The crystal structure of the unbound form of HIV-1 subtype A protease has been determined to 1.7 Å resolution. A detailed structural analysis and comparison of the unbound subtype A, B and C protease structures is presented. The results showed that although no inhibitor is present in the active site, the subtype A protease has flaps in the closed position.
Keywords: HIV-1 proteases, HIV-1 protease subtype A, unbound, crystal packing
Abstract
The crystal structure of the unbound form of HIV-1 subtype A protease (PR) has been determined to 1.7 Å resolution and refined as a homodimer in the hexagonal space group P61 to an R cryst of 20.5%. The structure is similar in overall shape and fold to the previously determined subtype B, C and F PRs. The major differences lie in the conformation of the flap region. The flaps in the crystal structures of the unbound subtype B and C PRs, which were crystallized in tetragonal space groups, are either semi-open or wide open. In the present structure of subtype A PR the flaps are found in the closed position, a conformation that would be more anticipated in the structure of HIV protease complexed with an inhibitor. The amino-acid differences between the subtypes and their respective crystal space groups are discussed in terms of the differences in the flap conformations.
1. Introduction
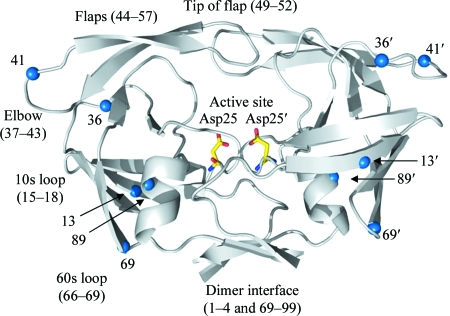
HIV protease (PR) is an aspartic PR that plays an essential role in the viral life cycle and infectivity by converting a non-infectious particle into a mature virion that is capable of infecting new target cells. The PR functions as an obligatory homodimer, with the active site located in the center and covered by two flexible loops called flaps. This overall structure is well conserved and is very similar for all HIV subtypes (Fig. 1 ▶).
Figure 1.
Ribbon diagram of the homodimer of the HIV-1 subtype A PR structure. Catalytic aspartic acids are shown as sticks. Blue spheres indicate amino acids that vary between subtype A PR and subtype B PR (numbers refer to amino-acid positions). Features of the PR structure are as labeled. This figure was generated using PyMOL (DeLano, 2002 ▶).
HIV is genetically very diverse and as such is classified into types, groups, subtypes, sub-subtypes and recombinant forms (Brodine et al., 1995 ▶; Fleury et al., 2003 ▶; Kantor & Katzenstein, 2004 ▶). Although subtype B, which is common in the United States, has been the subject of the majority of investigations, 90% of HIV-infected people worldwide harbor non-B-subtype variants of HIV-1 (Hemelaar et al., 2006 ▶, 2008 ▶; Osmanov et al., 2002 ▶; Taylor et al., 2008 ▶). In particular, subtypes A and C account for more than 85% of global infections and more than 95% of infections in South and Central Africa (Foulkes et al., 2006 ▶). Nevertheless, knowledge of resistance mutations in non-B subtypes and their clinical relevance is limited or controversial (Martinez-Cajas et al., 2009 ▶).
With the expanding access to antiretroviral therapy (ARV) for patients living with HIV/AIDS in developing countries (UNAIDS, 2008 ▶) and with the spread of non-B-subtype variants of HIV-1 in developed countries (Baldanti et al., 2008 ▶; Palma et al., 2007 ▶; Paraskevis et al., 2007 ▶), an important issue of concern is the behavior of non-B subtypes and their recombinant forms under the selective pressure of ARV regimens. There is an ongoing debate in the HIV field about how and to what extent naturally occurring polymorphisms (NOPs) may affect the emergence of ARV drug resistance in HIV-1 subtypes other than B. It has been argued that these polymorphisms might play roles such as increasing the catalytic activity of non-B-subtype PRs and enhancing virus viability (Krauchenco et al., 2008 ▶; Velazquez-Campoy et al., 2001 ▶), resulting in the development of diverse mutational pathways during ARV treatment (Dumans et al., 2004 ▶; Grossman et al., 2004 ▶), influencing the speed of acquisition of PR inhibitor (PI) related resistance mutations (Hirsch et al., 2000 ▶; Vergne et al., 2000 ▶) and contributing to resistance and/or maintenance of viral fitness once primary resistance mutations occur (Rose et al., 1996 ▶; Velazquez-Campoy et al., 2003 ▶). Several recent studies have argued that there are no substantial differences regarding known resistance-associated mutations and the newly emergent substitutions between non-B-subtype and B-subtype strains (Monno et al., 2009 ▶; Pillay et al., 2008 ▶). However, a recent study of the differences in the impact of several PI-selected mutations on subtypes B and G in 500 patients showed that the mutation L90M confers different levels of resistance in subtypes B and G (Santos et al., 2009 ▶). Another recent study indicated that even though in general drug selective pressure and resistance pathways are relatively similar in subtypes B and G some differences do occur, leading to subtype-dependent substitutions (Palma et al., 2009 ▶). With respect to the development of drug-resistance mutations (DRMs), a recent study showed that non-B subtypes tend to select for the same DRMs as described in subtype B in PR and reverse transcriptase (RT), although in distinct proportions (Dumans et al., 2009 ▶; Kantor et al., 2005 ▶). Also, an increasing body of evidence suggests that certain non-B subtypes are often more susceptible to specific ARV drugs. For example, the circulating recombinant form CRF02_AG PR presents a higher susceptibility to nelfinavir and ritonavir than subtypes C, F and G, whereas subtype G isolates are more susceptible to tipranavir (Abecasis et al., 2006 ▶). Unexpectedly, natural hypersusceptibility to lopinavir was also detected in subtypes C and G (Gonzalez et al., 2003 ▶, 2008 ▶). In an extensive review, Martinez-Cajas and coworkers searched 11 databases and retrieved 3486 citations on all aspects of non-B-subtype-related resistance research (Martinez-Cajas et al., 2008 ▶). They concluded that the genetic diversity of HIV-1 can affect the type of resistance mutations, the degree of resistance and the timing of the emergence of antiretroviral resistance. These observed differences in resistance pathways may impact on cross-resistance and the selection of second-line regimens with PIs.
These NOPs are found throughout the HIV genome, including PR and RT, which are the main targets of currently used anti-HIV therapeutic regimens. Thus, characterizing the structural differences among various non-B-subtype PRs may identify possible mechanisms that might cause the development of divergent mutational pathways against currently used PIs. Until recently, the only available crystal structures in the Protein Data Bank (PDB; Berman et al., 2000 ▶) were those of HIV subtype B PRs. More than 100 crystal structures of subtype B PRs are currently available and include both wild type and multidrug-resistant (MDR), both open and closed and both unbound and bound to various inhibitors. The first crystal structure of a non-B-subtype PR, subtype F, was solved in 2004 (Sanches et al., 2004 ▶) and a detailed analysis and comparison with subtype B followed (Sanches et al., 2007 ▶). A thorough biochemical characterization of subtype C PR has been published (Coman et al., 2007 ▶) and a detailed analysis of the crystal structure of subtype C PR followed in 2008 (Coman, Robbins, Goodenow et al., 2008 ▶). Solving the crystal structures of various non-B-subtype PRs has made structural comparisons and analyses of the NOP effects on the overall structure of the PR possible. It has previously been suggested that these differences might affect the binding of substrates/inhibitors through either long-range interactions or changes in the flexibility of the enzyme (Coman, Robbins, Fernandez et al., 2008 ▶; Gonzalez et al., 2008 ▶; Velazquez-Campoy et al., 2003 ▶). These studies showed that there are differences in the position of the outside loops, in which most NOPs are located. The presence of these polymorphic substitutions has been reported to cause a collapse of the elbow of the flap, resulting in the displacement of the main chain of this loop towards the loop 76–83, stabilizing the catalytic S1/S1′ pockets (Coman, Robbins, Goodenow et al., 2008 ▶; Sanches et al., 2007 ▶). Furthermore, it has also been observed that L89M can displace the 60s loop laterally and downwards and could participate in the formation of a more stable network of van der Waals interactions (Coman, Robbins, Goodenow et al., 2008 ▶). However, currently there is concern that the differences, which are observed especially in the outside loops of the enzyme, might be secondary not only to NOPs but also to different crystal contacts and different crystal packing.
Here, the first crystal structure of subtype A PR is described at 1.7 Å resolution and a detailed structural analysis and comparison of the unbound subtype A, B and C PR structures is presented. The results showed that although no inhibitor is present in the active site, the subtype A PR has flaps in the closed position and the crystal contacts are similar to those in other unbound and bound forms of PR crystallized in the hexagonal space group P61.
2. Materials and methods
2.1. Protein expression and purification
Protein expression and purification followed the protocol used for subtype C PR (Coman, Robbins, Fernandez et al., 2008 ▶). The HIV-1 PR DNA for the A subtype (obtained from the NIH Research Reagents Program, accession code p92RW009.6) was subcloned into pET23a expression vector (Novagen) and transformed into Escherichia coli strain BL21 Star DE3 plysS (Invitrogen). Protease expression was performed in M9 minimal media and was initiated when the OD600 reached 0.8 by the addition of 1 mM IPTG. After 3 h, the cells were harvested and lysed using an SLM Aminco French pressure cell. Inclusion bodies containing the PR were isolated by centrifugation through a 27% sucrose cushion. The inclusion bodies were dissolved in 8 M urea and refolded by dialysis against sodium phosphate buffer. The PR was purified by ammonium sulfate precipitation and gel-filtration chromatography as described previously (Coman, Robbins, Fernandez et al., 2008 ▶).
2.2. Crystallization
Crystals were grown using the hanging-drop vapor-diffusion method (McPherson, 1982 ▶). The purified subtype A PR was concentrated to 3.5 mg ml−1 with a buffer exchange to 50 mM sodium acetate pH 4.7. 2 µl protein solution was mixed with 2.5 µl reservoir solution (30 mM citric acid pH 5.0, 1 M NaCl) and 0.5 µl Anapoe-58 detergent (Hampton) on siliconized glass cover slides. The droplets were suspended over 0.5 ml reservoir solution and stored at room temperature. Hexagonal rod-like crystals formed within 2 d.
2.3. Data collection and reduction
A crystal of subtype A PR was ‘quick-dipped’ in 35% glycerol solution for 1 s and immediately flash-cooled to 100 K. X-ray diffraction data were measured using a MAR CCD 300 detector on the SER-CAT beamline at the Advanced Photon Source. The crystal-to-detector distance was 200 mm and a total of 240 diffraction data frames were measured using a 0.5° oscillation angle per frame. From these, a subset of 140 data frames was integrated and scaled with the HKL-2000 software (Otwinowski & Minor, 1997 ▶) to give a complete data set to 1.7 Å resolution.
Systematic absences in the diffraction data were consistent with either a 61 or 65 screw axis in the c direction. The data were merged and scaled separately in the hexagonal space groups P61 and P6122, yielding 19 874 and 10 865 unique reflections, respectively. Initial phasing was performed using data in space group P6122, but the lower symmetry space group was selected for the final refinement (see below). A complete summary of data statistics for processing in both space groups is included in Table 1 ▶.
Table 1. Crystal data and refinement.
Values in parentheses are for the highest resolution shell.
| Crystal data | ||
| Space group | P61 | P6122 |
| Unit-cell parameters (Å) | a = 62.2, c = 82.5 | a = 62.2, c = 82.5 |
| Molecules per ASU | 2 | 1 |
| VM (Å3 Da−1) | 2.14 | 2.14 |
| Solvent content (%) | 42.5 | 42.5 |
| Data collection | ||
| Resolution (Å) | 23.4–1.7 (1.76–1.7) | 23.4–1.7 (1.76–1.7) |
| Unique reflections | 19874 | 10865 |
| No. of frames | 140 | 140 |
| Oscillation per frame (°) | 0.5 | 0.5 |
| Rmerge† (%) | 8.9 (50.9) | 9.4 (54.8) |
| I/σ(I) | 12.3 (2.2) | 12.3 (2.2) |
| Completeness (%) | 99.6 (99.2) | 99.5 (99.9) |
| Data redundancy | 4.4 (3.7) | 8.0 (6.8) |
| Refinement statistics | ||
| No. of reflections used | 18760 | |
| Rcryst‡ (%) | 20.5 | |
| Rfree (%) | 24.4 | |
| Reflections used for Rfree (|Fo| > 0) (%) | 4.4 | |
| R.m.s.d. bonds (Å) | 0.008 | |
| R.m.s.d. angles (°) | 1.49 | |
| Ramachandran plot (%) | ||
| Most favored | 90.0 | |
| Allowed | 9.4 | |
| Generously allowed | 0.6 | |
| Model | ||
| Protein atoms | 1510 | |
| Waters | 111 | |
| Average temperature factors (Å2) | ||
| Main chain | 26.8 | |
| Side chain | 29.0 | |
| Waters | 30.4 |
R
merge = 
 , where Ii(hkl) is the intensity of an individual reflection and 〈I(hkl)〉 is the average intensity.
, where Ii(hkl) is the intensity of an individual reflection and 〈I(hkl)〉 is the average intensity.
R
cryst = 
 ; R
free is identical to R
cryst but was calculated for 5% of data omitted from refinement.
; R
free is identical to R
cryst but was calculated for 5% of data omitted from refinement.
From the unit-cell volume and the molecular weight of subtype A PR, a reasonable V M value (Matthews, 1968 ▶) of 2.14 Å3 Da−1 was calculated for one PR monomer per asymmetric unit in space group P6122 (P6522) or for a complete homodimer in P61 (P65). Additionally, the diffraction data scaled in P61 were examined using the twin-detection software in CNS (Brünger et al., 1998 ▶) and the Merohedral Twin Detector server (Padilla & Yeates, 2003 ▶; Yeates, 1997 ▶).
2.4. Rotation and translation search
Cross-rotation function and translation-function searches were performed in space group P6122, using X-ray diffraction data between 15.0 and 4.0 Å resolution, with the CNS package (Brünger et al., 1998 ▶). The molecular template used was the A chain of the HIV-1 subtype B homodimer in complex with atazanavir (PDB code 2aqu; Clemente et al., 2006 ▶), with the water molecules and the atazanavir inhibitor removed. A translation-function search in space group P6522 failed to produce a reasonable solution. Rigid-body refinement of the molecular-replacement solution using data between 50 and 3.5 Å resolution resulted in a model with R cryst = 28.4% and R free = 26.1%. Upon inspection of the resulting electron-density maps the flaps were found to be in the closed position and there was no density in the |F o| − |F c| difference map that would indicate an alternative interpretation. An OMIT map, based upon the final refined coordinates and centered on the active site, is shown in Fig. 2 ▶.
Figure 2.
Stereo 2|F o| − |F c| OMIT map centered on the active site of HIV-1 subtype A PR. The map is contoured at 1.3σ and includes a 4 Å radius buffer around flap residues and water positions. All atoms of the flap (residues 46–55 and 46′–55′), the active-site aspartates Asp25 and Asp25′ and two waters (depicted as red spheres) were omitted from the calculation of the map.
2.5. Refinement of the HIV-1 subtype A model
The problem of the proper choice of the space group for refinement of the structure of HIV PR has been faced repeatedly in the past (Wlodawer & Erickson, 1993 ▶; Sanches et al., 2007 ▶) and is a consequence of the possible presence of pseudo-symmetry, disorder, and/or twinning. An excellent discussion of the effects of pseudo-symmetry on twin detection has been given in the analysis of a difficult refinement of superoxide dismutase from Pyrobaculum aerophilum (Lee et al., 2003 ▶). Since the scaling parameters for subtype A PR were virtually identical in space groups P61 and P6122 (Table 1 ▶), the high-symmetry space group appeared to be the correct choice. However, the same result could be expected if the lower symmetry space group were correct but the data were fully twinned. Although twinning can normally be detected through analysis of statistical distribution of the intensities, this does not apply in this case since pseudo-symmetry restores the normal distribution of intensities. Indeed, this was tested by submitting the subtype A PR data scaled in space group P61 to the merohedral twin-detection server (Padilla & Yeates, 2003 ▶; http://nihserver.mbi.ucla.edu/pystats/). Not surprisingly, no twinning was detected, whereas the twin-detection protocol in CNS indicated a twin fraction of 0.48. Since the latter result is based on a comparison of the intensities of potential twin mates, it is not really applicable here. It may be noted that since only a very small part of the structure (a few residues) is responsible for the effects of disorder/twinning, its influence on the statistics of the intensities is necessarily very small, making it impossible to differentiate which of these two effects occurs in the crystal.
Atomic positional and temperature-factor refinements of the rigid-body model were performed in the CNS suite (Brünger et al., 1998 ▶), initially with data processed in space group P6122. The target values used for restraining the model geometry during refinement were from Engh & Huber (1991 ▶). A randomly selected subset of 5% of the reflections was withheld from the refinement cycles and was used to calculate R free. Replacement of amino acids which differed between subtype B PR and subtype A PR was performed in the molecular-graphics program Coot (Emsley & Cowtan, 2004 ▶). Interactive manual model refitting using Coot and electron-density maps calculated with 2|F o| − |F c| and |F o| − |F c| coefficients were used to guide manual refitting of the model. After several cycles, the resulting R cryst and R free values remained higher than expected and positive peaks at greater than 3.5σ persisted in the |F o| − |F c| difference electron-density map along the main chain of the monomer. At this stage a choice had to be made of either trying to model the asymmetry of the dimers as disorder or switching to the low-symmetry space group P61 with assumed twinning. Since the latter approach provides a simpler description of the model, it was selected.
Since the free R list of reflections differed between the data scaled in space groups P61 and P6122, the rigid-body model was used as the starting model. In space group P61 the asymmetric unit consists of the entire PR dimer, so the B monomer coordinates were calculated using the twofold symmetry in the graphics program Coot (Emsley & Cowtan, 2004 ▶). After appending to the A monomer, the coordinates were given the chain identifier B for refinement against the data that had been merged and scaled in P61. Refinement and model building in the lower symmetry space group improved the appearance of the electron-density maps even before correction for twinning. Subsequent refinements were performed with a twin law of k, h, −l and a twin fraction of 0.5 and the free R list of reflections was reselected so that pairs of twin-related reflections resided in either the working list or the cross-validation list. To verify the effects of twinned refinement, one round of positional refinement resulted in a decrease of 3.7% in R cryst and of 2.5% in R free when compared with an identical refinement cycle starting with the same molecular model but disregarding twinning. After several rounds of refinement and model building, the final model, consisting of two monomers and 111 water molecules, had an R cryst of 20.5% and an R free of 24.4%. A Ramachandran plot, as calculated in PROCHECK (Laskowski et al., 1993 ▶), showed that 90.0% of the amino acids were in the most favored region, 9.4% were in the additional allowed region and 0.6% were in the generously allowed region. A more complete summary of the final model of subtype A PR is included in Table 1 ▶.
Superimposition of the coordinates of the B monomer onto the A monomer allowed the comparison of side-chain conformations. Of a total of 99 amino acids, 20 have rotamer differences between the two monomers, including three valines, three isoleucines, Met46 and Phe53. The latter two amino acids are involved in crystal packing, but the electron density for the side chains of 53A and 53B are very weak in OMIT maps (Fig. 2 ▶). Packing effects would be expected to stabilize side chains, but in the case of Phe53 disorder between the two conformations may contribute to the weak density. In addition, although the flaps are in contact with flaps from symmetry-related molecules, in the uncomplexed PR they are separated by solvent cavities where inhibitors would normally bind in the closed conformation.
2.6. Comparison of unbound HIV PR structures
In order to compare the structure presented here with other crystal structures of unbound HIV PRs, 13 structures were retrieved from the PDB and superimposed onto the refined model of HIV subtype A PR (Fig. 3 ▶). The coordinates of the dimers from the structures refined in space groups P41 or P61 were used directly, whereas for those refined in P41212 the dimer coordinates were calculated using the twofold-symmetry operation between the monomers and were saved using Coot (Emsley & Cowtan, 2004 ▶). In two cases, the database holds identical or nearly identical PR structures, namely tethered subtype B PR [PDB codes 1lv1 (Kumar et al., 2002 ▶) and 1g6l (Pillai et al., 2001 ▶)] and an MDR form of B PR [PDB codes 1rpi (Logsdon et al., 2004 ▶) and 1tw7 (Martin et al., 2005 ▶)]. Of these two similar pairs of coordinates, only the two higher resolution structures, 1g6l and 1tw7, were included in the comparison.
Figure 3.
Stereo superposition of unbound PR homodimers. (a) Complete PR homodimers, (b) close-up of the flap regions shown in (a) viewed down the twofold axis. Color code: green, dimers in tetragonal space groups with semi-open flaps; red, dimers in tetragonal space groups with wide-open flaps; blue, dimers in the hexagonal space group P61 with closed flaps. Note that the tips of the flaps invert between the semi-open and closed conformations. PDB coordinates were as used in Fig. 5 ▶. This figure was generated using PyMOL (DeLano, 2002 ▶).
Nine of the remaining 11 unbound PR structures used for comparison crystallized in either space group P41212 or P41. They were the chemically synthesized HIV-1 subtype B PR (PDB code 3hvp; Wlodawer et al., 1989 ▶), wild-type (wt) subtype B PR (3phv; Lapatto et al., 1989 ▶), the F53L mutant subtype B PR (2g69; Liu et al., 2006 ▶), subtype B PR BRU isolate (1hhp; Spinelli et al., 1991 ▶), 6× mutant subtype B PR and two structures of the Q7K mutant B PR (2hb2, 1tw7 and 2pc0; Heaslet et al., 2007 ▶), the aforementioned MDR subtype B PR (1tw7; Martin et al., 2005 ▶) and subtype C PR (2r8n; Coman, Robbins, Goodenow et al., 2008 ▶).
The structure of subtype F PR (PDB code 2p3c; Sanches et al., 2007 ▶) with the inhibitor coordinates removed and the tethered unbound B PR structures (PDB code 1g6l; Pillai et al., 2001 ▶), which also crystallize in space group P61, were included in the comparison with subtype A PR. The Cα coordinates of all 11 structures were superimposed onto the subtype A coordinates using the LSQ superposition option in Coot (Emsley & Cowtan, 2004 ▶). In order to superimpose pairs of monomers, chain identifiers were removed and each amino-acid residue of the dimer was renumbered: 1001–1099 (A chain) and 2001–2099 (B chain). All 198 Cα atoms were used in the r.m.s.d. calculations, with the exception of 1hb4, in which two amino-acid residues of each flap were not modeled owing to weak electron density. The amino-acid sequences for all 12 structures used for superposition are listed in Fig. 4 ▶ and the pairwise root-mean-square displacements (r.m.s.d.s) are presented in Fig. 5 ▶. Figs. 4 ▶ and 5 ▶ introduce the color code also used in Figs. 6 and 7: red, wide-open flaps; green, semi-open flaps; blue, closed flaps.
Figure 4.
Sequences of HIV-1 PR structures.
Figure 5.
Pairwise r.m.s.d.s of Cα—Cα distances (Å).
2.7. Analysis of packing of selected unbound PR structures
The Chimera molecular-modeling software (Pettersen et al., 2004 ▶) was used to calculate the crystal-packing contacts for the subset of unbound PR dimers listed in Fig. 5 ▶ and shown in Fig. 6 ▶ with the refined subtype A PR. All symmetry-related dimer coordinates in a 3 × 3 × 3 grid of unit cells surrounding the reference molecule were calculated in order to ensure that all symmetry contacts were included. The Chimera default clash/contact cutoff distances were used to define hydrogen-bond and van der Waals interactions at the packing interfaces. Fig. 5 ▶ indicates whether or not each of the amino acids of the PR chain is involved in these contacts. No distinction was made between hydrophobic or hydrophilic contacts.
Figure 6.
Stereo figure showing the crystal contacts of HIV-1 subtypes. (a) Wide open (PDB code 2pc0; Heaslet et al., 2007 ▶), (b) semi-open (PDB code 2hb2; Heaslet et al., 2007 ▶) and (c) closed (PDB code 3ixo). PDB coordinates were as used in Fig. 7 ▶. Colored spheres represent the amino acids involved in contacts. A contact is calculated from the sum of atomic radii. This figure was generated using PyMOL (DeLano, 2002 ▶).
3. Results
3.1. Overall structure of subtype A PR
As expected, the HIV-1 subtype A PR structure is similar in overall shape and fold to subtype B, C and F PRs with inhibitor bound (Fig. 1 ▶). The major differences lie in the conformations of the flap region. The flaps in the crystal structures of the unbound subtype B PR and the one subtype C PR crystallized in tetragonal space groups are either semi-open or wide open. In the subtype A PR the flaps are in the closed position, the conformation that one would expect in a complex with inhibitor. Closed flaps were also seen in the crystal structures of two related unbound tethered subtype B PRs [PDB codes 1lv1 (Kumar et al., 2002 ▶) and 1g6l (Pillai et al., 2001 ▶)]. The authors of those reports suggested that crystal packing in the hexagonal space group was responsible for this unusual conformation of the flaps (Figs. 3 ▶ and 5 ▶).
Minor differences in the structure not related to the closed flap conformation are probably a consequence of the amino-acid differences which distinguish the A subtype from the B subtype: I13V, M36I, R41K, H69K and L89M. To examine the effects of these amino-acid changes, the refined model of subtype A PR was compared with the tethered subtype B PR crystal structure (PDB code 1g6l; Pillai et al., 2001 ▶). 1g6l also crystallizes in space group P61 and has only three mutations in the fused chains, namely C95M, presumably added to verify that the linker was intact in the crystal, and two N37S polymorphic mutations of the B subtype (Fig. 4 ▶).
The I13V and K20R changes do not induce significant differences between the two subtype structures. In contrast, the M36I change causes a 1 Å shift of Ile36 away from solvent and toward the side chains of Lys20 and, to a lesser extent, Ile15. This is presumably to compensate for the removed methionine S and methyl C atoms. The longer lysine side chain of the H69K substitution can form a hydrogen bond to the C-terminal carboxylate group of the twofold-related chain of the PR dimer, whereas the NE2 N atom of the histidine side chain in subtype B PR cannot form this interaction. Finally, in the L89M substitution the larger methionine side chain causes a side-chain repositioning of Ile64, although the separation between the Cα atoms of these residues does not increase significantly. The slight increase that is seen (∼0.5 Å) apparently arises from a local shift of several amino acids of the mid-60s portion of the chain with respect to those of the subtype B PR.
3.2. Comparison of subtype A PR with other unbound HIV PR subtypes
Fig. 5 ▶ summarizes the Cα pairwise superpositions of the other unbound PRs onto subtype A PR (Fig. 3 ▶). These distances are primarily a result of conformational differences between the flaps and, to a lesser extent, differences in the coordinates of the elbow region. It is clear that the table can be divided into three zones by comparing the ranges of r.m.s.d. values, one comprised of the first six subtype B PRs in the semi-open form, the next of three PRs in the wide-open form and the final three in the closed form (Fig. 5 ▶). The last three structures listed in Fig. 5 ▶, including subtype A PR, belong to space group P61, while the remainder were refined in either space group P41212 or P41.
An allosteric conformational change was originally invoked as the origin of the wide-open flaps in the MDR form of B PR (PDB code 1tw7; Martin et al., 2005 ▶). However, the wide-open conformation of the flaps has been shown to be only transiently populated during molecular-dynamics simulations (Layten et al., 2006 ▶; Lexa et al., 2009 ▶). It was concluded that crystal packing is the more likely source of the wide-open flap conformation. Caution against emphasizing the significance of the flap position was advised from the crystallographic point of view over ten years ago (Wlodawer & Vondrasek, 1998 ▶).
When comparing the closed forms in space group P61, the refined subtype A PR and the unbound tethered subtype B PR are both very similar in fold to the subtype F PR with inhibitor bound (Figs. 3 ▶ and 4 ▶). In contrast, the semi-open unbound B PR differs from numerous examples of inhibitor-bound B PR structures by a presumed hinge movement centered at the dimer interface of the β-sheet (Wlodawer & Vondrasek, 1998 ▶). This hinge movement varies by up to 2° and has the effect of tightening the inhibitor-bound conformation of the PR and is accompanied by a closing of the flaps onto the inhibitor. This inferred motion also results in many rearrangements of amino acids at the dimer interface.
When comparing the structures of unbound subtype B PR with the PR in inhibitor complexes crystallized in space groups P41 or P41212, the r.m.s.d. values for Cα atoms generally lie between 1 and 2 Å (data not shown). Similar values are found when comparing unbound semi-open with wide-open unbound subtype B PRs (Fig. 5 ▶). However, when comparing the unbound tethered subtype B PR (PDB code 1g6l; Pillai et al., 2001 ▶) with complexes of mutant subtype B PRs bound to inhibitors, a much higher Cα similarity is evident when they are all crystallized in space group P61. For example, the r.m.s.d. values between 1g6l and 1odx (Kervinen et al., 1996 ▶) is 0.3 Å, that between 1g6l and 1zpk (J. Duskova, T. Skalova, J. Dohnalek, H. Petrokova & J. Hasek, unpublished work) is 0.5 Å, and that between 1g6l and 1pyn (Kozisek et al., 2007 ▶) is 0.4 Å.
3.3. Important water interactions in subtype A PR
An important water molecule that forms a hydrogen-bonded bridge between the two PR flaps and two carbonyl O atoms of the inhibitors was noticed in the first structure of a complex of subtype B PR with a peptidomimetic inhibitor (Wlodawer et al., 1989 ▶) and was confirmed in other subsequently determined structures (Wlodawer & Erickson, 1993 ▶). Presumably this water, then called ‘water 301’, is also present when substrates are bound to the PR. In the refined subtype A PR the corresponding water 51 is located 2.9 Å from the amide N atom of Ile50B, but a more distant 3.7 Å from the same atom in Ile50A. In the subtype B tethered dimer (PDB code 1g6l; Pillai et al., 2001 ▶) nearly identical distances between a similar water and the flaps are seen. No other water molecules that would interact with Ile50 of subtype A PR or with water 51 are evident, but their absence might be related to the resolution of the diffraction data.
Water 51 is located 4.7 Å from the so-called catalytic water (numbered 36 in this structure), which lies between the side chains of the catalytic aspartates. This water molecule is closer to the carboxylate O atoms of Asp25B, at 2.7 and 3.3 Å, than to those of Asp25A, at 3.5 and 3.7 Å. The corresponding water molecule in the tethered subtype B PR (PDB code 1g6l; Pillai et al., 2001 ▶) is in a more symmetrical location, with distances of 2.9, 3.1, 3.1 and 3.2 Å from the four O atoms. Many other well ordered water molecules are also present in the subtype A active site, as well as diffuse chains of electron density that may be disordered water molecules.
3.4. Comparison of crystal-packing contacts between unbound subtype A PR and other PRs
From Fig. 7 ▶ it is evident that the packing contacts are grouped into the semi-open forms of unbound PRs, the wide-open forms and the closed forms so far only seen in space group P61. The lone outlier is the F53L mutant subtype B PR (PDB code 2g69; Liu et al., 2006 ▶), which crystallizes in a form with unique unit-cell parameters (a = b ≃ 61, c ≃ 56 Å) when compared with the range seen in the other tetragonal unbound PR structures (a = b ≃ 47, c ≃ 108 Å). Although its packing differs from that of 1hhp (Spinelli et al., 1991 ▶) and 2hb2 (Heaslet et al., 2007 ▶), especially in the 50s area of the flaps, all three contain semi-open flaps.
Figure 7.
Packing contacts (colored cells represent crystal-packing contacts).
Also evident in Figs. 6 ▶ and 7 ▶ are the large number of residues that are involved in crystal contacts in the wide-open forms of the unbound B and C PRs 2pc0 (Heaslet et al., 2007 ▶) and 2r8n (Coman, Robbins, Goodenow et al., 2008 ▶). The flap regions of these structures are extensively involved in crystal contacts. Apparently, the wide-open crystal forms are more tightly packed. This is borne out by the lower V M values for these crystals [2.56 and 2.52 Å3 Da−1, respectively, compared with 3.12 and 3.15 Å3 Da−1 for the semi-open forms 1hhp (Spinelli et al., 1991 ▶) and 2hb2 (Heaslet et al., 2007 ▶), respectively]. This close packing results in interactions between the elbow residues and the flap of a symmetry-related molecule, as has been pointed out previously (Layten et al., 2006 ▶).
4. Discussion
The crystal structure of HIV-1 subtype A PR has now been solved and compared with a set of 11 structures of other variants of HIV PR. The rather unexpected finding of a closed conformation of an unbound (apo) form of subtype A HIV PR set the basis for comparing four PR subtypes, A, B, C and F, crystallized in two crystal systems: tetragonal (P41 and P4122) and hexagonal (P61). Three conformations are compared in this paper, defined by the position of the flaps: semi-open, wide-open and closed. The goal of this work is to bring further understanding of the determinants for the different positions of the flaps.
As shown in Fig. 5 ▶, the r.m.s.d. value between the Cα positions of refined subtype A PR and subtype F PR in complex with the inhibitor TL-3 (PDB code 2p3c; Sanches et al., 2007 ▶) is only 0.6 Å. Such a small difference in overall fold is close to the experimental error of the diffraction experiments, which is typically 0.3–0.5 Å. For comparison, the r.m.s.d. value between the synthetic and wt B PRs, PDB entries 3hvp (Wlodawer et al., 1989 ▶) and 3phv (Lapatto et al., 1989 ▶), is 0.57 Å. One explanation might be that crystal-packing energies in the hexagonal space group induces flap closure and the unbound PR assumes the conformation of the inhibitor-bound form. Each flap in subtype A PR is sandwiched between the flap of its twofold-related monomer and the flap of a symmetry-related molecule. Another molecule, related by a different symmetry operation, is in contact with the opposite flap. The residues involved in the symmetry contact are Phe53 and Met46 from each flap; this contact is also present in those inhibitor complexes of B PR that crystallize in space group P61. Indeed, the superpositions of inhibitor-bound B-subtype PRs crystallized in space group P61 show the same crystal contacts as the two tethered B-subtype structures (1lv1 and 1g6l) and similar r.m.s.d. values for the superpositions that are evident in Fig. 5 ▶ (data not shown). If the argument that crystal packing is key to flap conformation has merit, perhaps it can be used to explain the difference between the semi-open and wide-open flap conformations in the tetragonal space groups P41 and P41212.
In conclusion, the conformation of the flaps in the different subtypes, bound and unbound, may be a ‘cause-and-effect’ observation. The question that needs to be addressed is: do the differences in flap/hinge amino-acid sequences and the associated dynamics affect the crystallization process or do the entropy-driven crystallization forces induce a conformation onto the flaps?
Supplementary Material
PDB reference: HIV-1 subtype A protease, 3ixo
Acknowledgments
We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT) located at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. This project was supported in part by NIH grant AI-28571 to BMD, in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (AW) and in part by Federal funds from the National Cancer Institute, NIH under Contract No. HHSN2612008000001E (ML). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government.
References
- Abecasis, A. B., Deforche, K., Bacheler, L. T., McKenna, P., Carvalho, A. P., Gomes, P., Vandamme, A. M. & Camacho, R. (2006). J. Antivir. Ther.11, 581–589. [PubMed]
- Baldanti, F., Paolucci, S., Ravasi, G., Maccabruni, A., Moriggia, A., Barbarini, G. & Maserati, R. (2008). J. Med. Virol.80, 947–952. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed]
- Brodine, S. K., Mascola, J. R., Weiss, P. J., Ito, S. I., Porter, K. R., Artenstein, A. W., Garland, F. C., McCutchan, F. E. & Burke, D. S. (1995). Lancet, 346, 1198–1199. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Clemente, J. C., Coman, R. M., Thiaville, M. M., Janka, L. K., Jeung, J. A., Nukoolkarn, S., Govindasamy, L., Agbandje-McKenna, M., McKenna, R., Leelamanit, W., Goodenow, M. M. & Dunn, B. M. (2006). Biochemistry, 45, 5468–5477. [DOI] [PMC free article] [PubMed]
- Coman, R. M., Robbins, A. H., Fernandez, M. A., Gilliland, C. T., Sochet, A. A., Goodenow, M. M., McKenna, R. & Dunn, B. M. (2008). Biochemistry, 47, 731–743. [DOI] [PubMed]
- Coman, R. M., Robbins, A. H., Goodenow, M. M., Dunn, B. M. & McKenna, R. (2008). Acta Cryst. D64, 754–763. [DOI] [PubMed]
- Coman, R. M., Robbins, A., Goodenow, M. M., McKenna, R. & Dunn, B. M. (2007). Acta Cryst. F63, 320–323. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Viewer. http://www.pymol.org.
- Dumans, A. T., Barreto, C. C., Santos, A. F., Arruda, M., Sousa, T. M., Machado, E. S., Sabino, E. C., Brindeiro, R. M., Tanuri, A., Duarte, A. J. & Soares, M. A. (2009). Infect. Genet. Evol.9, 62–70. [DOI] [PMC free article] [PubMed]
- Dumans, A. T., Soares, M. A., Machado, E. S., Hué, S., Brindeiro, R. M., Pillay, D. & Tanuri, A. (2004). J. Infect. Dis.189, 1232–1238. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Fleury, H., Recordon-Pinson, P., Caumont, A., Faure, M., Roques, P., Plantier, J. C., Couturier, E., Dormont, D., Masquelier, B. & Simon, F. (2003). AIDS Res. Hum. Retroviruses, 19, 41–47. [DOI] [PubMed]
- Foulkes, J. E., Prabu-Jeyabalan, M., Cooper, D., Henderson, G. J., Harris, J., Swanstrom, R. & Schiffer, C. A. (2006). J. Virol.80, 6906–6916. [DOI] [PMC free article] [PubMed]
- Gonzalez, L. M., Brindeiro, R. M., Tarin, M., Calazans, A., Soares, M. A., Cassol, S. & Tanuri, A. (2003). Antimicrob. Agents Chemother.47, 2817–2822. [DOI] [PMC free article] [PubMed]
- Gonzalez, L. M., Santos, A. F., Abecasis, A. B., Van Laethem, K., Soares, E. A., Deforche, K., Tanuri, A., Camacho, R., Vandamme, A. M. & Soares, M. A. (2008). J. Antimicrob. Chemother.61, 1201–1204. [DOI] [PubMed]
- Grossman, Z., Paxinos, E. E., Averbuch, D., Maayan, S., Parkin, N. T., Engelhard, D., Lorber, M., Istomin, V., Shaked, Y., Mendelson, E., Ram, D., Petropoulos, C. J. & Schapiro, J. M. (2004). Antimicrob. Agents Chemother.48, 2159–2165. [DOI] [PMC free article] [PubMed]
- Heaslet, H., Rosenfeld, R., Giffin, M., Lin, Y.-C., Tam, K., Torbett, B. E., Elder, J. H., McRee, D. E. & Stout, C. D. (2007). Acta Cryst. D63, 866–875. [DOI] [PubMed]
- Hemelaar, J., Gouws, E., Ghys, P. D. & Osmanov, S. (2006). AIDS, 20, W13–W23. [DOI] [PubMed]
- Hemelaar, J., Gouws, E., Ghys, P. D. & Osmanov, S. (2008). AIDS, 22, 322–323. [DOI] [PubMed]
- Hirsch, M. S., Brun-Vézinet, F., D’Aquila, R. T., Hammer, S. M., Johnson, V. A., Kuritzkes, D. R., Loveday, C., Mellors, J. W., Clotet, B., Conway, B., Demeter, L. M., Vella, S., Jacobsen, D. M. & Richman, D. D. (2000). J. Am. Med. Assoc.283, 2417–2426. [DOI] [PubMed]
- Kantor, R. et al. (2005). PLoS Med.2, e112. [DOI] [PMC free article] [PubMed]
- Kantor, R. & Katzenstein, D. (2004). J. Clin. Virol.29, 152–159. [DOI] [PubMed]
- Kervinen, J., Thanki, N., Zdanov, A., Tino, J., Barrish, J., Lin, P. F., Colonno, R., Riccardi, K., Samanta, H. & Wlodawer, A. (1996). Protein Pept. Lett.3, 399–406.
- Kozisek, M., Bray, J., Rezacova, P., Saskova, K., Brynda, J., Pokorna, J., Mammano, F., Rulisek, L. & Konvalinka, J. (2007). J. Mol. Biol.374, 1005–1016. [DOI] [PubMed]
- Krauchenco, S., Martins, N. H., Sanches, M. & Polikarpov, I. (2009). J. Enzyme Inhib. Med. Chem.24, 638–645. [DOI] [PubMed]
- Kumar, M., Kannan, K. K., Hosur, M. V., Bhavesh, N. S., Chatterjee, A., Mittal, R. & Hosur, R. V. (2002). Biochem. Biophys. Res. Commun.294, 395–401. [DOI] [PubMed]
- Lapatto, R., Blundell, T., Hemmings, A., Overington, J., Wilderspin, A., Wood, S., Merson, J. R., Whittle, P. J., Danley, D. E., Geoghegan, K. F., Hawrylik, S. J., Lee, S. E., Scheld, K. G. & Hobart, P. M. (1989). Nature (London), 342, 299–302. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291.
- Layten, M., Hornak, V. & Simmerling, C. (2006). J. Am. Chem. Soc.128, 13360–13361. [DOI] [PMC free article] [PubMed]
- Lee, S., Sawaya, M. R. & Eisenberg, D. (2003). Acta Cryst. D59, 2191–2199. [DOI] [PubMed]
- Lexa, K. W., Damm, K. L., Quintero, J. J., Gestwicki, J. E. & Carlson, H. A. (2009). Proteins, 74, 872–880. [DOI] [PMC free article] [PubMed]
- Liu, F., Kovalevsky, A. Y., Louis, J. M., Boross, P. I., Wang, Y.-F., Harrison, R. W. & Weber, I. T. (2006). J. Mol. Biol.358, 1191–1199. [DOI] [PubMed]
- Logsdon, B. C., Vickrey, J. F., Martin, P., Proteasa, G., Koepke, J. I., Terlecky, S. R., Wawrzak, Z., Winters, M. A., Merigan, T. C. & Kovari, L. C. (2004). J. Virol.78, 3123–3132. [DOI] [PMC free article] [PubMed]
- Martin, P., Vickrey, J. F., Proteasa, G., Jimenez, Y. L., Wawrzak, Z., Winters, M. A., Merigan, T. C. & Kovari, L. C. (2005). Structure, 13, 1887–1895. [DOI] [PubMed]
- Martinez-Cajas, J. L., Pai, N. P., Klein, M. B. & Wainberg, M. A. (2009). J. Int. AIDS Soc.12, 11. [DOI] [PMC free article] [PubMed]
- Martinez-Cajas, J. L., Pant-Pai, N., Klein, M. B. & Wainberg, M. A. (2008). AIDS Rev.10, 212–213. [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals. New York: Wiley.
- Monno, L., Scudeller, L., Brindicci, G., Saracino, A., Punzi, G., Chirianni, A., Lagioia, A., Ladisa, N., Lo Caputo, S. & Angarano, G. (2009). Antiviral Res.83, 118–126. [DOI] [PubMed]
- Osmanov, S., Pattou, C., Walker, N., Schwardländer, B. & Esparza, J. (2002). J. Acquir. Immune Defic. Syndr.29, 184–190. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Padilla, J. E. & Yeates, T. O. (2003). Acta Cryst. D59, 1124–1130. [DOI] [PubMed]
- Paraskevis, D. et al. (2007). J. Infect. Dis.196, 1167–1176. [DOI] [PubMed]
- Palma, A. C., Abecasis, A. B., Vercauteren, J., Carvalho, A. P., Cabanas, J., Vandamme, A. M. & Camacho, R. J. (2009). Infect. Genet. Evol., doi:10.1016/j.meegid.2009.06.019. [DOI] [PubMed]
- Palma, A. C., Araújo, F., Duque, V., Borges, F., Paixão, M. T. & Camacho, R. (2007). Infect. Genet. Evol.7, 391–398. [DOI] [PubMed]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. & Ferrin, T. E. (2004). J. Comput. Chem.25, 1605–1612. [DOI] [PubMed]
- Pillai, B., Kannan, K. K. & Hosur, M. V. (2001). Proteins, 43, 57–64. [DOI] [PubMed]
- Pillay, V., Pillay, C., Kantor, R., Venter, F., Levin, L. & Morris, L. (2008). AIDS Res. Hum. Retroviruses, 24, 1449–1454. [DOI] [PMC free article] [PubMed]
- Rose, R. E., Gong, Y. F., Greytok, J. A., Bechtold, C. M., Terry, B. J., Robinson, B. S., Alam, M., Colonno, R. J. & Lin, P. F. (1996). Proc. Natl Acad. Sci. USA, 93, 1648–1653. [DOI] [PMC free article] [PubMed]
- Sanches, M., Krauchenco, S., Martins, N. H., Gustchina, A., Wlodawer, A. & Polikarpov, I. (2007). J. Mol. Biol.369, 1029–1040. [DOI] [PubMed]
- Sanches, M., Martins, N. H., Calazans, A., de Moraes Brindeiro, R., Tanuri, A., Augusto Ceva Antunes, O. & Polikarpov, I. (2004). Acta Cryst. D60, 1625–1627. [DOI] [PubMed]
- Santos, A. F., Abecasis, A. B., Vandamme, A. M., Camacho, R. J. & Soares, M. A. (2009). J. Antimicrob. Chemother.63, 593–599. [DOI] [PubMed]
- Spinelli, S., Liu, Q. Z., Alzari, P. M., Hirel, P. H. & Poljak, R. J. (1991). Biochimie, 73, 1391–1396. [DOI] [PubMed]
- Taylor, B. S., Sobieszczyk, M. E., McCutchan, F. E. & Hammer, S. M. (2008). N. Engl. J. Med.358, 1590–1602. [DOI] [PMC free article] [PubMed]
- UNAIDS (2008). Report on the Global HIV/AIDS Epidemic 2008: Executive Summary. UNAIDS/08.27E/JC1511E. Geneva: UNAIDS.
- Velazquez-Campoy, A., Todd, M. J., Vega, S. & Freire, E. (2001). Proc. Natl Acad. Sci. USA, 98, 6062–6067. [DOI] [PMC free article] [PubMed]
- Velazquez-Campoy, A., Vega, S., Fleming, E., Bacha, U., Sayed, Y., Dirr, H. W. & Freire, E. (2003). AIDS Rev.5, 165–171. [PubMed]
- Vergne, L., Peeters, M., Mpoudi-Ngole, E., Bourgeois, A., Liegeois, F., Toure-Kane, C., Mboup, S., Mulanga-Kabeya, C., Saman, E., Jourdan, J., Reynes, J. & Delaporte, E. (2000). J. Clin. Microbiol.38, 3919–3925. [DOI] [PMC free article] [PubMed]
- Wlodawer, A. & Erickson, J. W. (1993). Annu. Rev. Biochem.62, 543–585. [DOI] [PubMed]
- Wlodawer, A., Miller, M., Jaskolski, M., Sathyanarayana, B. K., Baldwin, E., Weber, I. T., Selk, L. M., Clawson, L., Schneider, J. & Kent, S. B. (1989). Science, 245, 616–621. [DOI] [PubMed]
- Wlodawer, A. & Vondrasek, J. (1998). Annu. Rev. Biophys. Biomol. Struct.27, 249–284. [DOI] [PubMed]
- Yeates, T. O. (1997). Methods Enzymol.276, 344–358. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HIV-1 subtype A protease, 3ixo
PDB reference: HIV-1 subtype A protease, 3ixo