Variable packing interaction related to the conformational flexibility within the huntingtin-interacting protein 1 coiled coil domain.
Keywords: huntingtin-interacting protein 1, Huntington’s disease, coiled-coil domain
Abstract
Huntingtin-interacting protein 1 (HIP1) is an important link between the actin cytoskeleton and clathrin-mediated endocytosis machinery. HIP1 has also been implicated in the pathogenesis of Huntington’s disease. The binding of HIP1 to actin is regulated through an interaction with clathrin light chain. Clathrin light chain binds to a flexible coiled-coil domain in HIP1 and induces a compact state that is refractory to actin binding. To understand the mechanism of this conformational regulation, a high-resolution crystal structure of a stable fragment from the HIP1 coiled-coil domain was determined. The flexibility of the HIP1 coiled-coil region was evident from its variation from a previously determined structure of a similar region. A hydrogen-bond network and changes in coiled-coil monomer interaction suggest that the HIP1 coiled-coil domain is uniquely suited to allow conformational flexibility.
1. Introduction
Huntingtin-interacting protein 1 (HIP1) is a large multi-domain protein that functions in endocytosis and has been implicated in the pathogenesis of the neurodegenerative Huntington’s disease (Kalchman et al., 1997 ▶; Wanker et al., 1997 ▶). In endocytosis, HIP1 and the related protein HIP1R control the interaction of the vesicular coat protein clathrin with the actin cytoskeleton (Engqvist-Goldstein et al., 1999 ▶; Drubin et al., 2005 ▶). The association of HIP1 with the disease-related protein huntingtin is inversely related to polyglutamine expansion in huntingtin (Kalchman et al., 1997 ▶; Wanker et al., 1997 ▶). Additionally, HIP1 has been linked to caspase activation in conjuction with another binding partner, HIP1 protein interactor (HIPPI; Hackam et al., 2000 ▶; Gervais et al., 2002 ▶).
HIP1 can be divided into three major domains. The N-terminal ANTH domain binds phosphoinositides, the central coiled-coil domain mediates protein interactions and dimerization, and the C-terminal THATCH domain binds actin. The central coiled-coil domain (HIP1cc) is critical to HIP1 function (Wilbur et al., 2008 ▶). This region strongly promotes homodimerization and is not likely to heterodimerize with HIP1R (Wilbur et al., 2008 ▶). The coiled-coil domains of both HIP1 and HIP1R bind to a known regulatory region on clathrin light chain and that of HIP1 contains the binding site for HIPPI (Legendre-Guillemin et al., 2002 ▶, 2005 ▶; Chen & Brodsky, 2005 ▶; Gervais et al., 2002 ▶). HIP1 binding to clathrin light chain can regulate the self-assembly of clathrin; reciprocally, clathrin light chain regulates the ability of HIP1 to bind actin (Chen & Brodsky, 2005 ▶; Wilbur et al., 2008 ▶).
Despite the strong coiled-coil-forming propensity of the HIP1cc, structural and biochemical evidence suggests that the HIP1cc is conformationally dynamic (Ybe et al., 2007 ▶; Niu & Ybe, 2008 ▶; Wilbur et al., 2008 ▶). The structures of two fragments of the HIP1cc have shown canonical coiled-coil regions as well as expected coiled-coil regions that were split apart from each other (Ybe et al., 2007 ▶; Niu & Ybe, 2008 ▶). The HIP1cc structural data in combination with mutagenesis data identified regions that are important for binding HIPPI and suggested a model in which flexibility in the HIP1cc contributes to a ‘U’ or bent shape for HIP1 under certain circumstances (Niu & Ybe, 2008 ▶). Regions of relative flexibility and relative stability have been identified in the HIP1cc (Wilbur et al., 2008 ▶). The N-terminal half of the HIP1cc is the more stable, while the C-terminal half is more flexible. The flexibility of the C-terminal half of the HIP1cc allows a conformational change to occur to regulate actin binding (Wilbur et al., 2008 ▶). Identifying the interactions within the HIP1cc that allow such different properties within a single coiled-coil domain was the focus of the structural analysis reported here.
In order to elucidate the molecular mechanism in HIP1 that allows switching between the compact flexible coiled coil and the more canonical elongated coiled coil, we crystallized the most stable region of the HIP1cc. This allowed comparison with previously determined crystal structures that included the flexible region of the HIP1cc. Although crystallization of the complete HIP1cc was attempted, endogenous proteolysis of the HIP1cc occurred during crystallization and only the stable N-terminal region of the HIP1cc crystallized. We have determined a high-resolution structure of the N-terminal region of the HIP1cc which builds on the previously determined structures by improving the resolution of this region to 2.3 Å, confirming the flexible regions and identifying regions of plasticity within the core hydrophobic residues. This work is the first step in defining the molecular contacts that allow altered conformational dynamics in different regions of the HIP1cc.
2. Experimental methods
2.1. Protein expression and purification
Human HIP1 coiled coil (residues 361–637) was expressed in Escherichia coli using a pRSFDuet vector modified with a Gateway recombination cassette (Invitrogen). Purification was as described previously (Wilbur et al., 2008 ▶).
2.2. Crystallization and data collection
Attempts to crystallize HIP1 coiled-coil protein via sitting-drop vapor diffusion yielded crystals of a proteolytically cleaved fragment of the HIP1 coiled coil (residues 369–445). Crystallization occurred in 0.07 M MES pH 6.5 and 17.5% PEG 4000, 6000, 8000 or 10 000 over two to three months. Similar crystals could be grown within one week under the same conditions using the same starting material with the addition of attomolar concentrations of subtilisin protease. Crystal diffraction data were collected on Advanced Light Source beamline 8.3.1 at Lawrence Berkeley National Laboratory. Data-collection and refinement statistics are given in Table 1 ▶.
Table 1. Summary of data-collection and refinement statistics for HIP1 369–445.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 35.7, b = 57.0, c = 81.1, α = β = γ = 90 |
| Resolution (Å) | 50–2.30 (2.38–2.30) |
| Rmerge | 0.03 (0.17) |
| 〈I/σ(I)〉 | 32.7 (6.32) |
| Completeness (%) | 99.2 (96.8) |
| Redundancy | 3.9 (3.4) |
| Refinement | |
| Resolution (Å) | 40.6–2.30 |
| No. of reflections | 7331 |
| Rwork/Rfree (%) | 23/28 |
| No. of atoms | |
| Protein | 1281 |
| Water | 39 |
| B factor (mean) (Å2) | 50.3 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.36 |
2.3. Phasing, refinement and analysis
Data were processed to 2.3 Å resolution using HKL-2000 (HKL Research). A molecular-replacement solution was found with Phaser (McCoy et al., 2007 ▶) using a fragment of a single polyalanine α-helix from the coiled-coil protein cortexillin I (PDB code 1d7m; Burkhard et al., 2000 ▶) as a search model. Coot (Emsley & Cowtan, 2004 ▶) was used for model building and REFMAC (Murshudov et al., 1997 ▶) was used for refinement. TLS refinement was used in the last round of refinement. Hydrogen bonds were calculated using a distance cutoff of 3.0 Å in PyMOL. All images were generated using PyMOL (DeLano Scientific). Structures were aligned using the SSM function in Coot. Coiled-coil structures were analyzed for knob and hole packing with SOCKET (Walshaw & Woolfson, 2001 ▶) and variations in coiled-coil structure were analyzed with TWISTER (Strelkov & Burkhard, 2002 ▶).
3. Results and discussion
3.1. Crystallization
HIP1cc (residues 361–637) was recombinantly expressed in E. coli, purified as described previously and crystallization was initiated. Orthorhombic crystals grew within 2–3 months in a sitting-drop vapor-diffusion chamber. These crystals diffracted to 2.3 Å resolution. Mass-spectrometric analysis of the crystals suggested that the full-length HIP1cc was degraded prior to crystallization (data not shown). In an attempt to control the proteolysis of the HIP1cc prior to crystallization and improve the crystal-growth kinetics, a titration of subtilisin protease was added to the crystal drops of the complete HIP1cc at an extremely low concentration (see §2). This protease was chosen for its generally broad specificity and its known ability to generate a more stable N-terminal fragment upon cleavage of HIP1cc (Wilbur et al., 2008 ▶). In this case, orthorhombic crystals formed in under a week. Data were collected for both the endogenously degraded and the subtilisin-degraded crystals. Overall, the subtilisin-generated crystals were smaller and diffracted with weaker intensity. Although many of the subtilisin-degraded crystals diffracted to resolutions in the 2.8–4 Å resolution range, some diffracted as well as the endogenously degraded crystals (data not shown). The necessity for degradation of the HIP1cc before crystallization suggests that the flexible regions prevented crystallization. It is notable that direct addition of protease to the crystallization drop promoted crystal formation (data not shown).
3.2. HIP1cc structure and plasticity
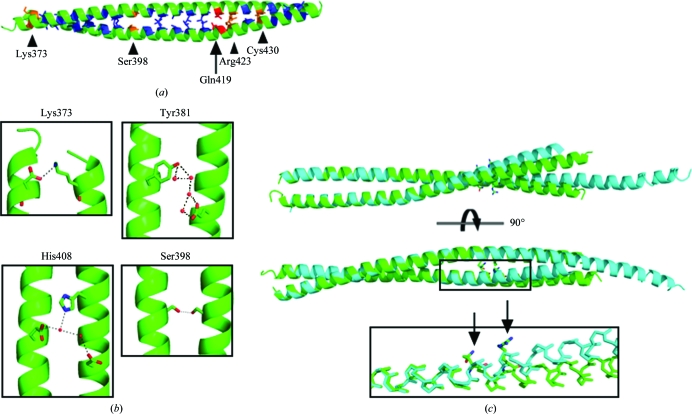
The crystal data of the HIP1cc degradation product were phased by molecular replacement using a polyalanine coiled coil (a fragment of PDB entry 1d7m). There is the equivalent of one molecule in the asymmetric unit, although the coiled-coil structure is formed along a twofold crystallographic symmetry axis. The structure of the coiled coil spans the N-terminal region including residues 367–445 of human HIP1 (referred to as HIP1coil). The heptad repeat of the HIP1cc is clearly defined, with the charged or polar amino acids Lys373, Ser398, Gln419, Arg423 and Cys430 occurring at the typically hydrophobic a or d positions (Fig. 1 ▶ a; Conway & Parry, 1990 ▶).
Figure 1.
Comparison of HIP1 coiled-coil structures. (a) The HIP1 coiled-coil structure determined in this work (residues 367–445; HIP1coil) is shown in green, with core amino acids in other colors. Hydrophobic residues are shown in blue and polar amino acids are shown in orange and red, where those in red have alternate packing in HIP1coil and HIP1split according to SOCKET. (b) Polar contacts (shown as black dashes) found in HIP1coil. The name of one amino acid involved in the polar contact is indicated for reference. (c) Overlay of HIP1coil, shown in green, and the previously determined structure of HIP1 (residues 371–472; HIP1split; PDB code 2qa7; Niu & Ybe, 2008 ▶), shown in cyan. Residues 371–425 were used for alignment. The inset shows the glutamine and arginine residues that occur in the core of the coiled coil and the point of deviation between the two determined structures. Arrows point to the glutamine and arginine side chains. The side chains of other residues have been removed for clarity.
To determine the differences between the structure determined in this work (HIP1coil) and the previously determined 2.8 Å resolution structure of HIP1 residues 371–472 (referred to as HIP1split; Niu & Ybe, 2008 ▶), the two structures were aligned using residues 371–425. The two structures are similar within this region, although the higher resolution of the HIP1coil structure allowed us to detect two previously unresolved hydrogen-bond networks within this region. Glu383 forms a large hydrogen-bonding network with Tyr381 and a series of water molecules (Fig. 1 ▶ b). His408 and Glu415 hydrogen bond via water molecules to Glu413 on the opposite coil (Fig. 1 ▶ b). Additional hydrogen bonding occurs between Lys373 and Asp374 and Ser398 and Ser398 on opposite chains, and was visible in both the HIP1coil and HIP1split structures (Fig. 1 ▶ b). Interestingly, Lys373 and Ser398 both occur in the expected hydrophobic positions within the heptad repeat but appear to contribute to the coiled-coil interaction through hydrogen bonding (Figs. 1 ▶ a and 1 ▶ b).
One helix within HIP1split is disordered starting at residue 440 (Niu & Ybe, 2008 ▶). This suggested a conformationally dynamic region in the coiled-coil structure between residues 440 and 472. A similar phenomenon was detected in the separately determined C-terminal structure (residues 482–586) of the HIP1cc (Ybe et al., 2007 ▶). This previous structural analysis showed that residues 482–539 split apart while residues 540–581 form a coiled coil. The fact that the HIP1coil crystals in this analysis formed after cleavage of the HIP1cc at residue 445, supports the idea of a conformationally dynamic region in the center of the HIP1cc.
The primary differences between the HIP1coil and HIP1split structures are between residues 419 and 445. The previously determined structures (Ybe et al., 2007 ▶; Niu & Ybe, 2008 ▶) suggest that residues 419–445 may facilitate major structural rearrangements in the coiled coil. The polar residues and a disulfide bond at heptad positions a and d can be expected to be hydrophobic and therefore may be destabilizing the core of the coiled coil (Figs. 1 ▶ a and 1 ▶ c). Comparison of the HIP1coil and HIP1split structures reveals an ∼8° and ∼13° shift in the axes of the aligned helices between the two structures (Fig. 1 ▶ c). These shifts are initiated in the region near residues Gln419 and Arg423, which occur at heptad positions d and a, respectively. Crystal-contact differences between the two crystallization conditions may have differentially stabilized the alternate conformations represented by these two structures. Despite this, each structure is expected to represent an accessible low-energy conformation (Kossiakoff et al., 1992 ▶). An analysis of each HIP1 structure via SOCKET (Walshaw & Woolfson, 2001 ▶), a program that evaluates the expected knob and hole packing within the core of coiled-coil structures, yields an additional core packing contact in the HIP1coil structure that occurs at Gln419 and does not occur in HIP1split (Fig. 1 ▶ a). This contact, which is found in the HIP1coil structure but not in the HIP1split structure, suggests that this polar residue is related to the deviation between the two structures and supports the idea that the N-terminal region of the coiled coil of HIP1 may rearrange packing interactions to accommodate structural deviations in the more flexible regions.
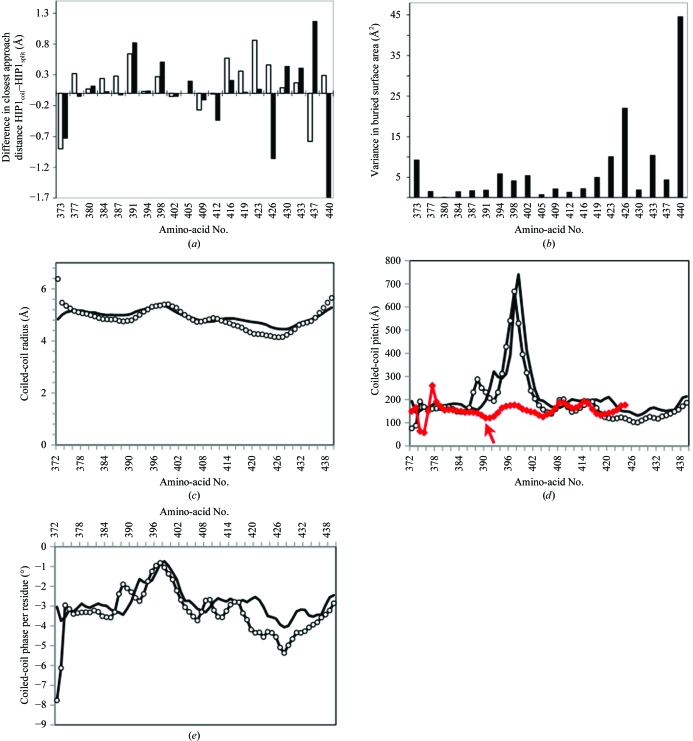
HIP1coil has a slightly larger distance between Cα atoms of heptad core residues than observed for the HIP1split structure (Fig. 2 ▶ a). This causes a slight increase in the overall radius of the HIP1coil structure (Fig. 2 ▶ c). The greater distance between Cα atoms may be accommodated for by variability in the side-chain positions. Some core-residue side chains are closer together in the HIP1coil structure than in the HIP1split structure, while others are further apart (Fig. 2 ▶ b). Residues in heptad positions a and d that are C-terminal to residue Gln419 show a greater variance in the amount of surface area buried between HIP1coil and HIP1split (residues at the end of the chains were excluded from this analysis). The greater variability in Cα position and side-chain buried surface area that occurs near the region of structural flexibility suggests greater plasticity in the coiled-coil interaction in this region (Figs. 2 ▶ a and 2 ▶ b). The differences in coiled-coil pitch and the phase of each residue, as analyzed by TWISTER (Strelkov & Burkhard, 2002 ▶), confirm regions of structural variability when comparing the two structures (Figs. 2 ▶ d and 2 ▶ e). Although both structures have a major change in coiled-coil pitch near residue Ser398, which occurs in an expected hydrophobic position, a majority of the differences in pitch, phase and radius of the coiled coils occur in the more C-terminal region, supporting the idea that changes in the overall structure in the C-terminal region are likely to be accommodated for by plasticity in the side-chain positions (Fig. 2 ▶). The major change in coiled-coil pitch near Ser398 appears to be unique to HIP1. Analysis of tropomyosin and GIT1 coiled coils [PDB codes 2efr (S. Minakata, Y. Nitanai, K. Maeda, N. Oda, K. Wakabayashi & Y. Maeda, unpublished work) and 2w6a (Schlenker & Rittinger, 2009 ▶), respectively], which contain hydrogen-bonded serine residues within the core of the coiled coil, do not have the same change in coiled-coil pitch (Fig. 2 ▶ d and data not shown). It is known that mutation of a core residue to serine can switch the oligomerization state of the GCN4 coiled coil (Akey et al., 2001 ▶). It is likely that the combination of the Ser398 hydrogen bond and surrounding sequence determinants are responsible for forcing a structural rearrangement that results in the dramatic change in the coiled-coil pitch in HIP1. We hypothesize that a combination of the structural features noted above, including various polar residues within the coiled-coil core, plasticity in side-chain interactions and changes in the coil radius, pitch and phase, are intimately linked to the ability of HIP1 to dynamically change conformation. While further structural and functional studies will be necessary to shed light on this hypothesis, it is clear that the structural variations identified by TWISTER suggests two distinct HIP1 coiled-coil structures, which may have further implications for the mechanism of action of this protein.
Figure 2.
Variations in core-residue packing between HIP1 coiled-coil structures. (a) The distance between the Cα atoms of corresponding residues within each HIP1 coiled-coil homodimer was measured and the difference between the structures was plotted (white bars). The same procedure was used for the difference in closest distances between corresponding side chains within each structure (black bars). (b) The buried surface area for each of the heptad core residues was calculated (each residue occurs four times: twice in each structure). The variance of the mean was plotted for each heptad core residue. (c) Coiled-coil radius, (d) coil pitch and (e) coiled-coil phase per residue relative to residue number as determined using TWISTER (Strelkov & Burkhard, 2002 ▶). HIP1coil, solid black line; HIP1split, open circles; the GIT1 coiled-coil structure (which contains a hydrogen-bonded serine in the coiled-coil core; PDB code 2w6a), red line. GIT1 residue numbering is shifted by −55 for representation on the plot; the arrow points to Ser445.
Overall, the structure of HIP1coil confirms the previously determined regions of large structural variability suggested by the HIP1split structure (Niu & Ybe, 2008 ▶). The HIP1coil structure further indicates that plasticity in the coiled-coil core from residues 419 to 440 could accommodate or may otherwise be related to structural rearrangements C-terminal to residue 440 while maintaining strong dimerization of the coiled coil.
Supplementary Material
PDB reference: Huntingtin-interacting protein 1 coiled-coil domain, 3i00
Acknowledgments
This work was supported by NIH grant GM038093 to FMB. We thank Dr Pascal Egea for helpful discussions relating to X-ray crystallography.
References
- Akey, D. L., Malashkevich, V. N. & Kim, P. S. (2001). Biochemistry, 40, 6352–6360. [DOI] [PubMed]
- Burkhard, P., Kammerer, R. A., Steinmetz, M. O., Bourenkov, G. P. & Aebi, U. (2000). Structure, 8, 223–230. [DOI] [PubMed]
- Chen, C. Y. & Brodsky, F. M. (2005). J. Biol. Chem.280, 6109–6117. [DOI] [PubMed]
- Conway, J. F. & Parry, D. A. (1990). Int. J. Biol. Macromol.12, 328–334. [DOI] [PubMed]
- Drubin, D. G., Kaksonen, M., Toret, C. & Sun, Y. (2005). Novartis Found. Symp.269, 35–42. [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Engqvist-Goldstein, A. E., Kessels, M. M., Chopra, V. S., Hayden, M. R. & Drubin, D. G. (1999). J. Cell Biol.147, 1503–1518. [DOI] [PMC free article] [PubMed]
- Gervais, F. G., Singaraja, R., Xanthoudakis, S., Gutekunst, C. A., Leavitt, B. R., Metzler, M., Hackam, A. S., Tam, J., Vaillancourt, J. P., Houtzager, V., Rasper, D. M., Roy, S., Hayden, M. R. & Nicholson, D. W. (2002). Nature Cell. Biol.4, 95–105. [DOI] [PubMed]
- Hackam, A. S., Yassa, A. S., Singaraja, R., Metzler, M., Gutekunst, C. A., Gan, L., Warby, S., Wellington, C. L., Vaillancourt, J., Chen, N., Gervais, F. G., Raymond, L., Nicholson, D. W. & Hayden, M. R. (2000). J. Biol. Chem.275, 41299–41308. [DOI] [PubMed]
- Kalchman, M. A., Koide, H. B., McCutcheon, K., Graham, R. K., Nichol, K., Nishiyama, K., Kazemi-Esfarjani, P., Lynn, F. C., Wellington, C., Metzler, M., Goldberg, Y. P., Kanazawa, I., Gietz, R. D. & Hayden, M. R. (1997). Nature Genet.16, 44–53. [DOI] [PubMed]
- Kossiakoff, A. A., Randal, M., Guenot, J. & Eigenbrot, C. (1992). Proteins, 14, 65–74. [DOI] [PubMed]
- Legendre-Guillemin, V., Metzler, M., Charbonneau, M., Gan, L., Chopra, V., Philie, J., Hayden, M. R. & McPherson, P. S. (2002). J. Biol. Chem.277, 19897–19904. [DOI] [PubMed]
- Legendre-Guillemin, V., Metzler, M., Lemaire, J. F., Philie, J., Gan, L., Hayden, M. R. & McPherson, P. S. (2005). J. Biol. Chem.280, 6101–6108. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Niu, Q. & Ybe, J. A. (2008). J. Mol. Biol.375, 1197–1205. [DOI] [PMC free article] [PubMed]
- Schlenker, O. & Rittinger, K. (2009). J. Mol. Biol.386, 280–289. [DOI] [PubMed]
- Strelkov, S. V. & Burkhard, P. (2002). J. Struct. Biol.137, 54–64. [DOI] [PubMed]
- Walshaw, J. & Woolfson, D. N. (2001). J. Mol. Biol.307, 1427–1450. [DOI] [PubMed]
- Wanker, E. E., Rovira, C., Scherzinger, E., Hasenbank, R., Walter, S., Tait, D., Colicelli, J. & Lehrach, H. (1997). Hum. Mol. Genet.6, 487–495. [DOI] [PubMed]
- Wilbur, J. D., Chen, C.-Y., Manalo, V., Hwang, P. K., Fletterick, R. J. & Brodsky, F. M. (2008). J. Biol. Chem.283, 32870–32879. [DOI] [PMC free article] [PubMed]
- Ybe, J. A., Mishra, S., Helms, S. & Nix, J. (2007). J. Mol. Biol.367, 8–15. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Huntingtin-interacting protein 1 coiled-coil domain, 3i00
PDB reference: Huntingtin-interacting protein 1 coiled-coil domain, 3i00