Abstract
Comparison of mutant KIR3DL1*015 substituted at natural positions of variation showed that tryptophan/leucine dimorphism at position 283 uniquely changes receptor conformation and can strongly influence binding of the A24nef tetramer. Dimorphic motifs at positions 2, 47 and 54 in D0 and 182 and 283 in D1+D2 distinguish the two 3DL1 lineages, typified by 3DL1*005 and 3DL1*015. The inter-lineage recombinant, KIR3DL1*001, combines D0 of 3DL1*005 with D1+D2 of 3DL1*015 and binds A24nef more strongly than either parent. In contrast, the reciprocal recombinant with D0 from 3DL1*015 and D1+D2 from 3DL1*015 cannot bind A24nef. Thus D0 polymorphism directly affects the avidity of the KIR3DL1 ligand-binding site. From these observations, multiple sequence alignment and homology modeling, we constructed structural models for KIR3DL1 and its complex with A24nef. In these models D0, D1 and D2 come together to form a binding surface for A24nef, which is contacted by all three Ig-like domains. A central pocket binds arginine 83, the only Bw4 motif residue essential for KIR3DL1interaction, similarly to the binding of lysine 80 in HLA-C by KIR2DL1. Central to this interaction is a salt bridge between arginine 83 of Bw4 and glutamate 282 of 3DL1, which juxtaposes the functionally influential dimorphism at position 283. Further 3DL1 mutants were tested and shown to have A24nef binding properties consistent with the models. A24nef was not bound by KIR3DS1, the activating counterpart of KIR3DL1. Moreover, introducing any one of three residues specific to KIR3DS1: serine 163, arginine 166 or leucine 199, into 3DL1*015, abrogated A24nef binding.
Keywords: Cell surface molecules, Human, MHC, Natural Killer cells
INTRODUCTION
Killer-cell immunoglobulin-like receptors (KIR) comprise a variable family of activating and inhibitory lymphocyte receptors that recognize polymorphic epitopes of MHC class I (1). In higher primates, KIR are principally expressed by NK cells, for which the variegated expression and immunological functions of KIR are remarkably similar to those of the structurally unrelated Ly49 family of mouse NK cell receptors (2). During development, interactions between inhibitory KIR or Ly49 and their cognate self -MHC class I ligands determine the strength with which the mature NK cells can respond to unhealthy cells in which MHC class I expression is perturbed by infection or malignancy (3). The interactions of KIR with MHC class I have been mainly studied in the human species, where HLA-A, B and C all provide ligands for inhibitory KIR. In contrast, ligands and functions for the activating KIR are poorly understood (4, 5).
HLA-C is recognized by inhibitory KIR2DL. Crystallographic structures of HLA-C bound to KIR2DL show that D1 and D2, the two extracellular Ig-like domains, form a binding site that interacts with a part of the face of MHC class I that binds T-cell receptors (1). Binding loops of D1 and D2 contact the C-terminal part of the α1 helix, the C-terminal part of the bound peptide, and the N-terminal part of the α2 helix (1). At the interface between KIR2D and HLA-C, residue 44 of KIR2D and residue 80 of HLA-C determine specificity. Thus, methionine 44 containing KIR2DL1 is specific for HLA-C with lysine 80, whereas lysine 44 containing KIR2DL3 is specific for HLA-C with asparagine 80. The two inhibitory KIR3DL that recognize HLA-A and B have a third Ig-like domain, D0, in addition to D1 and D2. KIR3DL2 has narrow specificity, resembling a T-cell receptor, for complexes of an Epstein-Barr virus peptide bound to HLA-A*03 or HLA-A*11 (6). KIR3DL1 has a broad specificity, more like those of KIR2DL, for the Bw4 epitopes carried by subsets of HLA-A and HLA-B allotypes. The Bw4 epitope is defined by polymorphic sequence motifs at positions 79-83 of the α1 helix. The essential residue for 3DL1 recognition of Bw4 is arginine 83 (7), contrasting with the importance of residue 80 in HLA-C for KIR2DL recognition. Progress in visualizing the molecular details of the interaction between 3DL1 and Bw4, and the contribution of the D0 domain, has been slow because of difficulties in expressing soluble 3DL1 in native conformation.
Mutagenesis experiments have shown that D0 is important for the folding and cell surface expression of 3DL1 (8), and that it contributes to binding HLA class I (9). Deletion of residues 50 and 51 from D0, a natural deletion in the chimpanzee counterpart of 3DL1, increased binding-site avidity as did alanine substitutions at positions 49-52 (9). Independent evidence for the functional importance of sites in D0 was obtained from examining the natural variation in 3DL1, and related KIR in non-human primates, for the signature of natural selection. Strong evidence for natural selection on all three Ig domains was obtained (10). In D1 and D2 these sites largely correspond to those that contact bound HLA class I in the crystal structures of KIR2DL bound to HLA-C, consistent with the results of mutagenesis in these domains (9). Four sites of strong positive selection in D0 (positions 5, 31, 32, and 51) were predicted to form a cluster on the D0 surface, providing a good candidate for a functional binding site, because residue 51 was known to influence ligand-binding (9). The fifth positively selected residue, position 20, was also predicted to be on the D0 surface, but far apart from the four-residue cluster (10).
To explore further the role of D0, we used mutagenesis and binding of antibodies and HLA class I tetramers to determine the influence of natural variation on 3DL1 binding to Bw4+ HLA class I. By combining these approaches with multiple sequence alignment and homology modeling we built a structural model of KIR3DL1 bound to Bw4.
MATERIALS AND METHODS
Antibodies and flow cytometric analyses
PE-labeled anti-KIR3DL1 monoclonal antibodies DX9, Z27 and 177407 were obtained commercially (DX9; BD Biosciences, USA; Z27: Beckman Coulter, USA; 177407: R&D Systems, USA). Additional antibodies reactive with KIR3DL1 were 5.133 (a gift from M. Colonna Washington University School of Medicine, USA) and 177406, 177407, 177409, 177410, and 177412 (R&D Systems, USA), which were conjugated to phycoerythrin (PE) using Zenon technology (Invitrogen, USA) and the Phycolink kit (Prozyme) as prescribed by the manufacturers. Anti-KIR3DL2 antibody DX31 was from L. Lanier (University of California San Francisco, USA), anti-KIR2DS4 antibody FES172 was purchased from Beckman Coulter (Fullerton, CA, USA).
Human subjects, blood samples and KIR typing
Blood samples were donated by six healthy individuals; informed consent was as approved by the Stanford University IRB on Human Subjects. PBMC were prepared with Ficoll-Paque PLUS (Amersham Biosciences, Piscataway, NJ). KIR3DL1 types were determined by sequencing cDNA and/or SSP-PCR as described (11). KIR3DL1 phenotypes of peripheral blood NK cells were determined as described (11). KIR3DL2-positive cells were eliminated from the analysis using anti-KIR3DL2 antibody (DX31) because antibodies 5.133 and 177407 cross-react with KIR3DL2.
KIR3DL1-EGFP mutant constructs
Chimeric KIR3DL1*015-EGFP, comprising the coding region of KIR3DL1*015 fused in frame with the coding sequence of EGFP, was subcloned into pcDEFIII (12) and used as the template for site-directed mutagenesis performed with the Quik Change Mutagenesis kit (Stratagene, La Jolla, CA). Error-free clones were identified by automated sequencing (ABI 377 instrument) using primers spanning the entire KIR3DL1-EGFP region (1Fa: 5’-ccacagaaaaccttccct-3’, 1Ra: 5’-atctgaccaacattgcag-3’, 2Fa: 5’-cgtgaccttgtcctgtag-3’, 2Ra: 5’- gagcctacgttcatgggc-3’, 2Rb: 5’-cgcactgcagggagcctacg-3’, 3Fa: 5’-ctcctcttctttctcctt-3’, 3Ra: 5’-ctgttctgttccctgcag-3’, 4Fa: 5’-accttgtcctgtagctcc-3’, 5Fa: 5’-agatccaaagttgtctcc-3’, Fe1-3c: 5’-tcttggtccagagggcc-3’, pcDEF-R: 5’-gtggcaccttccagggtcaa-3’). DNA was prepared using the QIAGEN Endofree Maxiprep Kit (Qiagen, USA) and resuspended in TE to a concentration of 1μg/μl. Using similar methods, a recombinant chimeric cDNA construct encoding the extracellular Ig-like domains (D0, D1, D2) of 3DS1*013 and the stem (ST) and cytoplasmic (CYT) domains of 3DL1*015 was made with an in-frame C-terminal EGFP tag and subcloned into the expression vector pcDEFIII.
Cell culture and transfection
The Jurkat T cell line was cultured at 37°C with 5% CO2 in RPMI 1640 medium (Invitrogen, USA) supplemented with 10% (v/v) bovine calf serum, 1% L-glutamine (Invitrogen, USA), and 1X Penicillin-Streptomycin (Invitrogen, USA). Genomic HLA class I typing by the LABType® method (One Lambda Inc, Canoga Park, CA) showed Jurkat cells have the HLA-A*03/A*03, -B*07/B*35, and -Cw*04/Cw*07 alleles, none of which encodes a Bw4 epitope. Absence of Bw4 on the Jurkat cell surface was confirmed by flow cytometry with the anti-Bw4 antibody (One Lambda Inc, Canoga Park, CA). Neither did Jurkat cells endogenously express KIR3DL1/S1 or other KIR at the cell surface, as assessed by flow cytometry with anti-KIR monoclonal antibodies.
Transfection of Jurkat cells with 3DL1 constructs was performed using AMAXA nucleofection technology (AMAXA, Germany). 2 × 106 Jurkat cells in the exponential (log) phase of growth were washed twice in RPMI-1640, resuspended in 100 μl of AMAXA Nucleofector solution V and mixed with 4 μg of construct DNA. Cells were electroporated using the G-10 pulsing parameter and were immediately transferred to pre-warmed culture medium in a 6-well culture plate. Transfected cells were grown for 72 hours before staining with anti-KIR3DL1 antibodies. All transfection experiments were performed in triplicate and results were averaged and expressed as fraction of antibody binding with 3DL1*015. The NKL cell line was maintained in RPMI 1640 medium supplemented with 10% FBS (Invitrogen, USA), 2mM glutamine (Invitrogen, USA), 100U/ml penicillin/streptomycin (Invitrogen, USA) and 100 U/ml rIL-2 (NIH, NCI, Preclinical Repository, USA). Transfection of NKL cells with 3DL1 constructs was performed using the AMAXA nucleofection system as described above for the Jurkat cell line with the exception that pulsing parameter O-17 was used.
Flow cytometric analyses
Transfected Jurkat cells were washed in PBS with 1% FCS and stained with PE-conjugated monoclonal antibodies. Live cells, detected by exclusion of propidium iodide (PI), were analyzed for PE and EGFP fluorescence using a FACScan flow cytometer (BD Biosciences, USA). Mean PE fluorescence intensity (mfi) was normalized against mfi of EGFP to determine the relative cell surface expression of KIR3DL1 protein as described previously (12).
KIR3DL1 on the cell surface of CD56+CD3- NK cells was measured using monoclonal antibodies and flow cytometry. Phycoerythrin conjugates of anti-NKB1 (DX9) (BD Biosciences, CA), anti-3DL1 (177407, R&D systems, USA), and anti-3DL1/3DS1 (Z27, Beckman-Coulter, USA) were used in combination with anti-CD3-Peridinin chlorophyll protein conjugates (SK7, BD Biosciences, USA) and anti-CD56-fluorescein (NCAM16.2, BD Biosciences, USA) to stain PBMC. Three-color flow cytometry used a FACScan instrument and CellQuest software (BD Biosciences, USA). Mean fluorescence intensity (mfi) within the population of KIR3DL1-expressing cells was used for comparison between KIR3DL1 allotypes.
Tetramer analyses
HLA class I tetramers were made as described by Altman et al. (13). In brief, the HLA class I heavy chain cDNA was modified by substitution of the exons encoding the transmembrane and cytosolic regions with a sequence encoding the BirA biotinylation enzyme recognition site. The modified HLA heavy chain and ß2-microglobulin, were synthesized in a prokaryotic expression system (pET, R&D Systems), purified from bacterial inclusion bodies, solubilized in the presence of denaturant (4M urea) and allowed to refold by dilution with the synthetic peptide of choice. Refolded monomeric complexes were purified by FPLC and biotinylated using BirA (Avidity), then combined with phycoerythrin (PE)-labeled streptavidin (Sigma) at a 4:1 molar ratio to form tetrameric HLA-peptide complexes.
Stable Jurkat transfectants expressing KIR3DL1 allotypes and mutants were generated and maintained in culture medium containing G418 (2mg/ml). For staining followed by flow cytometric analysis, 0.5 × 106 transfected Jurkat cells were incubated with 1μg of PE-conjugated tetramer for 1 hr at 4°C. Cells were washed twice in PBS and analyzed by FACS. Specificity of tetramer binding was confirmed by blocking with antibody DX9 (4°C, 30’) prior to addition of tetramer. The principal tetramer (A24nef) used in this study comprised HLA-A*2402 loaded with a peptide (RYPLTFGW) derived from the nef2 protein of HIV. Additional tetramers used in this study as controls were HLA-A*0201 CMV (NLVPMVATV), HLA-A*0203 CMV (NLVPMVATV), HLA-A*0207 FluM (GILGFVFTL), HLA-A*2901 gp160 (FNCGGEFFY), HLA-A*0301 gp41 and (RLRDLLLIVTR) (6, 14)
Computational structural analyses
Sequences homologous to the D0 domain of 3DL1*015 were obtained from the Protein Data Bank (PDB) and Swiss-Prot databank using Blast (15) on the Swiss Institute of Bioinformatics (SIB) server (http://www.ch.embnet.org/software/bBLAST.html). Multiple sequence alignments were performed with ClustalW (16) on the EMBL-EBI server (http://www.ebi.ac.uk/clustalw/) and then displayed using ESPript2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The amino acid sequence of the D1+D2 domains of 3DL1*015 shows 72% identity (identical residue) and 77% similarity (residue with similar physico-chemical properties) to D1+D2 of 2DL1 and 2DL2. D0 shows 36% identity and 55% similarity to D1 of 2DL2, and 38% identity and 50% similarity to D1 of 2DL1. The online software for PDB, Blast, ClustalW, and ESPript2.2 were all accessed in May, 2007.
Homology models for D0 and D1+D2 of 3DL1*015 were built separately using the program MODELLER version 9v1 (http://www.salilab.org/modeller/about_modeller.html) (17, 18). The model for D1+D2 of 3DL1 was based on the D1+D2 domains in the three-dimensional structure of KIR2DL1 bound to HLA-C*04 (PDB file 1IM9) (19). The HLA-bound forms of KIR2DL were preferred for modeling, because our aim was to analyze the effects of 3DL1 polymorphism on HLA binding and maximal KIR-HLA interactions were recovered from the complex. D1 of 2DL2 bound to HLA-C*03 (PDB file 1EFX) (20) was chosen for modeling D0 of 3DL1 rather than D1 of 2DL1, because the latter exhibited a larger gap in the multiple sequence alignment.
Two constraints used in making the model were: first, that D0 was positioned relative to D1+D2 so that residues methionine 2 and isoleucine 54 interact with HLA class I ligand as indicated by the functional experiments described in this paper; second, that typtophan 13 of D0 was positioned close to tyrosine 281 of D2 in order to create a hydrophobic interaction between the two domains and stabilize the relative position of D0. Using the loop refinement feature of MODELLER, favorable conformations were generated for the linker region between D0 and D1 of 3DL1. The stereochemical quality of the 3DL1 model was checked with program Procheck (http://www.biochem.ucl.ac.uk/~roman/procheck/procheck.html) for which the online software was accessed in July, 2007 (21). HLA-contacting residues of domains D1+D2 of 3DL1 were determined by superimposing our generated model on domains D1+D2 of the complex of 2DL1 and HLA-C*04 (PDB file 1IM9) (19) and by superimposing the structure of HLA-A*2402 (PDB file 2BCK) (22), a ligand for 3DL1*015, onto HLA-Cw4. The configuration of the 3DL1*015-A*2402 complex was optimized by energy minimization using the Gromacs package version 3.3.1 (http://www.gromacs.org) and the Gromos force field. Images of the model were made with Pymol (http://www.pymol.org). Separate electropotential surface maps of 3DL1*015 and A*2402 were produced using the APBS software package (23). Solvent accessibility scores for amino acid residues were calculated using the program DSSP (24) and displayed in the alignment with ESPript2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Relative solvent accessibility was determined against a fully extended Gly-X-Gly tripeptide as reference. Buried residues are defined as those having <7% of the maximum solvent-accessible surface area.
RESULTS
Dimorphism at position 283 in the D2 domain changes KIR3DL1 conformation
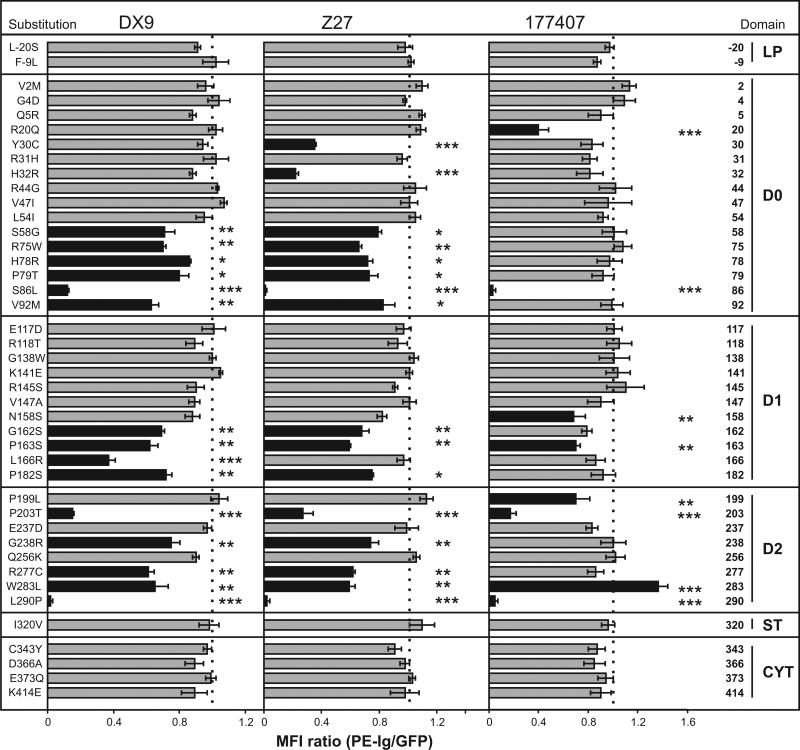
KIR3DL1*015 is the most prevalent form of 3DL1 and has the consensus sequence (10). Using the 3DL1*015-EGFP construct as template, 42 different point mutants were made, each corresponding to a natural substitution present in another form of 3DL1 or in 3DS1, the allotypic activating receptor. Mutant cDNA were transiently transfected into Jurkat T cells, which express neither KIR nor any 3DL1-binding HLA class I allotype. The transfected cells were assessed for binding to three phycoerythrin-conjugated monoclonal antibodies: DX9, specific for 3DL1 (25); Z27 that binds strongly to 3DL1 and weakly to 3DS1 (26); and 177407 that was raised against 3DL1, but also cross-reacts with KIR3DL2 (data not shown). The binding data obtained with these antibodies was normalized to the abundance of KIR3DL1, as measured by EGFP fluorescence (12) (Figure 1).
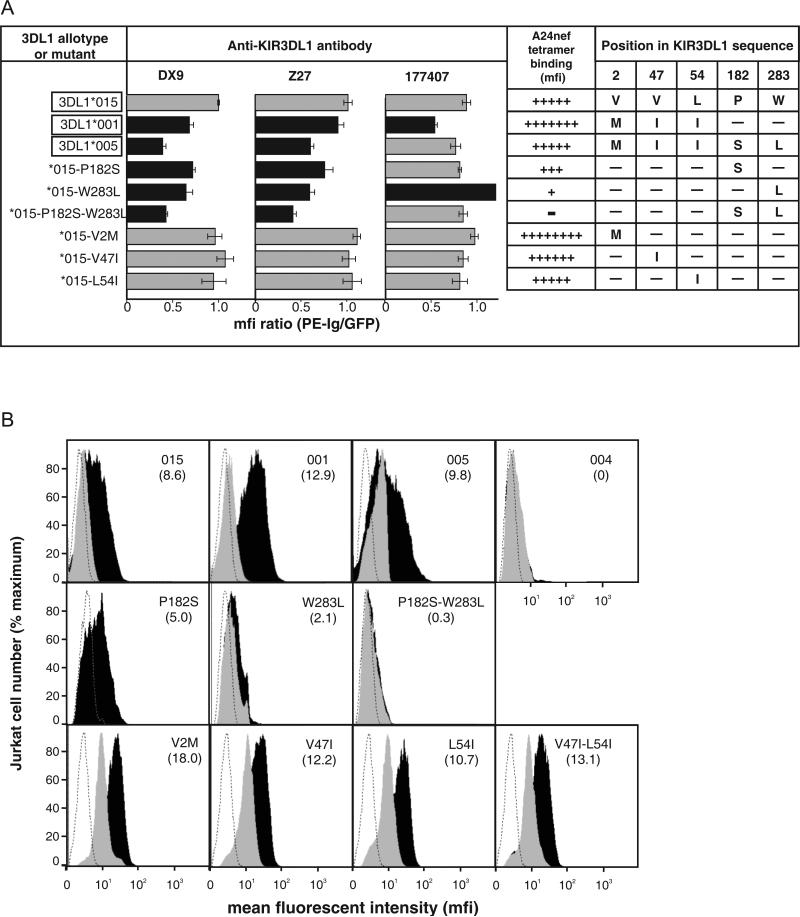
Figure 1. Interaction of monoclonal antibodies with KIR3DL1*015 point mutants.
Each 3DL1*015 mutant has a substitution that occurs naturally in another KIR3DL1/S1 allotype. Jurkat cells transiently transfected with each mutant were tested for binding to three monoclonal antibodies: DX9, Z27 and 177407. To correct for differential expression of the mutants, the phycoerythrin (PE) fluorescence due to antibody binding was normalized to the fluorescence due to the green fluorescent protein attached to the cytoplasmic tail of each 3DL1. The binding was then normalized to that obtained for ‘wild type’ 3DL1*015. Shaded black are mutants giving statistically significant differences in binding compared to 3DL1*015. Error bars represent SEM derived from three independent experiments. Statistical significance of differences in binding was calculated using the Student's t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
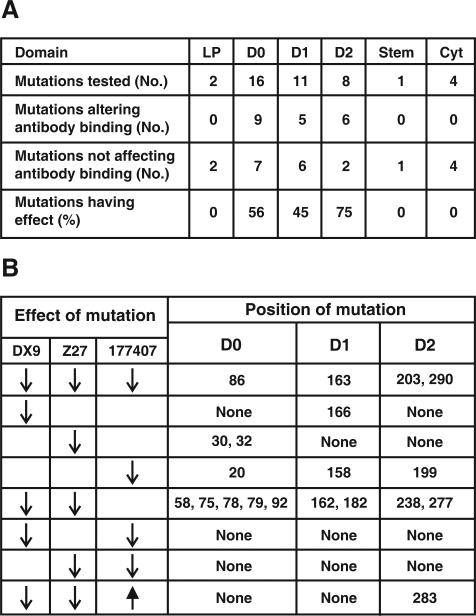
Twenty mutations affected binding to one or more of the anti-KIR3DL1 antibodies; all were in the extracellular domains, with greater abundance in D0 and D2, than D1 (Figure 2A). Mutation at three positions greatly reduced binding to all three antibodies (Figure 2B), indicating that threonine 203 in 3DL1*037 and proline 290 in 3DS1*045 likely reduce cell surface expression of these rare variants, as previously shown for serine 86 in 3DL1*004 (12, 27). Overall, DX9 and Z27 exhibited similar reaction patterns with the panel of mutants, although clear differences were observed for the position 30, 32 and 166 mutants. In contrast, antibody 177407 exhibited a very different specificity: only eight mutations reduced 177407 binding, compared to 15 for DX9 and 16 for Z27, and the mutations affecting 177407 binding are largely non-overlapping with those that affect DX9 and Z27 binding (Figure 2B). These results point to considerable overlap between the epitopes recognized by DX9 and Z27, but little overlap between them and the epitope recognized by 177407.
Figure 2. Mutation at position 283 in D2 uniquely changes the capacity of KIR3DL1*015 to bind anti-KIR3DL1 monoclonal antibodies.
Panel A shows the proportion of mutations that affect antibody binding and their distribution between the various domains of KIR3DL1*015. Panel B summarizes the effects of mutation on the binding of antibodies DX9, Z27 and 177407, and shows that only mutation at position 283 both increase (177407) or decrease (DX9 and Z27) the binding of different antibodies. Reduction and increase of antibody binding are indicated by downward and upward pointing arrows respectively. Absence of an arrow denotes no effect on antibody binding.
Mutation at position 283 in the D2 domain gave a unique pattern of antibody binding: substitution of leucine for tryptophan increased the binding to 177407 while reducing the binding to DX9 and Z27 (Figures 1 and 2B). Thus tryptophan 283 favors binding to DX9 and Z27, whereas leucine 283 favors binding to 177407. Position 283 in 3DL1 corresponds to position 188 in KIR2DL, an invariant tryptophan in all KIR2DL. Crystallographic structures of KIR2DL1, 2 and 3 showed tryptophan 188 is one of eleven hydrophobic residues forming an inter-domain core that affects the hinge angle between the D1 and D2 domains (1). These residues are conserved in all KIR2DL and KIR3DL1, the only exception being position 283 in 3DL1 where leucine is the only alternative to the otherwise constant tryptophan.
That the constituent residues of the hydrophobic core are conserved in 3DL1*015 predict it has a core like that of KIR2DL. In the 3DL1*015-W283L mutant, replacement of the bulky, aromatic tryptophan at position 283 by the smaller, aliphatic leucine is predicted to alter the hinge angle and change the relative orientation of the D1 and D2 domains. Such a conformational difference could account for the observed differences in antibody binding between *015 and *015-W283L. Because residue 283 is buried and surface inaccessible in the 2DL2 structure, the tryptophan/leucine polymorphism in 3DL1 is more likely to exert its effects on antibody binding in an indirect manner rather than contributing directly to antibody contact.
Dimorphism at position 283 distinguishes the two KIR3DL1 lineages
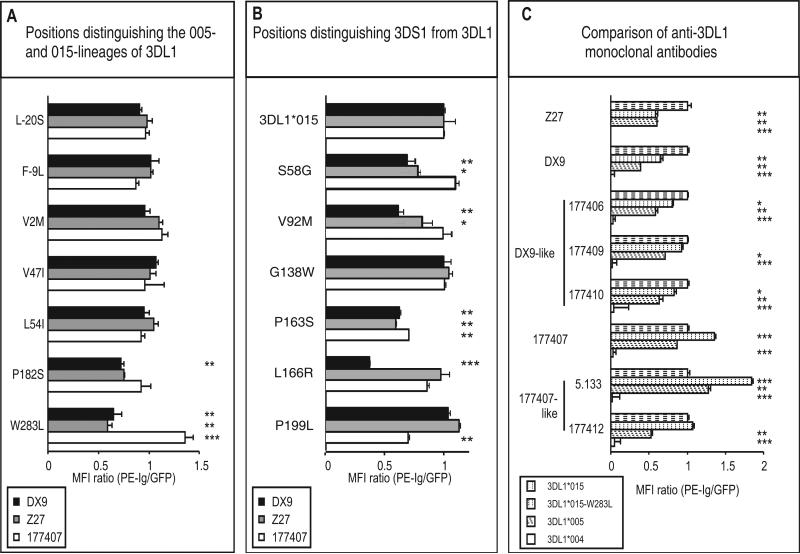
Tryptophan and leucine at position 283, are both at high frequency in human populations, and comprise one of seven dimorphisms that distinguish the 015- and 005-lineages of 3DLl allotypes (10). These positions are distributed throughout the extracellular domains, but only substitution at position 182 in D1 and 283 in D2 had significant effects upon antibody binding (Fig. 3A). Whereas substitution of tryptophan (015-lineage) to leucine (005-lineage) increased 177407 binding and decreased DX9 and Z27 binding, substitution of proline (015-lineage) to serine (005-lineage) at position 182 reduced DX9 and Z27 binding but had little effect on 177407 (Fig. 3A). Thus both the serine 182 and leucine 283 residues present in 3DL1*005 have the effect of reducing interaction with DX9 and Z27 while preserving or increasing the interaction with 177407. In each case the effects are quantitative and for neither position 182 or 283 did the lineage-specific substitution abrogate binding of any antibody.
Figure 3. Mutation at lineage-specific sites and comparison of anti-KIR3DL1 antibodies.
Panel A shows the effects on antibody binding of substitution at the seven sites that distinguish the *005 and *015 KIR3DL1 lineages. In each 3DL1*015 mutant one of the sites has been changed to the residue present in 3DL1*005. Panel B shows the effects on antibody binding of substitution at the six sites that distinguish KIR3DS1 from KIR3DL1. In each 3DL1*015 mutant one of the sites has been changed to the residue present in 3DS1*013. Panel C compares the binding of eight different monoclonal anti-KIR3DL1 antibodies to three natural variants -- 3DL1*004, *005, *015 -- and the 3DL1*015-W283L mutant. 3DL1*004, which is not expressed at cell surfaces, is a negative control (12). The methods of data collection and analysis are as described in the legend to Figure 1.
At the seven positions which distinguish the two 3DL1 lineages, 3DS1 has four identical to 3DL1*015 (-20, 47, 182 and 283) and three identical to 3DL1*005 (-9, 2 and 54). That 3DS1 is not significantly more like one 3DL1 lineage than the other, is consistent with 3DS1 and 3DL1 having diverged before 3DL1 split into the 005- and the 015-lineages (10). Also consistent with this evolutionary model, 3DS1 is distinguished from both 3DL1 lineages at six positions (58, 92, 138, 163, 166 and 199) that have no overlap with those distinguishing the two 3DL1 lineages. When the 3DS1 residues at these positions were individually introduced into 3DL1*015, the resulting mutants retained substantial reactivity with all three antibodies, which bound to levels >50% of that for 3DL1*015, except for mutation at position 166 which reduced DX9 binding by ~60% (Fig. 3B). Thus the very low Z27 binding and the lack of DX9 binding that characterize 3DS1 (26, 28, 29) cannot be attributed to the effect of a single substitution. Consequently, these properties likely arise from the concerted effect of two or more of the six 3DS1-specific substitutions.
We also assessed five additional anti-KIR3DL1 monoclonal antibodies for binding to 3DL1*015, *015-W283L, *005 and *004. This comparison revealed no novel specificity: three antibodies were shown to be similar to DX9, two were similar 177407, and none resembled Z27 (Fig. 3C).
Conformational change caused by position 283 dimorphism does not determine the high- and low-binding phenotypes of 3DL1 allotypes
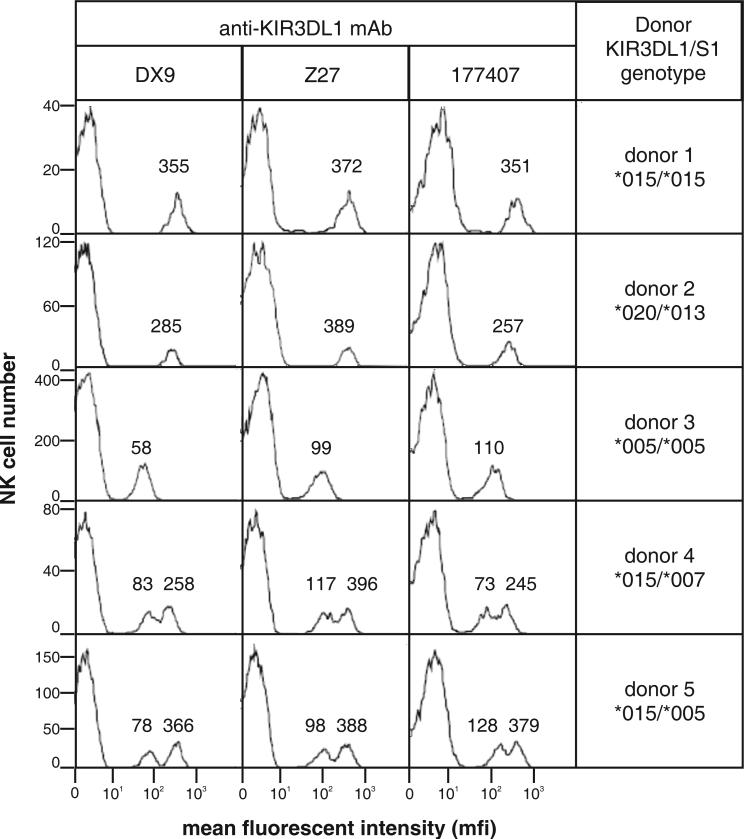
Our previous studies have shown that peripheral blood NK cells expressing particular KIR3DL1 allotypes can be distinguished by the amount of DX9 or Z27 antibody they bind (11, 30, 31). For example, 3DL1*015 binds these antibodies to high level, 3DL1*005 to low level. To see if these differences correlated with the conformational change conferred by polymorphism at position 283, we examined the binding of 177407 to peripheral blood NK cells of defined KIR3DL1/S1 genotype and compared the results to those obtained with DX9 and Z27 (Fig. 4). If conformational change was the basis for the difference, the expectation was that 3DL1*005-expressing NK cells would bind more 177407 than 3DL1*015-expressing NK cells. That was not the case: all three antibodies distinguished high binding (3DL1*015 and *020) from low binding (*005 and *007) allotypes. Although antibody 177407 is different from DX9 and Z27 in conformational sensitivity, it too can distinguish between the high- and low-expressing 3DL1 allotypes. Thus this major distinction, between high- and low-binding allotypes, is not correlated with either the different epitope specificity of the antibodies or the presence at position 283 of either tryptophan (*015, *020 and *007) or leucine (*005). It can therefore be attributed to varying levels of cell-surface expression of KIR3DL1, as was previously concluded (11, 30, 31).
Figure 4. Like DX9 and Z27, the 177407 antibody distinguishes KIR3DL1 allotypes expressed on the surface of peripheral blood NK cells.
NK cells were isolated from the blood of donors of known KIR3DL1/S1 genotype and analyzed for expression of 3DL1/S1 using the DX9, Z27 and 177407 antibodies and flow cytometric analysis as described (11). KIR3DL1 allotypes are expressed at different levels on the NK cell surface and bind the DX9 and Z27 antibodies to high level (for example, 3DL1*015 and *020) or to low level (for example, 3DL1*005 and *007) Because minority subsets of NK cells express KIR3DL1, in heterozygous donors there are different subsets of NK cells expressing the two alleles (as well as a small number of cells expressing both alleles). If the donor is homozygous for a high-expressing allele (donor 1) then a unimodal distribution of 3DL1-expressing cells is observed, with a relatively high level of antibody bound. The value for the mean fluorescent intensity (mfi) of cells within each peak is given above the peak: 355 with DX9 of donor 1, for example. Similarly, a donor homozygous for a low-expressing allele (donor 3) gives a unimodal distribution with relatively low mfi. Heterozygote donors with a high- and a low-expressing allele (donors 4 and 5) give a bimodal distribution because of the two subsets of NK cells, one expressing the high allele and the other the low allele. The very low binding of 3DS1*013 to Z27 (26, 28) is seen in donor 2 as slight shoulder on the peak of KIR-negative NK cells (the majority) with mfi <1.
One consistent difference was that *005 bound more 177407, but less DX9 and Z27, than *007 (Fig. 4). Thus *005, which has leucine 283, binds better to 177407, whereas *007, which has tryptophan 283, bound better to DX9 and Z27, an observation consistent with the comparison of *015 and *015-W283L (Fig.1). Thus this difference in antibody binding could reflect conformational change between *005 and *007, rather than their relative abundance at the cell surface. However, the effect is small and elevates neither *005 nor *007 into the range observed for high-binding allotypes such as *015.
Dimorphisms in D0, D1 and D2 combine to alter avidity of KIR3DL1 for A24nef
3DL1*015 and *005 differ by five substitutions: three in D0 (positions 2, 47 and 54), one in D1 (position 182) and one in D2 (position 283). These substitutions are recombined in the common 3DL1*001 allotype, which has D0 derived from *005 and D1+D2 derived from *015. Previous qualitative analysis showed that the A24nef tetramer, comprising HLA-A*2402 and a peptide derived from the nef2 protein of HIV, binds to 3DL1*001, *005, and *015 (14). Here, we quantified the binding of A24nef to Jurkat cells expressing 3DL1*001, *005, *015 and selected mutants (Figure 5). The results are summarized in Fig. 5A and representative examples of the flow cytometric analysis are shown in Figure 5B. As controls, the levels of 3DL1 cell-surface expression were assessed using the anti-KIR3DL1 antibodies (Figure 5A). The observed binding was highly specific for the A24nef tetramer, as no binding was seen with five other tetramers tested (see Materials and Methods).
Figure 5. Interactions between D0 polymorphisms and D1+D2 polymorphisms determine the binding of 3DL1 to the A24nef tetramer.
Panel A summarizes the binding of anti-KIR3DL1 antibodies and the A24nef tetramer to natural and mutant 3DL1 and the amino acid substitutions that distinguish them. Antibody binding was determined as described in the legend to figure 1. The names of the natural 3DL1 variants are boxed. The bars corresponding to levels of antibody bound that are significantly different from 3DL1*015 are shaded black. The extent to which the A24nef tetramer bound to natural and mutant 3DL1 expressed by transiently transfected Jurkat cells is indicated by the number of crosses (+), which reflect the mfi values calculated from the flow cytometric data shown in panel B. Panel B shows the A24nef tetramer staining of individual mutants transfected into Jurkat cells. The dotted line is staining of untransfected Jurkat cells, the gray curve is A24nef staining of Jurkat transfectants pre-incubated with DX9, and the black curve is A24nef staining of Jurkat transfectants without anti-body blocking. The identity of the transfectant is shown in the upper right-hand corner of each plot and the number below indicates the mfi of the A24nef stained cells.
Jurkat cells transfected with 3DL1*001 bound A24nef to a higher level (mfi:12.9) than cells transfected with either 3DL1*005 (9.8) or *015 (8.6). This hierarchy of binding did not correspond to that observed with the anti-KIR3DL1 antibodies, all of which bound less to 3DL1*001 than *015; and 177407 bound less to 3DL1*001 than *005 (Figure 5A). Thus the elevated binding of A24nef to 3DL1*001 compared to*005 and *015 reflects superior avidity for A24nef rather than greater abundance at the cell surface.
These results demonstrate that in 3DL1*001, the combination of the 3DL1*005 D0 with the D1+D2 of 3DL1*015 binds more effectively to A24nef than the ‘parental’ domain combinations in either 3DL1*015 or 3DL1*005. The reciprocal recombinant to 3DL1*001 combines the D0 of 3DL1*015 with D1+D2 of 3DL1*005. Such an allotype has not been identified in any human population (10), but is identical in structure to the *015-P182S-W283L mutant. Although we found this mutant was well expressed at the cell surface as detected by antibodies, it was unable to bind A24nef (Figure 5). Thus the combination of the 3DL1*015 D0 and the 3DL1*005 D1+D2 is far inferior to either 3DL1*001, *005 or *015 in binding A24nef. These results clearly demonstrate the important role of complementary polymorphisms in D0 and D1+D2 for the functional interaction of KIR3DL1 with HLA class I ligands. In particular, the inter-lineage recombination that produced 3DL1*001 combined features of 3DL1*015 and *005 that synergize to increase the avidity of 3DL1*001 for the A24nef tetramer. Since its origin in one individual, 3DL1*001 has spread throughout the world, being now present in all major population groups except Amerindians (10). These results also demonstrate that the ‘enhancing’ effect D0 has on the binding of HLA class I to KIR3DL1 does not depend on interaction between two cell membranes and formation of an immunological synapse, one of several models we considered previously (9), as the effect is clearly seen here for the interaction of tetrameric HLA class I ligand with cell-surface associated receptor.
Comparing the double mutant at positions 182 and 283 with the single mutants showed that the inability of *015-P182S-W283L to bind A24nef was principally due to leucine 283, with a minor contribution from serine 182 (Figure 5). The loss of A24nef binding by *015-W283L is paralleled by increased binding to antibody 177407. Thus the conformation of 3DL1 that favors binding to 177407 appears less permissive to A24nef binding. Although the introduction of serine 182 and leucine 283 into 3DL1*015 abrogates binding to A24nef, the presence of these residues in *005 is permissive for A24nef binding. Because residues 182 and 283 are the only differences in D1+D2 between 3DL1*015 and *005, these observations imply that the substitutions distinguishing 3DL1*005 from *015 in D0 affect the conformation of D1+D2 to make 3DL1*005 permissive for A24nef binding. Highest binding is, however, achieved by the combination present in 3DL1*001 of D0 from *005 with D1+D2 from *015 (Figure 5). As 3DL1*001 only differs from *015 at positions 2, 47 and 54 in D0, its superiorA24nef binding must arise from one or more of these substitutions. Single mutations at each of these positions bound A24nef like 3DL1*015, as did a double mutant at positions 47 and 54. Either all three substitutions are necessary for superior binding, or it is the combination of methionine 2 with either isoleucine 47 or isoleucine 54. In 3DL1*005 these same substitutions compensate for the presence of serine 182 and leucine 283, permitting this allotype to bind A24nef at a level comparable to that of 3DL1*015.
Model for the structure of KIR3DL1*015 and its interaction with A*2402
Previously, alanine substitutions at positions 49, 50, 51 and 52 in D0 increased Bw4+ HLA-B binding by 3DL1, but not at twelve other positions examined (9). Alanine at either position 50 or 51 could also compensate for the reduced binding to 3DL1 caused by mutating residue 76 in HLA-B from glutamate to alanine (9). Residues 49-52 are invariant in 3DL1, but they are flanked by the dimorphisms at residues 47 and 54 shown here to modulate D0 function. Positions 5, 20, 31, 32 and 51 in D0 have also been identified as sites of positive natural selection (10). Of these, 5, 31, 32 and 51 are predicted to cluster on the D0 surface, whereas 20 lies apart, but also on the surface. Together these functional and phylogenetic analyses suggest that residues 47-54 mark a functional site of interaction between the D0 domain of 3DL1 and bound Bw4+ HLA class I ligand. In the absence of a crystallographic structure for 3DL1, we explored this possibility by using multiple sequence alignment, homology modeling, and the functional data to construct structural models for 3DL1*015 and its interaction with A24nef.
Model of the 3DL1*015 structure
D1+D2 of 3DL1*015 were modeled on D1+D2 in the crystallographic structure of 2DL1 bound to HLA-C*04 (PDB file 1IM9) (19). Based upon similarities observed in the multiple sequence alignment (Figure 6), the D0 domain of 3DL1*015 was modeled on D1 in the crystallographic structure of 2DL2 bound to HLA-C*03 (see Materials and Methods). The models can be viewed at http://csb.stanford.edu/karine/3DL1. The high sequence identity between D1 and D2 of KIR3DL1 and their KIR2DL counterparts (~77% and ~88%, respectively), gives confidence in the validity of the structural models for 3DL1 D1 and D2. In contrast, D0 of 3DL1 has only ~38% sequence identity with D2 of 2DL2. However, the structure of a protein with >30% sequence identity to a known structure can often be predicted with accuracy equivalent to a low-resolution X-ray structure (32). Consequently, detection of secondary structures elements like α-helices and β-sheets is more reliable, whereas attribution of length and conformation for loops are less accurate. Similarly for the position of D0 relative to D1/D2 which was inferred on the basis of functional properties and structural constraints. Shifts of D0 with respect to D1-D2 might occur, because D0 is not predicted to interact strongly with D1, and the linker between them appears flexible. We should stress that the model of 3DL1*015 presented here has limitations in its structural foundation, but is consistent with current knowledge of the function and polymorphism of KIR3DL1/S1.
Figure 6. Multiple sequence alignment for the extracellular ligand-binding domains domains (D0, D1 and D2) of human KIR.
Included in the alignment were representative KIR of all four human lineages: 2DL4*003 of lineage I (Q99706); 3DL1*015 (P43529), 3DS1*013 (Q14943), and 3DL2*001 (P43630) of lineage II; 2DL1*001 (P43626) and 2DL2*001 (P43627) of lineage III, and KIR3DL3*001 (Q8N743) of lineage V. Note that 2DL4 has only D0 and D2 domains, but no D1 domain. Above the alignment are shown the elements of secondary structure in the KIR3DL1 model and the template used (2DL2 for D0 and 2DL1 for D1-D2). The arrows indicate positions of the β strands and T the residues implied in bulge conformation. Below the alignment: each amino acid is assessed to be exposed (black), buried (white) or intermediate (gray) according to their accessible solvent area (asa); large stars indicate the sites of mutation studied here, rectangles the mutations that gave a significant difference in antibody binding compared to 3DL1*015. Above the alignment: + denotes residues in the electropositive site predicted for D0 and D1; * indicates residues that contribute to the hydrophobic binding pocket; squares indicate residues involved in predicted salt bridges and hydrogen bonds; and gray lines show the position of loops that form the predicted HLA-binding site, or are close to it.
Whereas the interaction of D1 with D2 in the model of 3DL1*015 appears to be dominated by a core of eleven hydrophobic residues, sequence comparison revealed no equivalent core of hydrophobic residues between D0 and D1 (Figure 6): only one hydrophobic interaction, between tryptophan 13 of D0 and glycine 138 of D1, being predicted. Constraining the position of D0 with respect to D1+D2 is the length of the linker (residues 96-103) between D0 and D1. Consequently, the only possible position for D0 that permitted contact with HLA class I ligand bound to D1+D2, as deduced from previous (9) and present studies in mutagenesis, was in the space between D1 and D2.
In this orientation three flexible loops of D0 (residues R48-L54, Y30-N35, and H78-V92) and the N-terminal region (H1-P9) are positioned close to the ligand-binding site. To model the rotational orientation of D0 with respect to D1+D2 we applied four additional constraints. Firstly, the N-terminal region could not be buried through interdomain interactions, because of its hydrophilicity and potential flexibility. Secondly, our functional analysis implicated residues 2, 47 and/or 54 in the enhanced binding of ligand. The positioning of residue 2 in the N-terminal region was limited by the constraint to keep this region accessible; residue 47 is located in a β-sheet and could not be placed near the binding site without stretching the D0-D1 linker; and so residue 54 was placed as close to the ligand-binding site as possible. Thirdly, residue 86 was made surface accessible because it is implicated in chaperone interactions (12). Fourthly, we required that the interaction of D0 with the other two domains be stabilized by hydrophobic interaction. Of the possible hydrophobic contacts examined, only those involving tryptophan 13 of D0 were compatible with the other three constraints. Moreover, tryptophan 13 provided an excellent candidate for fitting into the hydrophobic core of D1+D2, notably by making stacking interactions with tyrosine 281 of D2. In the deduced model, the D0 domain is seen to interact intimately with both D1 and D2, forming a compact, rather than an elongated, structure (Figure 7).
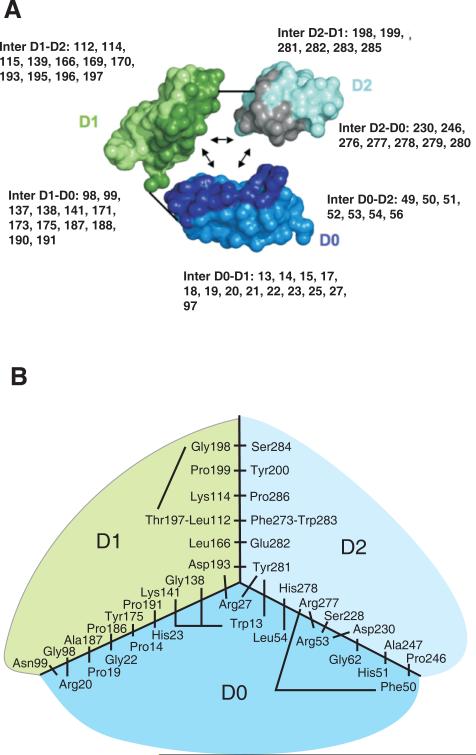
Figure 7. Model for the interaction of the D0, D1 and D2 domains in KIR3DL1.
Panel A shows the interactions between D0, D1 and D2 in the structural model. KIR3DL1 is represented in a simplified way with each amino acid being represented by two balls, one corresponding to the Cα atom the other to the center of the side-chain. The three domains have been separated so as to show their mutually interacting surfaces, which are shaded more darkly. The short straight lines show where the polypeptide chain connects D0 to D1 and D1 to D2. In each domain, the residues that contribute to interdomain interactions are shown. Here panel B shows a schematic view of the interacting residues that bring the three domains together. The view is from the top of the ligand-binding site in a two dimensional projection. The central part of the diagram corresponds to the junction of the three domains D0, D1 and D2. Connecting lines represent interactions between pairs of residues. Not all hydrophobic residues from the hydrophobic core are shown; those absent are valine 145, alanine 169, glycine 170, valine 196.
Of the non-covalent interactions between domains, those between D0 and D1 are fewest in number in the model. Hydrophobic interactions are made by residues 187, 188 and 191, part of a stretched loop maintained by the functionally influential proline 182 (Figure 5). Arginine 20, which defines the smaller of the two sites of positive natural selection on the D0 domain surface (10), interacts with residues 96-103 in the hinge between D0 and D1. Substitution of arginine 20 for glutamine may alter conformation, as detected by reduced 177407 antibody binding to *015-R20Q (Figures 1 and 2), by affecting the angle between the two domains. Although D0 and D2 are not directly connected in the polypeptide chain, the model predicts they interact strongly through hydrophobic contacts, a salt bridge and polar interactions. As well as the hydrophobic interaction between tyrosine 281 in D2 and tryptophan 13 in D0, arginine 277 in D2 is surrounded by four residues of D0: isoleucine 49, phenylalanine 50, histidine 51 and glycine 52 and interacts with phenylalanine 50 and arginine 53.
Model for the interaction of KIR3DL1*015 with HLA-A*2402
The crystallographic structure of A*2402 (PDB file 2BCK) (22) was first docked onto the D1+D2 domains of 3DL1*015 using the complex of HLA-C*04 bound to KIR2DL1 as the model (19). The configurations of certain side chains were then changed manually, either to avoid steric incompatibility between the two proteins or to optimize polar or salt bridge interactions. Finally, the energy of the system was minimized to optimize the contacts between 3DL1*015 and A*2402. Clearly, the limitations to the model of 3DL1*015 will extend to this model of the complex formed by 3DL1*015 and A*2402, which has additional potential for inaccuracy that derives from using the complex of HLA-C*04 and KIR2DL1 as template for docking HLA-A*2402 on KIR3DL1.
In the model the D0, D1 and D2 domains of 3DL1*015 come together to form a flat surface predicted to contact A*2402 using eight loops: two in D0 (residues 30-34 and 51-55); three in D1 (residues 115-118, 138-142 and 162-169); and three in D2 (198-202, 225-230 and 275-282) (Figure 8A). Consistent with this contribution of D0, the larger site of positive natural selection on the D0 domain surface (10) comprises residues from both the loops (31, 32 and 51) as well as position 5 from the amino-terminal region (which is near the binding site). Importantly, leucine 54 in D0 appears to contact the essential arginine 83 in the Bw4 motif, and arginine 31 in D0 contacts asparagine 86, also in the helix of the A*2402 α1 domain. As in the complexes of KIR2DL with HLA-C (19, 20), the binding loops of the D1 and D2 domains of 3DL1*015 are predicted to interact with the α1 and α2 helices of A*2402, respectively.
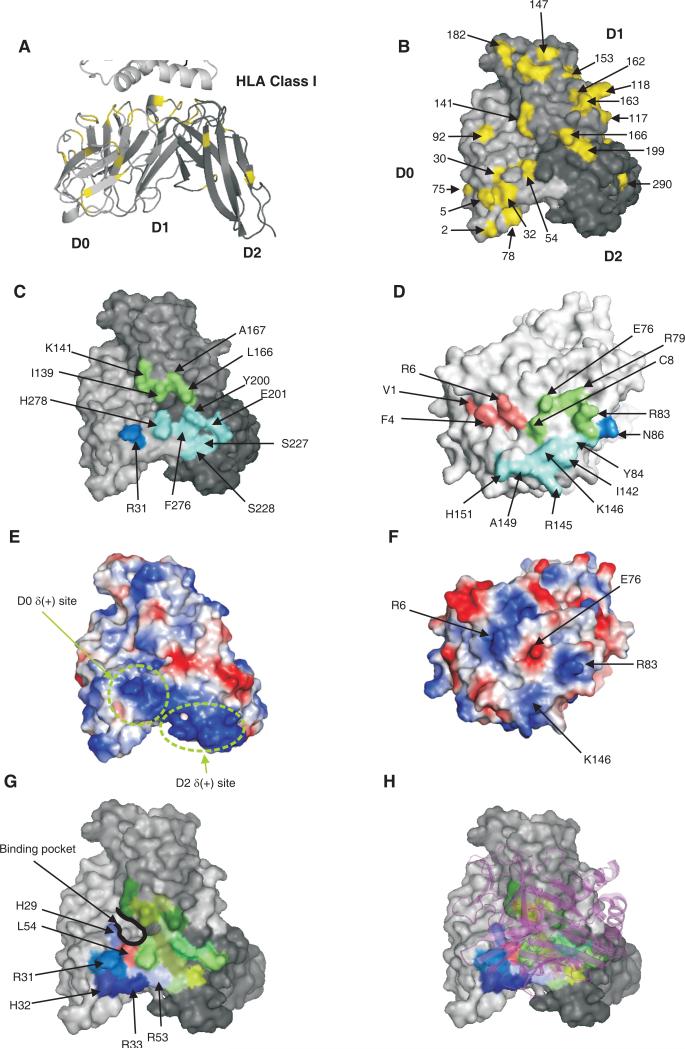
Figure 8. Model for the binding of HLA-A*2402 to 3DL1*015.
A shows a cartoon representation of the KIR3DL1 model. B, C, E, G, and H are surface representations of KIR3DL1 viewed from the bottom of the D1-D2 domain elbow. D and F are surface representations of HLA-A viewed from the top of the peptide-binding groove. A and B show polymorphic amino acids of 3DL1/S1 mapped in yellow onto the D0, D1 and D2 domains of the 3DL1*015 model. C shows the footprint of A*2402 on the surface of 3DL1*015. Blue, green and cyan denote respectively the D0, D1 and D2 domains buried by HLA-A. D shows the footprint of 3DL1*015 on the surface of A*2402. Blue, green and cyan denotes the A*2402 residues contacted respectively by the D0, D1 and D2 domains. Peptide contacting residues are colored in pink. E and F show the electropotential surfaces of 3DL1*015 and A*2402. Red and blue colors represent negative and positive electrostatic potential, respectively. The junction of the electropositive regions of the D0 [D0δ(+)] and D2 [D2δ(+)] domains result in a large electropositive area. Basic residues of A*2402 (arginine 145 and arginine 83) interact with acidic residues of 3DL1*015 (glutamate 201 and glutamate 282) to form salt bridges. The interaction between arginine 83 of HLA-A is the basis for the Bw4 specificity of KIR3DL1 (7). G shows the binding pocket defined by domains D0, D1 and D2 of KIR3DL1. The electropositive stretch of amino acids (29-32) is shown in shades of blue. Residue 54 at the base of binding pocket is shown in red. H shows the HLA ribbon structure in purple superimposed on the binding pocket.
A striking feature of the model is the predominance of hydrophobic interactions between 3DL1*015 and A*2402, and loss of the conserved electrostatic bonds that characterize the binding of KIR2DL to HLA-C (19, 20). For example, the two salt bridges common to the interaction of KIR2DL with HLA-C and involving the D1 domain (D135-R145 and D183-K146) are absent from the modeled complex of 3DL1*015 bound to A*2402. Instead, there are two salt bridges involving D2 (E282-R83 and E201-R145), which make a greater contribution to interactions with HLA class I than observed for KIR2DL. However, the majority of the binding is mediated by hydrophobic contacts with D1 (L54-R83, I139-R83, K141-R83, L166-E76/ pCys8, M165-E76, F276-I142 and H278-I142) including one between leucine 166 and the cysteine at position 8 in the peptide bound to A*2402.
Arginine 83 of A*2402 is predicted to bind to a specific pocket of 3DL1*015
Interaction of KIR2D with HLA-C involves specificity-determining residues at position 44 in the D1 domain of the KIR and at position 80 of HLA-C. For example, the lysine 80 of HLA-C*04 is bound in a pocket by 2DL1, making hydrophobic interaction with methionine 44 of 2DL1, as well as a hydrogen bond to serine 184 and a salt bridge with glutamate 187. Whereas the specificity-determining residue of the HLA-C is position 80, the residue essential for Bw4 binding to 3DL1 is arginine 83 (7, 33). In the model, arginine 83 can make an analogous set of interactions to those observed for lysine 80 in the structure of 2DL1 bound to HLA-C*04 (18). Thus it forms a hydrophobic interaction with isoleucine 139 (equivalent to methionine 44 in 2DL1), a hydrogen bond with serine 279 (equivalent to serine 184 in 2DL1) and a salt bridge with glutamate 282 (equivalent to glutamate 187 in 2DL1).
The arginine 83 binding pocket of 3DL1*015 is formed at the junction of the D0, D1 and D2 domains and can be divided into three strata: upper, intermediate and lower. Upper residues are hydrophobic (L54, I139 and L166) or positively charged (K141 and H278), intermediate residues are polar (S11, H29, Q56, G138, R277, S279 and E282), and lower residues are components of the hydrophobic core (W13, A169, G170, P280 and Y281). The salt bridge that arginine 83 makes with glutamate 282 is part of a rigid loop structured by tyrosine 281 and tryptophan 283, residues deeply anchored in the hydrophobic core of 3DL1. Substitution of leucine for tryptophan at position 283 of 3DL1*015 is predicted to change the position of glutamate 282 in the binding pocket, consistent with the differential antibody reactivity and loss of A24nef binding by mutant 3DL1*015-W283L. Inability to form the salt bridge between arginine 83 of A*2402 and glutamate 282 of 3DL1*015 could account for the complete loss of A24nef binding.
Functional tests of the structural model
To test key features of the modeled interaction between 3DL1*015 and A24nef, additional mutants were made and, in combination with selected mutants from the first set, examined for binding to A24nef (Figure 9). The combination of serine 182 and leucine 283 is permissive for A24nef binding in the presence of one D0 motif (M2, I47, I54 – as in *005) but not the other (V2, V47, L54 – as in *015-P182S-W283L). The model suggested this difference was caused by altered flexibility of the N-terminal region (residues 1-8). With the non-permissive motif, this region was predicted to be flexible, whereas with the permissive motif it was predicted to be tethered, with methionine 2 being buried between phenylalanine 9 and tyrosine 30. There the juxtaposition of methionine 2 and leucine 54 reinforced the hydrophobicity and strength of the interaction between D0 and A*2402. Consistent with this model, mutant *005-Y30C, in which tyrosine 30 was substituted for the smaller cysteine, failed to bind A24nef (Figure 9).
Figure 9. Binding of A24nef to mutants of 3DL1*015 and 3DL1*005.
Mutant KIR3DL1 were transfected into Jurkat cells and tested for binding A*24nef as described in the legend to figure 5. From the structural model all the mutations were predicted to perturb the interaction between 3DL1 and A24nef. The substitutions at positions 163, 166 and 199 abrogate A24nef binding, but not the binding of anti-3DL1 antibodies (Figure 3B) are all ones present in KIR3DS1.
The model predicts that a cluster of electropositive residues on the D0 surface, comprising arginine 31, histidine 32, arginine 33 and arginine 53, is important for ligand binding. Consistent with this hypothesis, increasing the electropositivity by substituting arginine for histidine at position 32, gave *005-H32R stronger avidity for A24nef than *005, *015 and *001. The only hydrophobic interaction predicted between D0 and D1 involves tryptophan 13 (D0) and glycine 138 (D1). Glycine 138 is also predicted to form hydrophobic interactions with tyrosine 281 of D2 and arginine 83 of A*2402. Both tryptophan 13 and glycine 138 are conserved in 3DL1, but glycine 138 is replaced by tryptophan in 3DS1 (10). Although mutant *015-G183W showed no perturbation in antibody binding (Figure 1), binding to A24nef was severely reduced (Figure 9).
The model predicts that arginine 277 in D2 has hydrophobic interactions with the loop comprising residues 47-56 of D0 and electrostatic interactions with aspartate 230 of D2. Proximal to arginine 277 is a flexible loop containing phenylalanine 276, histidine 278 and serine 279 of KIR3DL1 that are seen to interact with residues isoleucine 142, lysine 146 and isoleucine 80 of A*2402. Thus residue 277 has a network of interactions with residues in D0, D2 and A*2402. In four natural allotypes (3DL1*006, *022, *034 and *035) arginine 277 is replaced by cysteine. Introducing cysteine 277 into 3DL1*015 abrogated A24nef binding (Figure 9) and caused some reduction in antibody binding. In contrast, replacement of arginine 277 with histidine, the residue distinguishing the rare 3DS1*048 allotype (10), preserved the capacity to bind A24nef. This indicates the importance of the network of electrostatic interactions around arginine 277 for ligand binding, as predicted by the model.
Three 3DS1-specific residues abrogate A24nef binding when introduced into 3DL1
We examined the capacity of KIR3DS1, the activating counterpart of KIR3DL1, to bind A24nef. Jurkat cells tranfected with 3DS1*013 failed to bind A24nef (Figure 9), although cell-surface 3DS1 was detected with the Z27 antibody. A similarly negative result was obtained with 3DL1*004, which is sequestered inside cells and does not reach the cell surface (12). That KIR3DS1 fails to bind A24nef, is consistent with previous failure to detect interaction of 3DS1 with Bw4+ HLA class I (34). Mutant *015-G183W is an example where the substitution is to a 3DS1-specific residue and this led to reduction in A24nef binding (Figure 9). Three other mutants containing a single 3DS1-specific residue at either position 163, 166 and 199, were shown to be incapable of binding A24nef.
In the model, proline 163 and leucine 166 are part of a flexible HLA-binding loop comprising residues 161-166 that connects strands E and F in the D1 domain. Proline 163 is predicted to structure the conformation of this loop while leucine 166 forms hydrophobic contacts with glutamate 76 and isoleucine 80 of A*2402, and also with the cysteine at position 8 of the bound nef peptide. Proline 163 and leucine 166 are completely conserved in 3DL1, but in 3DS1are replaced by serine 163 and arginine 166. Mutants *015-P163S and *015-L166R were b oth expressed at the cell surface, the former giving reduced binding with all three antibodies, the latter only with DX9 (Figure 3). Despite cell-surface expression, neither mutant bound A24nef, consistent with an essential role for proline 163 and leucine 166 in ligand binding (Figure 9).
Proline 199, in a flexible HLA-binding loop of D2, is predicted to make hydrophobic contact with alanine 150 of A*2402 and with cysteine 8 of the nef peptide. Proline 199 is present in all 3DL1 (except the rare 3DL1*040) but is replaced by leucine in 3DS1. A24nef binding of *015 was abrogated when proline 199 was replaced by leucine, whereas DX9 and Z27 antibodies bound more strongly and binding of the177407 antibody was decreased by ~20%. Although having no major effect on the expression and conformation of 3DL1, this substitution profoundly affected the functional interaction with HLA class I. Tryptophan 138, serine 163, arginine 166 and leucine 199 are all substitutions that distinguish 3DS1 from 3DL1; individually, three of them (positions 163, 166 and 199) abrogate A24nef binding and the fourth (position 138) effects a reduction. Although we have only examined a single combination of Bw4+ HLA class I and peptides, our results illustrate the potential for the 3DS1-specific residues to reduce avidity for HLA class I. (Figure 9).
In summary, the properties of this series of site-directed mutants are consistent with the models of 3DL1*015 and its complex with A*2402. Although the results of such analysis cannot prove the validity of the models, they show their value as tools for the design of future experiments until such time as crystallographic structures for KIR3DL1 and 3DS1 are achieved.
DISCUSSION
The highly variable KIR that regulate human NK cells through recognition of MHC class I, originated with a single KIR3D gene (35). Today, HLA-A and –B epitopes are still recognized by KIR3D having extracellular D0, D1 and D2 domains, but the more recently evolved HLA-C is recognized by KIR2D having only D1 and D2. The genes encoding KIR2D clearly evolved from KIR3D, because they all retain a pseudoexon 3 encoding a D0 domain that is not expressed in the mRNA (36). Crystallographic structures of HLA-C bound to KIR2D have shown precisely how they interact and the critical role that dimorphisms at position 44 in KIR2D and 80 in HLA-C play in determining the specificity of the interactions (1). These studies, however, could not address the function of the D0 domain in KIR3DL1 and how it contributes to NK cell recognition of the Bw4 epitope carried by HLA-A and HLA-B. The results of the study presented here have addressed these important questions.
D0 and D1+D2 of KIR3DL1 both play critical roles in binding Bw4 ligand
Although KIR3DL1 alleles are numerous, the simple and fundamental basis for the variation lies with two dimorphic motifs, one at positions 2, 47 and 54 in D0, the other at positions 182 in D1 and 283 in D2. The two ancient lineages of KIR3DL1 (10), typified by 3DL1*015 and 3DL1*005, differ at both these motifs, whereas 3DL1*001 is a modern recombinant that combines the D0 motif of 3DL1*005 with the D1+D2 motif of 3DL1*015. We find that the A24nef tetramer, comprising a defined HIV peptide bound to A*2402, binds with markedly higher avidity to 3DL1*001 than to either its 3DL1*015 or 3DL1*005 parents. In contrast to 3DL1*001, a mutant form of 3DL1 that combines the D0 of 3DL1*015 with the D1+D2 of 3DL1*005 does not bind A24nef at all, despite being well expressed at the cell surface and able to bind anti-3DL1 monoclonal antibodies. These properties demonstrate that both D0 and D1+D2 play critical, and complementary roles in the binding of HLA class I ligands to KIR3DL1. Thus different combinations of the natural forms of D0 with the natural forms of D1 and D2 can give binding sites that have high avidity, moderate avidity or no avidity for the A24nef ligand.
The influence of the D0 motif on the dimorphism at position 283 in D2 is particularly striking: in 3DL1*005 the natural combination of its D0 motif and leucine 283 are permissive to A24nef binding, whereas when leucine 283 combines with the D0 motif of 3DL1*015 D0 in the 3DL1*015-W283L mutant, A24nef binding is almost obliterated. This functional modulation associated with the leucine/tryptophan dimorphism at position 283 correlates with a conformational difference detected by monoclonal anti-KIR3DL1 antibodies. However, these conformational differences are not the basis for the high and low antibody-binding phenotypes of peripheral blood NK cells expressing different KIR3DL1 alleles (11, 30, 31). Monoclonal antibodies biased towards either conformation detect the different phenotypes, consistent with them being the consequence of different levels of cell-surface expression.
The striking effect we observe with A24nef is unlikely to be peculiar to this ligand. In cellular cytotoxicity assays, where the Bw4+ HLA-B*5801 ligand was associated with heterogeneous peptides, KIR3DL1*001 exhibited stronger inhibitory function than 3DL1*005, *007, *015, and *020 (11). And in similar assays using HLA-A*3201 and B*1513 as the ligands, 3DL1*001 gave stronger responses than 3DL1*015 (37). Previously, when examining unnatural mutations in D0 of KIR3DL1, we observed increased avidity of ligand-binding. Because the effects were quantitative, we described the function of D0 as enhancing the binding achieved by D1+D2 (9). Here, by studying the interaction between natural polymorphisms in D0 and D1+D2, we discovered more dramatic, qualitative effects raising the possibility that D0 contributes directly to the ligand-binding site and to making contact with the HLA class I ligand.
3DS1 has acquired several substitutions that individually abrogate A24nef binding
The KIR3DL1/S1 locus comprises three lineages that have been maintained by balancing selection for >3 million years. Illustrating this balance, the lineage defining 3DL1*005, 3DL*015 and 3DS1*013 alleles are the only alleles represented in all modern human populations (10). In the context of A24nef binding, balancing selection has maintained weaker and stronger versions of the dimorphic motifs in both D0 and D1+D2. 3DL1*005 has the stronger D0 and weaker D1+D2, 3DL1*015 the weaker D0 and stronger D1+D2. As a consequence of these differences 3DL1*005 and 3DL1*015 bind A24nef at comparable levels. The more recently formed recombinant 3DL1*001 combines the stronger versions of both motifs and binds A24nef more strongly than either 3DL1*005 or 3DL1*015. The high allele frequencies (5-30%, excluding Amerindians) that 3DL1*001 has reached in modern human populations suggest it has been selected for its superior strength, but given the past history of balancing selection, this is likely to have a cost that will prevent 3DL1*001 from replacing either 3DL1*015 or 3DL1*005. Furthermore, the absence of a natural KIR3DL1 allotype that combines the D0 of 3DL1*015 with the D1+D2 of 3DL1*005 suggests that its inability to bind A24nef reflects a wider deficiency in binding Bw4+ HLA class I ligands.
Unlike polymorphic KIR3DL1, the closely related activating receptor KIR3DS1 is conserved and has not been shown to engage HLA class 1 in cellular assays (28, 38). Consistent with these observations, we found that cells transfected with 3DS1 do not bind A24nef. Moreover, introducing a single 3DS1-specific residue at either position 163, 166 or 199 of 3DL1*015 was sufficient to abrogate A24nef binding; an effect not caused by reduced cell-surface expression. Thus 3DS1 has successively acquired mutations that either reduce avidity for MHC class I or narrow the specificity to complexes of MHC class I and peptide that are present at undetectable levels on the surfaces of the cells that have been studied. It remains, however, possible that 3DS1 is specific for complexes of peptide and Bw4+ HLA class I that are expressed at detectable levels on cells either infected with particular pathogens or subject to other pathologies, such as malignant transformation. The conservation and prevalence of 3DS1 point to these changes being the result of selection. Although 3DS1 cannot bind the complex of A*2402 with the HIV nef peptide, the combination of 3DS1 with Bw4 having isoleucine 80 (as is the case for A*2402) is associated with decreased progression to AIDS in HIV-infected individuals (39) and 3DS1-expressing NK cells are preferentially expanded in individuals having Bw4 with isoleucine 80. Moreover, in healthy 3DS1+ individuals the frequency of 3DS1+ NK cells is higher in Bw4+ than Bw4- individuals (38).
Modeling the interaction of KIR3DL1 with A24nef
Previous comparison of human and chimpanzee KIR3DL, showed that deletion or substitution at residues 50 and 51 in a D0 improved the avidity of binding for the Bw4 epitope (9). Residue 51 was also identified as a site of positive natural selection and predicted to cluster on the surface of D0 with three other positively-selected residues: 5, 31 and 32 (10). Residues 50 and 51 are conserved in 3DL1 and predicted to be part of a conserved loop that is flanked by residues 47 and 54 of the polymorphic D0 motif. Similarly, residue 5 is conserved and is part of the amino-terminal region containing residue 2 of the polymorphic D0 motif. Thus in identifying functionally important sites in D0 the results obtained here are both complementary and concordant with the earlier studies. These results provided the foundation for modeling the interaction of KIR3DL1 with A24nef, which was strengthened by the demonstration that arginine 83 is the only residue in the Bw4 motif (residues 76-83) that is essential for binding to KIR3DL1 (7). This contrasted with the importance of residue 80 for HLA-C interactions with KIR2D (40), anticipating differences between KIR3D and KIR2D in binding their cognate ligands.
Our model for the structure of A24nef bound to KIR3DL1*015 was based on the crystallographic structures of KIR2D, HLA-A*2402, and KIR2D bound to HLA-C (19, 20, 22). This model was compatible with the experimental data obtained prior to its construction and to the functional properties of additional mutants that were designed to test particular aspects of the model. In the model the D0, D1 and D2 domains mutually interact to form a flat surface where loops of all three domains contact A24nef. At a central region the three domains form a pocket that interacts with arginine 83 of A*2402. Within this pocket arginine 83 of A*2402 is predicted to make very similar specificity-determining interactions with the D1 and D2 domains of 3DL1 to those made by lysine 80 of C*04 with the D1 and D2 domains of 2DL1. A salt bridge with glutamate 282, is augmented by a hydrogen bond with serine 184 and hydrophobic interaction with isoleucine 139. These residues correspond to glutamate 187, serine 184 and methionine 44 in 2DL1, respectively. Although the specificity-determining interactions seem similar, overall the KIR3DL1 interaction with Bw4+ HLA-A and –B, appears largely based upon hydrophobic interactions, whereas that between HLA-C and KIR2DL is characterized by electrostatic interactions (1).
Several lines of evidence favor an evolutionary model in which the KIR2D interactions with HLA-C that dominate the KIR-mediated regulation of human NK cell responses are of relatively recent origin and evolved from KIR3D recognition of HLA -A and B (5). Our studies suggest that all three Ig-like domains of KIR3DL1 interact with the Bw4 epitope of HLA-A and -B in a mutually dependent manner that focuses on residue 83 in the α1 helix. In the evolution of HLA -C and its interactions with KIR2DL this register was adjusted, position 80 in the α1 helix became the focus of attention and D0 was no longer needed. In human KIR2D genes the exon encoding D0 remains, although it is not incorporated into mRNA, whereas in chimpanzees some HLA -C specific KIR retain D0 although its removal has no obvious affect on function. For KIR3DL1, however, the D0 domain remains essential for both the receptor's folding and its functional interactions with Bw4+ HLA class I ligands (8).
ACKNOWLEDGEMENTS
We would like to thank Michael Levitt for his support of the collaboration by K.B. on this work.
Footnotes
This work was supported by NIH Grant AI064520 to FMB and PP. KB is a recipient of Marie Curie fellowship MOIF-2005-022323.
REFERENCES
- 1.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 2.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 3.Jonsson AH, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol. 2009;101:27–79. doi: 10.1016/S0065-2776(08)01002-X. [DOI] [PubMed] [Google Scholar]
- 4.Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 5.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 6.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, Braud VM. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 7.Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293–6300. doi: 10.4049/jimmunol.181.9.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rojo S, Wagtmann N, Long EO. Binding of a soluble p70 killer cell inhibitory receptor to HLA-B*5101: requirement for all three p70 immunoglobulin domains. Eur J Immunol. 1997;27:568–571. doi: 10.1002/eji.1830270231. [DOI] [PubMed] [Google Scholar]
- 9.Khakoo SI, Geller R, Shin S, Jenkins JA, Parham P. The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J Exp Med. 2002;196:911–921. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 11.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pando M, Gardiner CM, Gleimer M, McQueen K, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in the Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 13.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 14.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, Dong T, Rowland-Jones S. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 18.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 19.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 20.Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 22.Cole DK, Rizkallah PJ, Gao F, Watson NI, Boulter JM, Bell JI, Sami M, Gao GF, Jakobsen BK. Crystal structure of HLA-A*2402 complexed with a telomerase peptide. Eur J Immunol. 2006;36:170–179. doi: 10.1002/eji.200535424. [DOI] [PubMed] [Google Scholar]
- 23.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 25.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180:537–543. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 27.Thomas R, Yamada E, Alter G, Martin MP, Bashirova AA, Norman PJ, Altfeld M, Parham P, Anderson SK, McVicar DW, Carrington M. Novel KIR3DL1 Alleles and Their Expression Levels on NK Cells: Convergent Evolution of KIR3DL1 Phenotype Variation? J Immunol. 2008;180:6743–6750. doi: 10.4049/jimmunol.180.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trundley A, Frebel H, Jones D, Chang C, Trowsdale J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur J Immunol. 2007;37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 30.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 31.Gumperz JE, Valiante NM, Parham P, Lanier LL, Tyan D. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J Exp Med. 1996;183:1817–1827. doi: 10.1084/jem.183.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang Z. Advances in homology protein structure modeling. Curr Protein Pept Sci. 2006;7:217–227. doi: 10.2174/138920306777452312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL, Parham P. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 34.Gillespie GM, Bashirova A, Dong T, McVicar DW, Rowland-Jones SL, Carrington M. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res Hum Retroviruses. 2007;23:451–455. doi: 10.1089/aid.2006.0165. [DOI] [PubMed] [Google Scholar]
- 35.Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59:517–522. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- 36.Vilches C, Pando MJ, Parham P. Genes encoding human killer-cell Ig-like receptors with D1 and D2 extracellular domains all contain untranslated pseudoexons encoding a third Ig-like domain. Immunogenetics. 2000;51:639–646. doi: 10.1007/s002510000184. [DOI] [PubMed] [Google Scholar]
- 37.Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, Graef T, McQueen KL, Guethlein LA, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Tyan D, Verity DH, Vaughan RW, Davis RW, Fraser PA, Riley EM, Ronaghi M, Parham P. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morvan M, Willem C, Gagne K, Kerdudou N, David G, Sebille V, Follea G, Bignon JD, Retiere C. Phenotypic and functional analyses of KIR3DL1+ and KIR3DS1+ NK cell subsets demonstrate differential regulation by Bw4 molecules and induced KIR3DS1 expression on stimulated NK cells. J Immunol. 2009;182:6727–6735. doi: 10.4049/jimmunol.0900212. [DOI] [PubMed] [Google Scholar]
- 39.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 40.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]