of their healthy relatives (15). Healthy first degree relatives of SLE patients had significantly higher serum IFN-α than healthy unrelated subjects, and there was strong clustering of the high IFN-α trait in both affected and unaffected first degree relatives of high IFN-α SLE patients. This familial clustering was highly statistically significant (λ1st=5.2, p=2.4×10−7), and the two most common patterns in the pedigrees were that of a “high IFN-α family” in which the SLE-affected person and at least one healthy family member had high IFN-α, and a “low IFN-α family” in which all family members including the SLE-affected individual had low IFN-α. 30 SLE spouses were also studied, and none of them had high serum IFN-α, suggesting that environmental factors were not driving this familial correlation. These data taken together suggest that high serum IFN-α is a heritable risk factor for SLE. The heritability of IFN-α in SLE families set the stage for genetic studies to determine which genes were involved in the regulation of this risk factor.
Age- and Gender-Related Patterns of Serum IFN-α Activity in SLE Families
SLE is characterized by a striking 9:1 female to male ratio of disease incidence (28), which is not well understood currently. The peak of disease incidence for women is in the early reproductive years (ages 20-30), and peak disease incidence may be later (ages 40-50) for men (29). This large gender differential and disparate age of peak disease incidence in SLE hold great promise for understanding disease pathogenesis. A number of theories have been proposed to explain the female-male differential in SLE, including the influence of sex hormones (30, 31), X-chromosomal influences (32), or microchimerism in women who have been pregnant (33). While some supportive data exists for each of these hypotheses, it seems likely that multiple factors will contribute, and age- or gender-specific cytokine differences could play a role in the observed age-and gender-related incidence patterns observed in SLE.
Given the data supporting IFN-α as a causal agent in human lupus, we examined IFN-α levels in the family cohort in the context of age and gender in both healthy and affected family members (34). In this study, age was inversely correlated with serum IFN-α in female SLE patients as well as healthy first degree female relatives. In male patients and relatives, there was a non-significant trend toward an inverse relationship between age and serum IFN-α activity. Thus, all subgroups of family members showed some evidence of a pattern in which serum IFN-α was higher in younger people. When looking at the raw data, it appeared that these relationships were complex and not well-captured by a simple correlation analysis.
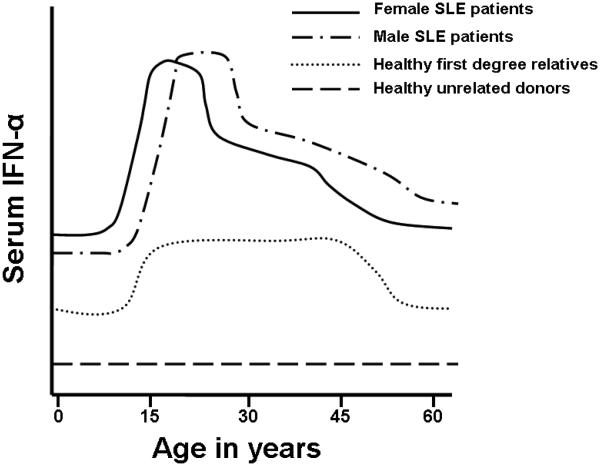
To address this issue, we used a sliding window analysis technique to determine which age ranges were characterized by significant increases in serum IFN-α. Age ranges which showed a significant increase in serum IFN-α were compiled for the patient, relative and healthy donor groups. Multivariate testing was then used to establish the boundaries of the high IFN-α age groups which best fit each data set. This pattern-finding algorithm revealed significantly increased IFN-α activity between the ages of 12 and 22 for female SLE patients and between the ages of 16 and 29 for male SLE patients. Both male and female healthy first degree relatives had significantly decreased IFN-α activity after the age of 50 (Figure 1). Overall there were no major gender-related differences in the cohort, and the healthy relatives and healthy unrelated donors are shown unstratified by gender in Figure 1. The age ranges of highest serum IFN-α corresponded well with the age ranges of peak SLE incidence for each gender. These data once again place high serum IFN-α at the scene of the crime in the disease initiation phase of human SLE.
Figure 1.
Diagram of mean serum IFN-α at different ages in female and male SLE patients, healthy family members, and unrelated healthy controls. Y-axis represents relative serum IFN-α levels measured as a functional activity, and X-axis shows age in years of the subject at the time of donation of the serum sample.
Osteopontin Polymorphisms Regulate Serum IFN-α and Serum Osteopontin in an Age- and Gender-Specific Manner
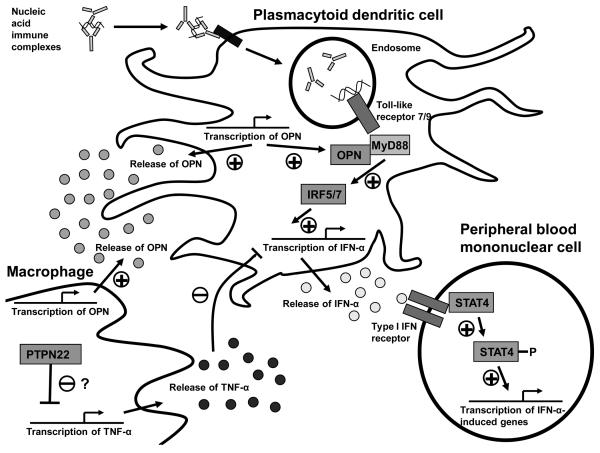
Osteopontin/secreted phosphoprotein 1 (OPN) is a phosphorylated extracellular matrix protein with a variety of functions, including wound healing, bone formation and remodeling (35), as well as immunological functions such as T-cell activation, Th1 differentiation, B-cell activation (36), and macrophage activation and chemotaxis (37). Studies have demonstrated high levels of OPN in biopsies of inflamed tissues in SLE and other autoimmune diseases (38, 39). OPN interacts with the MyD88 adaptor protein in murine dendritic cells and is required for downstream IFN-α production in the setting of Toll-like receptor 9 ligation, providing a link between this cytokine and IFN-α (40) (see Figure 2 for an illustration).
Figure 2.
Schematic diagram illustrating proposed functional locations of SLE-risk loci described and their proposed influence upon cytokine production and responses in SLE. Nucleic acid immune complexes can activate endosomal Toll-like receptors, and downstream signaling via the MyD88 adaptor leads to IFN-α production. This TLR/MyD88 signaling could be augmented by increases in OPN, as determined by OPN risk genotype. TNF-α can inhibit IFN-α production in dendritic cells, and PTPN22 alleles could influence IFN-α production via modulation of TNF-α production or TNF-α responses in macrophages or dendritic cells. After IFN-α has been produced, the STAT4 autoimmune disease risk allele results in increased sensitivity and downstream signaling following type I IFN receptor ligation.
A number of candidate gene studies have demonstrated an association between variants of the OPN gene and SLE (41-43). Interestingly, the largest genetic study to date found that the association of OPN variants with SLE was particular to male patients, who composed only 10% of patients in the study (43). This result is surprising as previous genetic studies which found evidence of association did not separate males and females for analysis and had similar proportions of male and female patients (42). Few studies have examined OPN levels in SLE patient serum or the potential influence of OPN genetic variants upon serum OPN levels. One previous study found that the SLE-associated rs9138 C allele in the 3′ untranslated region was associated with increased serum OPN levels in healthy controls, however this association was not seen in the SLE patients (42).
To investigate the role of the OPN gene in SLE, we studied SLE patients from multiple ancestral backgrounds to determine whether OPN genetic variants were linked to altered cytokine profile (7). Given the previous results suggesting a gender-specific association, we separated male and female patients for these analyses. The most consistently replicated SLE-associated SNP (rs9138 C) in the 3′ untranslated region of the gene was associated with increased serum OPN in male patients in a dose response fashion. Surprisingly, female patients as a group did not share this relationship. OPN functions in long bone remodeling (35), and OPN expression can be induced by estrogen through a non-classical estrogen receptor-related-α pathway (44). We hypothesized that age-related regulation of OPN in female patients may be present. When the female patients were stratified by age, younger patients (age <23) showed a dose-response increase in serum OPN in relation to the rs9138 C allele. This relationship was clearly not present in older patients (age ≥23), and risk allele carriers showed a dramatic difference in serum OPN levels by age. The same age- and gender- related patterns were present in all ancestral backgrounds.
Serum IFN-α levels were correlated with serum OPN levels in simultaneous samples, and the relationship between IFN-α and rs9138 genotype mirrored the relationships described above for serum OPN. Taken in the context of murine data which suggests an essential role for OPN in IFN-α production, we hypothesize that OPN may regulate IFN-α in SLE patients. We favor the hypothesis that both intracellular and extracellular OPN levels are differentially regulated by the SLE-associated SNP in the 3′ untranslated region of the gene. This SNP could function by altering mRNA poly-adenylation or stability, leading to increased protein translation. Other influences such as promotion of gene expression due to increased estrogen receptor-related-α signaling could then cooperate with increased mRNA stability to result in the higher OPN protein levels we detected. Increased OPN levels in the setting of the risk variant could subsequently augment downstream signaling from endosomal Toll-like receptors (Figure 2), resulting in greater IFN-α production, and the correlation between serum IFN-α and serum OPN which was observed. The OPN gene in SLE provides an example of a genetic risk locus which influences serum levels of two distinct but related inflammatory cytokines. Additionally, genetic variation at this locus should contribute to the age-related pattern we observed in serum IFN-α in the female SLE cohort described above.
PTPN22 SLE-Risk Allele is Associated with Increased Serum IFN-α and Decreased TNF-α
The C1858T polymorphism (rs2476601) in protein tyrosine phosphatase non-receptor type 22 (PTPN22) has been associated with risk of SLE, as well as multiple other autoimmune diseases including autoimmune thyroid disease, juvenile idiopathic arthritis, rheumatoid arthritis, and type I diabetes (45). IFN-α has been convincingly implicated in the pathogenesis of these autoimmune disorders (46-48) with the exception of rheumatoid arthritis, although emerging evidence suggests that IFN-α is expressed in rheumatoid arthritis joint tissue in humans and is associated with inflammation (49, 50). Interestingly, the PTPN22 1858 T autoimmune disease risk allele is not associated with multiple sclerosis (45), and is associated with protection from Crohn's disease (51). The mechanism by which the risk variant of PTPN22 predisposes to particular autoimmune diseases and protects from at least one is unknown. It is striking that the 1858 T risk allele associates with a number of IFN-α-related diseases, and not with multiple sclerosis which is commonly treated by administering IFN-β, a type I interferon closely related to IFN-α. In Crohn's disease, TNF-α is an important pathogenic cytokine (52), and there is no established role for IFN-α.
The PTPN22 C1858T polymorphism results in an arginine to tryptophan coding change in the Lyp protein, and work in lymphocytes suggests that the mutation results in decreased T- and B-cell responsiveness, as well as alterations in cytokine production in lymphocytes in vitro (53, 54). The exact molecular mechanism of the PTPN22 1858 T risk allele is not currently well described, and it is possible that relationships between this allele and cytokine profile could be secondary to other immune system alterations. While work has thus far focused on lymphocytes, the Lyp protein could presumably alter function in myeloid and dendritic cells, and little is known about in vivo human cytokine profiles related to this allele.
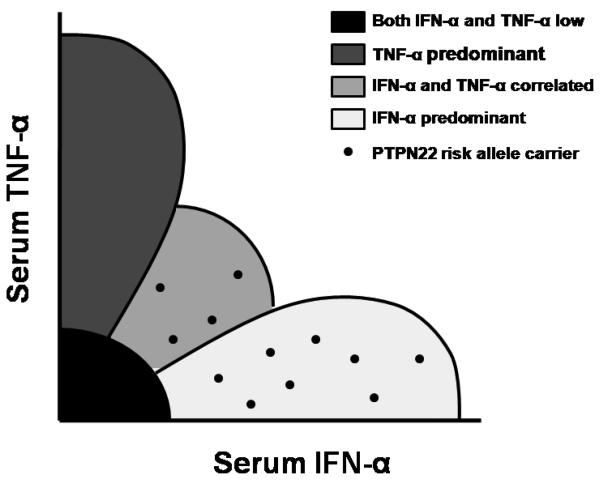
When we examined cytokine profiles in SLE patients carrying the PTPN22 1858 T risk allele, these patients had both higher serum IFN-α activity and lower TNF-α levels. When patients were grouped into categories of cytokine expression using a clustering algorithm, risk allele carriers much more commonly showed either an IFN-α predominant or an IFN-α and TNF-α correlated cytokine profile, instead of a TNF-α predominant or both cytokines low pattern (Figure 3). Interestingly, 25% of male patients carried the risk allele, compared to 10% of female patients (p=0.024), however cytokine skewing was similar in both sexes. This sex-skewing of PTPN22 allele frequencies had not been previously reported in SLE, although the small number of male SLE patients often makes such studies difficult. In rheumatoid arthritis, a similar finding of increased PTPN22 risk allele carriage in male patients as compared to female patients has been described (55).
Figure 3.
Diagram of serum IFN-α and TNF-α when measured simultaneously in SLE serum showing an overall inverse correlation between the two cytokine levels and the typical location of PTPN22 risk allele carriers within this continuum.
The strong influence of the PTPN22 1858 T allele upon both IFN-α and TNF-α levels in serum is interesting, as multiple lines of evidence suggest that these two cytokines can cross-regulate each other. In vitro, TNF-α can inhibit the production of IFN-α in dendritic cells (47), and in humans therapeutic blockade of TNF-α can lead to increased serum IFN-α (56, 57). The skewed cytokine profile we observe in the setting of the PTPN22 1858 T risk variant suggests that the PTPN22 gene could participate in this cross-regulation (Figure 2), which is not currently well understood at the molecular level. IFN-α and TNF-α are both cytokines which are largely produced by antigen presenting cells of the innate immune system such as dendritic cells and macrophages, respectively. The influence of PTPN22 variants upon both of these cytokines could suggest a function for PTPN22 in cells of the innate immune system, in addition to those which have been demonstrated in T and B lymphocytes (53). As noted above, TNF-α is important in Crohn's disease pathogenesis, and therapeutic blockade of TNF-α has been a highly effective disease treatment (52). It is interesting that the PTPN22 1858 C allele which is not associated with SLE and is linked to higher TNF-α in our study associates with Crohn's disease susceptibility. The cytokine patterns we observe in the setting of different PTPN22 alleles could help to explain the paradox of opposite alleles of the same gene predisposing to different autoimmune diseases.
SLE-Associated STAT4 Variant Confers Increased Sensitivity to IFN-α Signaling
The autoimmune disease risk alleles described above have all been associated with increased serum IFN-α in one fashion or another, and higher circulating levels of IFN-α will presumably alter immune system homeostasis, shifting responses to more pro-inflammatory antigen presentation. One could also envision the scenario in which a genetic polymorphism does not predispose to higher levels of circulating cytokine, but instead predisposes to an increased downstream responsiveness to a given amount of cytokine. The signal transducer and activator of transcription 4 (STAT4) gene can be activated and phosphorylated upon ligation of the type I IFN receptor by IFN-α (58), and subsequently induce downstream transcription of IFN-α-induced genes (Figure 2).
STAT4 variants are associated with susceptibility to SLE and rheumatoid arthritis (59, 60), as well as Sjogren's syndrome (61), and juvenile idiopathic arthritis (62). IFN-α is an important cytokine in Sjogren's syndrome, some types of juvenile idiopathic arthritis, and SLE as previously mentioned (47, 63, 64). While IFN-α has not classically been implicated in rheumatoid arthritis pathogenesis, recent studies suggest a role for IFN-α in rheumatoid synovial inflammation (49, 50). In SLE patients, the STAT4 risk allele is associated with nephritis and antibodies to double-stranded DNA, and these two clinical features are linked to disease severity and disease activity, respectively (65).
We examined serum IFN-α in SLE patients in the context of the STAT4 risk allele (66), and the risk allele was associated with lower serum IFN-α in SLE patients. This was intriguing, and we hypothesized that the STAT4 risk allele may predispose to a greater sensitivity to IFN-α. In this scenario, higher serum IFN-α in subjects with the non-risk genotype of STAT4 could indicate that more IFN-α was required to result in SLE susceptibility for these subjects. If the risk of SLE due to IFN-α is dose-dependent, then subjects who develop SLE in the setting of a STAT4 non-risk genotype would have higher serum IFN-α activity when compared to the general SLE cohort, and conversely subjects with the STAT4 risk genotype may develop SLE at lower serum levels of IFN-α. Alternatively, the non-risk genotype of STAT4 may result in impaired feedback regulation of IFN-α, and decreased sensitivity to IFN-α could result in over-production. To explore this further, we examined serum IFN-α activity as well as IFN-α-induced gene expression in peripheral blood mononuclear cells from the same sample in SLE patients. We then compared the upstream serum IFN-α signal with the amount of downstream IFN-α-induced gene expression in subjects with different STAT4 genotypes. This approach allowed for an in vivo measurement of IFN-α sensitivity in human SLE patients in the context of different STAT4 variants (66). We found that the risk allele of STAT4 was associated with increased IFN-α-induced gene expression despite simultaneously being associated with lower serum levels of IFN-α, confirming that this polymorphism was associated with increased IFN-α sensitivity (66).
Conclusions
Autoimmune diseases such as SLE involve complex immune system dysregulation which is polygenic in nature, and likely results from synergy at a number of loci relevant to the immune system. In these studies we have shown examples of alleles that are associated with increased circulating IFN-α in SLE, and one allele that is associated with increased sensitivity to IFN-α in SLE. It is likely that these two categories of risk factors act synergistically to result in the in vivo pathway dysregulation that predisposes to human disease. By performing genetic studies on an individual feature of the immune system such as circulating cytokine levels, one can detect much larger genetic effects than those observed in classical case-control studies, and begin to ascertain the functional significance of disease-associated polymorphisms in human disease in vivo. This is critical, as there is a great deal of genetic diversity in the immune system between species, and model organisms may not be adequate for the study of the subtle genetic changes which result in human susceptibility. The immune system has been under selective pressure from micro-organisms for all of human history, and there is a good deal of variability in autoimmune disease risk loci between different historical continental human populations. Progress in human autoimmunity will require these types of studies which both embrace and define the diversity present in human complex disease.
Clinical trials in SLE have largely been unsuccessful, and underlying heterogeneity in the molecular pathogenesis of disease has likely contributed to this lack of success. Anti-cytokine therapies have been highly successful in other autoimmune diseases, for example anti-TNF-α are widely used in rheumatoid arthritis and Crohn's disease. Data from mouse models has suggested that TNF-α may actually have a protective effect in SLE (67), and many have been wary to try this therapy in human SLE patients. Small open label trials have reported success with TNF-α blockade in SLE (68), and it is possible that genetic and/or cytokine analysis may assist in identifying appropriate patients for this therapy. A phase I trial of anti-IFN-α antibodies in SLE has been completed (69), and we are currently awaiting the results of phase II trials. Anti-osteopontin antibodies have been studied in murine models of inflammatory arthritis (70), and it seems likely that a human therapeutic targeting this cytokine will be developed. It is possible that in the near future genetics could also be leveraged in the prediction of response/non-response to anti-cytokine agents in SLE, building upon the work described herein.
Acknowledgments
Grant Support and Disclosures: SN Kariuki – none; TB Niewold – NIH K08 AI083790, NIAID Clinical Research Loan Repayment AI071651, Arthritis National Research Foundation Eng Tan Scholar Award, University of Chicago CTSA Core Subsidy Grant and Collaborative University of Chicago/Northshore University Health System Translational Research Pilot Grant from UL1 RR024999
Abbreviations
- SLE
systemic lupus erythematosus
- IFN-α
interferon alpha
- TNF-α
tumor necrosis factor alpha
- OPN
osteopontin
- MHC
major histocompatibility complex
- PTPN22
protein tyrosine phosphatase non-receptor type 22
- STAT4
signal transducer and activator of transcription 4
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chun HY, Chung JW, Kim HA, Yun JM, Jeon JY, Ye YM, et al. Cytokine IL-6 and IL-10 as biomarkers in systemic lupus erythematosus. J Clin Immunol. 2007;27(5):461–6. doi: 10.1007/s10875-007-9104-0. [DOI] [PubMed] [Google Scholar]
- 2.Liu TF, Jones BM. Impaired production of IL-12 in system lupus erythematosus. II: IL-12 production in vitro is correlated negatively with serum IL-10, positively with serum IFN-gamma and negatively with disease activity in SLE. Cytokine. 1998;10(2):148–53. doi: 10.1006/cyto.1997.0269. [DOI] [PubMed] [Google Scholar]
- 3.Grondal G, Gunnarsson I, Ronnelid J, Rogberg S, Klareskog L, Lundberg I. Cytokine production, serum levels and disease activity in systemic lupus erythematosus. Clin Exp Rheumatol. 2000;18(5):565–70. [PubMed] [Google Scholar]
- 4.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358(9):929–39. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 5.Kariuki SN, Crow MK, Niewold TB. The PTPN22 C1858T polymorphism is associated with skewing of cytokine profiles toward high interferon-alpha activity and low tumor necrosis factor alpha levels in patients with lupus. Arthritis Rheum. 2008;58(9):2818–2823. doi: 10.1002/art.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Studnicka-Benke A, Steiner G, Petera P, Smolen JS. Tumour necrosis factor alpha and its soluble receptors parallel clinical disease and autoimmune activity in systemic lupus erythematosus. Br J Rheumatol. 1996;35(11):1067–74. doi: 10.1093/rheumatology/35.11.1067. [DOI] [PubMed] [Google Scholar]
- 7.Kariuki SN, Moore JG, Kirou KA, Crow MK, Utset TO, Niewold TB. Age- and gender-specific modulation of serum osteopontin and interferon-alpha by osteopontin genotype in systemic lupus erythematosus. Genes Immun. 2009 doi: 10.1038/gene.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301(1):5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 9.Niewold TB. Interferon alpha-induced lupus: proof of principle. J Clin Rheumatol. 2008;14(3):131–2. doi: 10.1097/RHU.0b013e318177627d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crow MK. Type I interferon in systemic lupus erythematosus. Curr Top Microbiol Immunol. 2007;316:359–86. doi: 10.1007/978-3-540-71329-6_17. [DOI] [PubMed] [Google Scholar]
- 11.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100(5):2610–5. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairhurst AM, Wandstrat AE, Wakeland EK. Systemic lupus erythematosus: multiple immunological phenotypes in a complex genetic disease. Adv Immunol. 2006;92:1–69. doi: 10.1016/S0065-2776(06)92001-X. [DOI] [PubMed] [Google Scholar]
- 13.Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36(8):481–90. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- 14.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54(6):1906–16. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 15.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landolt-Marticorena C, Bonventi G, Lubovich A, Ferguson C, Unnitahn T, Su J, et al. Lack of association between the interferon-alpha signature and longitudinal changes in disease activity in Systemic Lupus Erythematosus. Ann Rheum Dis. 2008 doi: 10.1136/ard.2008.093146. [DOI] [PubMed] [Google Scholar]
- 17.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10(5):373–9. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harley IT, Kaufman KM, Langefeld CD, Harley JB, Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10(5):285–90. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8(6):907–22. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 20.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52(5):1491–503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 21.Ytterberg SR, Schnitzer TJ. Serum interferon levels in patients with systemic lupus erythematosus. Arthritis Rheum. 1982;25(4):401–6. doi: 10.1002/art.1780250407. [DOI] [PubMed] [Google Scholar]
- 22.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58(8):2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronnblom LE, Alm GV, Oberg KE. Possible induction of systemic lupus erythematosus by interferon-alpha treatment in a patient with a malignant carcinoid tumour. J Intern Med. 1990;227(3):207–10. doi: 10.1111/j.1365-2796.1990.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 24.Niewold TB, Swedler WI. Systemic lupus erythematosus arising during interferon-alpha therapy for cryoglobulinemic vasculitis associated with hepatitis C. Clin Rheumatol. 2005;24(2):178–81. doi: 10.1007/s10067-004-1024-2. [DOI] [PubMed] [Google Scholar]
- 25.Tsao BP, Grossman JM, Riemekasten G, Strong N, Kalsi J, Wallace DJ, et al. Familiality and co-occurrence of clinical features of systemic lupus erythematosus. Arthritis Rheum. 2002;46(10):2678–85. doi: 10.1002/art.10519. [DOI] [PubMed] [Google Scholar]
- 26.Ramos PS, Kelly JA, Gray-McGuire C, Bruner GR, Leiran AN, Meyer CM, et al. Familial aggregation and linkage analysis of autoantibody traits in pedigrees multiplex for systemic lupus erythematosus. Genes Immun. 2006;7(5):417–32. doi: 10.1038/sj.gene.6364316. [DOI] [PubMed] [Google Scholar]
- 27.Ioannou Y, Isenberg DA. Current evidence for the induction of autoimmune rheumatic manifestations by cytokine therapy. Arthritis Rheum. 2000;43(7):1431–42. doi: 10.1002/1529-0131(200007)43:7<1431::AID-ANR3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Petri M. Epidemiology of systemic lupus erythematosus. Best Pract Res Clin Rheumatol. 2002;16(5):847–58. doi: 10.1053/berh.2002.0259. [DOI] [PubMed] [Google Scholar]
- 29.Lopez P, Mozo L, Gutierrez C, Suarez A. Epidemiology of systemic lupus erythematosus in a northern Spanish population: gender and age influence on immunological features. Lupus. 2003;12(11):860–5. doi: 10.1191/0961203303lu469xx. [DOI] [PubMed] [Google Scholar]
- 30.Szyper-Kravitz M, Zandman-Goddard G, Lahita RG, Shoenfeld Y. The neuroendocrine-immune interactions in systemic lupus erythematosus: a basis for understanding disease pathogenesis and complexity. Rheum Dis Clin North Am. 2005;31(1):161–75, x. doi: 10.1016/j.rdc.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 31.van Vollenhoven RF, Engleman EG, McGuire JL. An open study of dehydroepiandrosterone in systemic lupus erythematosus. Arthritis Rheum. 1994;37(9):1305–10. doi: 10.1002/art.1780370906. [DOI] [PubMed] [Google Scholar]
- 32.Chagnon P, Schneider R, Hebert J, Fortin PR, Provost S, Belisle C, et al. Identification and characterization of an Xp22.33;Yp11.2 translocation causing a triplication of several genes of the pseudoautosomal region 1 in an XX male patient with severe systemic lupus erythematosus. Arthritis Rheum. 2006;54(4):1270–8. doi: 10.1002/art.21733. [DOI] [PubMed] [Google Scholar]
- 33.Stevens AM, Tsao BP, Hahn BH, Guthrie K, Lambert NC, Porter AJ, et al. Maternal HLA class II compatibility in men with systemic lupus erythematosus. Arthritis Rheum. 2005;52(9):2768–73. doi: 10.1002/art.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niewold TB, Adler JE, Glenn SB, Lehman TJ, Harley JB, Crow MK. Age- and sex-related patterns of serum interferon-alpha activity in lupus families. Arthritis Rheum. 2008;58(7):2113–9. doi: 10.1002/art.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denhardt DT, Noda M. Osteopontin expression and function: role in bone remodeling. J Cell Biochem Suppl. 1998;30-31:92–102. [PubMed] [Google Scholar]
- 36.Lampe MA, Patarca R, Iregui MV, Cantor H. Polyclonal B cell activation by the Eta-1 cytokine and the development of systemic autoimmune disease. J Immunol. 1991;147(9):2902–6. [PubMed] [Google Scholar]
- 37.Scatena M, Liaw L, Giachelli CM. Osteopontin: a multifunctional molecule regulating chronic inflammation and vascular disease. Arterioscler Thromb Vasc Biol. 2007;27(11):2302–9. doi: 10.1161/ATVBAHA.107.144824. [DOI] [PubMed] [Google Scholar]
- 38.Masutani K, Akahoshi M, Tsuruya K, Tokumoto M, Ninomiya T, Kohsaka T, et al. Predominance of Th1 immune response in diffuse proliferative lupus nephritis. Arthritis Rheum. 2001;44(9):2097–106. doi: 10.1002/1529-0131(200109)44:9<2097::AID-ART360>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 39.Chabas D, Baranzini SE, Mitchell D, Bernard CC, Rittling SR, Denhardt DT, et al. The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science. 2001;294(5547):1731–5. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- 40.Shinohara ML, Lu L, Bu J, Werneck MB, Kobayashi KS, Glimcher LH, et al. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol. 2006;7(5):498–506. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forton AC, Petri MA, Goldman D, Sullivan KE. An osteopontin (SPP1) polymorphism is associated with systemic lupus erythematosus. Hum Mutat. 2002;19(4):459. doi: 10.1002/humu.9025. [DOI] [PubMed] [Google Scholar]
- 42.D'Alfonso S, Barizzone N, Giordano M, Chiocchetti A, Magnani C, Castelli L, et al. Two single-nucleotide polymorphisms in the 5′ and 3′ ends of the osteopontin gene contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2005;52(2):539–47. doi: 10.1002/art.20808. [DOI] [PubMed] [Google Scholar]
- 43.Han S, Guthridge JM, Harley IT, Sestak AL, Kim-Howard X, Kaufman KM, et al. Osteopontin and systemic lupus erythematosus association: a probable gene-gender interaction. PLoS ONE. 2008;3(3):e0001757. doi: 10.1371/journal.pone.0001757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanacker JM, Delmarre C, Guo X, Laudet V. Activation of the osteopontin promoter by the orphan nuclear receptor estrogen receptor related alpha. Cell Growth Differ. 1998;9(12):1007–14. [PubMed] [Google Scholar]
- 45.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG, Nath SK, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases--a meta-analysis. Rheumatology (Oxford) 2007;46(1):49–56. doi: 10.1093/rheumatology/kel170. [DOI] [PubMed] [Google Scholar]
- 46.Mavragani CP, Danielides S, Niewold TB, Kirou KA, Moutsopoulos HM, Crow MK. Activation of the type I interferon pathway in autoimmune thyroid disease. Arthritis Rheum. 2007;56(suppl):S229. [Google Scholar]
- 47.Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci U S A. 2005;102(9):3372–7. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devendra D, Eisenbarth GS. Interferon alpha--a potential link in the pathogenesis of viral-induced type 1 diabetes and autoimmunity. Clin Immunol. 2004;111(3):225–33. doi: 10.1016/j.clim.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Walker JG, Ahern MJ, Coleman M, Weedon H, Papangelis V, Beroukas D, et al. Characterisation of a dendritic cell subset in synovial tissue which strongly expresses Jak/STAT transcription factors from patients with rheumatoid arthritis. Ann Rheum Dis. 2007;66(8):992–9. doi: 10.1136/ard.2006.060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roelofs MF, Wenink MH, Brentano F, Abdollahi-Roodsaz S, Oppers-Walgreen B, Barrera P, et al. Type I interferons might form the link between Toll-like receptor (TLR) 3/7 and TLR4-mediated synovial inflammation in rheumatoid arthritis (RA) Ann Rheum Dis. 2009;68(9):1486–93. doi: 10.1136/ard.2007.086421. [DOI] [PubMed] [Google Scholar]
- 51.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40(8):955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutgeerts P, Vermeire S, Van Assche G. Biological therapies for inflammatory bowel diseases. Gastroenterology. 2009;136(4):1182–97. doi: 10.1053/j.gastro.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Vang T, Miletic AV, Bottini N, Mustelin T. Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity. 2007;40(6):453–61. doi: 10.1080/08916930701464897. [DOI] [PubMed] [Google Scholar]
- 54.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179(7):4704–10. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 55.Pierer M, Kaltenhauser S, Arnold S, Wahle M, Baerwald C, Hantzschel H, et al. Association of PTPN22 1858 single-nucleotide polymorphism with rheumatoid arthritis in a German cohort: higher frequency of the risk allele in male compared to female patients. Arthritis Res Ther. 2006;8(3):R75. doi: 10.1186/ar1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dastmalchi M, Grundtman C, Alexanderson H, Mavragani CP, Einarsdottir H, Helmers SB, et al. A high incidence of disease flares in an open pilot study of infliximab in patients with refractory inflammatory myopathies. Ann Rheum Dis. 2008;67(12):1670–7. doi: 10.1136/ard.2007.077974. [DOI] [PubMed] [Google Scholar]
- 57.Mavragani CP, Niewold TB, Moutsopoulos NM, Pillemer SR, Wahl SM, Crow MK. Augmented interferon-alpha pathway activation in patients with Sjogren's syndrome treated with etanercept. Arthritis Rheum. 2007;56(12):3995–4004. doi: 10.1002/art.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyler DR, Persky ME, Matthews LA, Chan S, Farrar JD. Pre-assembly of STAT4 with the human IFN-alpha/beta receptor-2 subunit is mediated by the STAT4 N-domain. Mol Immunol. 2007;44(8):1864–72. doi: 10.1016/j.molimm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357(10):977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sigurdsson S, Nordmark G, Garnier S, Grundberg E, Kwan T, Nilsson O, et al. A risk haplotype of STAT4 for systemic lupus erythematosus is over-expressed, correlates with anti-dsDNA and shows additive effects with two risk alleles of IRF5. Hum Mol Genet. 2008;17(18):2868–76. doi: 10.1093/hmg/ddn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korman BD, Alba MI, Le JM, Alevizos I, Smith JA, Nikolov NP, et al. Variant form of STAT4 is associated with primary Sjogren's syndrome. Genes Immun. 2008;9(3):267–70. doi: 10.1038/gene.2008.1. [DOI] [PubMed] [Google Scholar]
- 62.Prahalad S, Hansen S, Whiting A, Guthery SL, Clifford B, McNally B, et al. Variants in TNFAIP3, STAT4, and C12orf30 loci associated with multiple autoimmune diseases are also associated with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60(7):2124–2130. doi: 10.1002/art.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bave U, Nordmark G, Lovgren T, Ronnelid J, Cajander S, Eloranta ML, et al. Activation of the type I interferon system in primary Sjogren's syndrome: a possible etiopathogenic mechanism. Arthritis Rheum. 2005;52(4):1185–95. doi: 10.1002/art.20998. [DOI] [PubMed] [Google Scholar]
- 64.Niewold TB, Rivera TL, Buyon JP, Crow MK. Serum type I interferon activity is dependent on maternal diagnosis in anti-SSA/Ro-positive mothers of children with neonatal lupus. Arthritis Rheum. 2008;58(2):541–6. doi: 10.1002/art.23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, et al. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS Genet. 2008;4(5):e1000084. doi: 10.1371/journal.pgen.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: Autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009;182(1):34–8. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacob CO, McDevitt HO. Tumour necrosis factor-alpha in murine autoimmune ‘lupus’ nephritis. Nature. 1988;331(6154):356–8. doi: 10.1038/331356a0. [DOI] [PubMed] [Google Scholar]
- 68.Aringer M, Smolen JS. The role of tumor necrosis factor-alpha in systemic lupus erythematosus. Arthritis Res Ther. 2008;10(1):202. doi: 10.1186/ar2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yao Y, Richman L, Higgs BW, Morehouse CA, de los Reyes M, Brohawn P, et al. Neutralization of interferon-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60(6):1785–96. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 70.Fan K, Dai J, Wang H, Wei H, Cao Z, Hou S, et al. Treatment of collagen-induced arthritis with an anti-osteopontin monoclonal antibody through promotion of apoptosis of both murine and human activated T cells. Arthritis Rheum. 2008;58(7):2041–52. doi: 10.1002/art.23490. [DOI] [PubMed] [Google Scholar]