Abstract
Previous studies have shown that microgroove-initiated contact guidance can induce bone formation in osteoprogenitor cells (OPGs) and produce changes in the cell proteome. For proteomic analysis, differential in-gel electrophoresis (DIGE) can be used as a powerful diagnostic method to provide comparable data between the proteomic profiles of cells cultured in different conditions. This study focuses on the response of OPGs to a novel nanoscale pit topography with osteoinductive properties compared with planar controls. Disordered near-square nanopits with 120 nm diameter and 100 nm depth with an average 300 nm centre-to-centre spacing (300 nm spaced pits in square pattern, but with ±50 nm disorder) were fabricated on 1×1 cm2 polycaprolactone sheets. Human OPGs were seeded onto the test materials. DIGE analysis revealed changes in the expression of a number of distinct proteins, including upregulation of actin isoforms, beta-galectin1, vimentin and procollagen-proline, 2-oxoglutarate 4-dioxygenase and prolyl 4-hydroxylase. Downregulation of enolase, caldesmon, zyxin, GRASP55, Hsp70 (BiP/GRP78), RNH1, cathepsin D and Hsp27 was also observed. The differences in cell morphology and mineralization are also reported using histochemical techniques.
Keywords: nanotopography, nanobioscience, osteoprogenitor cells, tissue engineering, differential proteomics
1. Introduction
The use of surface topography to direct stem cell differentiation has been proposed as a new and innovative strategy in bone tissue engineering. Different features of nanoscale topography can influence a number of cell types in various ways. For example, well-defined nanopillar arrays of polyethylene glycol were used to modulate the adhesion and growth of cardiomyocytes (Kim et al. 2005); ridge/grooves fabricated in polystyrene can induce contact guidance and produce oriented growth of glioma cells along defined directions (Zhu et al. 2004); the morphology and proliferation of smooth muscle cells was altered when cultured on nanogrooved surfaces (Yim et al. 2005); microneedle-like posts have been used as mechanical sensors to control cell adhesion and it was found that RhoA had a vital role in this mechanism (Tan et al. 2003); and nanoporous alumina membranes with different sizes can alter the molecular responses of smooth muscle cells (Nguyen et al. 2007). These, and similar, studies indicate a role for nanotopography in the modulation of tissue formation.
Recent work from our laboratories has also implicated roles for nanoscale topography in skeletal cell differentiation and implications therein for bone tissue engineering (Dalby et al. 2006a,b, 2007c). When considering nanopits (120 nm diameter and 100 nm depth) fabricated by electron-beam lithography (EBL), it was seen that the changes in pit spacing by as little as ±20 nm could strongly influence bone osteoid formation in vitro. Pits in an absolute square arrangement with 300 nm centre-to-centre spacing showed little production of bone osteoid, whereas by adding degrees of controlled disorder (near square) strongly induced osteoid formation to an optimum of ±50 nm (NSQ50). Totally random pit placing resulted in a low degree of bone matrix formation (Dalby et al. 2007c). This shows the exquisite level of control that can be elicited on progenitor and stem cells by control and manipulation of their nanoenvironment.
The same pits were also seen to strongly affect the formation of focal adhesions in human osteoblasts. The square arrangement leads to a shift in the distribution of focal complexes (less than 2 μm), focal adhesions (2–5 μm) and super long adhesions (more than 5 μm) towards the transient focal complexes. However, the near-square topography caused a shift in the opposite direction towards an increased density of the super long adhesions (Biggs et al. 2007b). A study in fibroblasts cultured on the absolute square pits used immunotransmission electron microscopy to show that the cells could not form adhesions over the pits, and thus the surface area available to the cells to form adhesions was reduced (Dalby et al. 2007a). However, the results of studies of the adhesion length (Biggs et al. 2007b) suggest this is otherwise in cells cultured on the near-square pits, i.e. the pits rather induce adhesion formation in bone cells.
Larger pit systems, more similar in size to osteoclast resorption pits, produced by photolithography, with pits of 362 μm depth and 40 μm diameter, have also been observed to promote the induction of bone osteoid and the expression of the osteoblast-specific extracellular matrix proteins (osteocalcin and osteopontin; Dalby et al. 2006b). Nanopits have also been shown to affect the integrin expression in human foetal osteoblast cells and to induce differences in the expression of integrin-mediated cell signalling molecules, e.g. focal adhesion kinase (FAK) in the osteoblastic cells (Lim et al. 2007). These results suggest roles for biomimicry in the fabrication of biomaterial topographies. That is, when techniques such as EBL are employed to produce nanosurfaces, perhaps a biological level of disorder must be included, or perhaps natural features (such as resorption pits in bone) should be incorporated.
In this study, the novel near-square nanopit arrays embossed in the biodegradable polymer polycaprolactone (PCL, approved for use in the body) has been further considered with respect to their influence on osteoprogenitor proteomes. Coomassie blue and alizarin red have been used to observe the cellular morphology of the human osteoprogenitors. Further to this, differential in-gel electrophoresis (DIGE) has been used to probe differences in the proteomes of osteoprogenitor cell (OPG) populations cultured on the near-square material compared with planar control.
Human OPGs were chosen as they contain a mixture of cells from primitive mesenchymal stem cells to osteoprogenitor and mature osteoblasts with cells at different stages of commitment. This heterogeneous cell mix has the potential to differentiate into mature osteoblasts and is representative of the adherent cells of the bone marrow (Triffitt et al. 1998; Oreffo et al. 2005).
DIGE is a technique that facilitates comparative proteomic analysis by the separation of protein extracts on two-dimensional gels, after labelling with spectrally resolvable fluorophores. Owing to the small areas of topography used, a saturation labelling protocol was adopted in which all the cysteine residues were fluorescently labelled to maximize the sensitivity of protein detection (Marouga et al. 2005). Mass spectrometry was employed to identify proteins that were observed to be differentially expressed in the protein identification and assignation of up- or downregulation.
2. Materials and methods
2.1. Fabrication
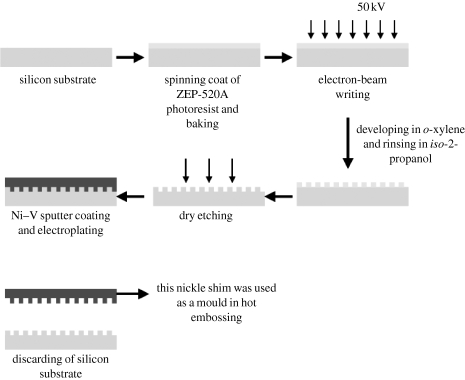
Samples were made in a three-step process of EBL, nickel dye fabrication and hot embossing. Silicon substrates were coated with ZEP-520A resist to a thickness of 100 nm. After the samples were baked for a few hours at 180°C, they were exposed in a Leica LBPG 5-HR100 beamwriter at 50 kV. We have developed an efficient way to pattern a 1 cm2 area with 1–10 billion pits. Three different pit sizes were made using different spot sizes. An 80 nm spot size was used, finally resulting, after embossing, in pits with a diameter of 120 nm. The pitch between the pits was set to an average of 300 nm with a ±50 nm error (in x and y) written into the placement of the pits at the centre of the square. After exposure, the samples were developed in o-xylene at 23°C for 60 s and rinsed in copious amounts of iso-2-propanol.
Nickel dyes were made directly from the patterned resist samples. A thin (50 nm) layer of Ni–V was sputter coated on the samples. This layer acted as an electrode in the subsequent electroplating process. The dyes were plated to a thickness of approximately 300 μm. A diagram of the EBL process is shown in figure 1.
Figure 1.
The diagram of NSQ50 fabrication using EBL.
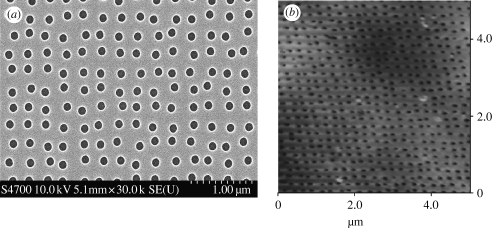
Polymeric replicas were made in PCL (Sigma-Aldrich) sheets. A PCL sheet was cut into 1 cm2 squares before being cleaned with 75 per cent ethanol followed by deionized water and blown dry with cool air. The PCL substrates were heated by light until they started to melt. Either the nickel dye or the planar slides were embossed onto the PCL substrates. The PCL substrates were cooled down and the moulds were removed. The samples were denoted as being near square ±50 nm (NSQ50; figure 2).
Figure 2.
SEM and AFM images of NSQ50 nanopitted substrates. (a) Near-square nanopits with 120 nm diameter, 100 nm depth and with average 300 nm centre-to-centre spacing (300 nm spaced pits in square pattern, but with ±50 nm disorder) on poly(carbonate) substrates produced by EBL and embossing were viewed by SEM. (b) Embossed PCL sheet was analysed by atomic force microscopy to confirm that the nanoscale topography was successfully transferred.
2.2. Cell culture
Human OPGs were obtained from haematologically normal patients undergoing routine surgery. Only tissue that would have been discarded was used with the approval of the Southampton and South West Hants Local Research Ethics Committee. Primary cultures of bone marrow cells were established as described previously (Yang et al. 2003).
Human OPGs including the osteoblasts were cultured in 75 cm2 tissue culture flasks at passage 2. The culture was maintained in basal medium (α-MEM containing 10% FCS and 2% antibiotics) at 37°C, supplemented with 5 per cent CO2. Confluent cell sheets were trypsinized and 1×105 cells were seeded onto NSQ50 and control flat PCL sheets. The tissue culture was maintained in a static culture, and the medium was changed twice per week.
2.3. Histology
Coomassie blue was used to monitor cell morphology at two time points (one and three weeks, selected to allow viewing of individual cells and nascent bone nodules). The PCL sheets with cultured cells were fixed in 4 per cent formaldehyde in PBS for 15 min at each time point. Cell staining was performed using 5 per cent Coomassie blue in 40 per cent methanol and 10 per cent acetic acid for 5 min. The stained materials were washed twice in tap water. The samples were viewed by brightfield microscopy. Pictures were taken with a greyscale digital camera (Scion Corporation Model CFW-1310M).
2.4. Scanning electron microscopy
Cells were fixed with 1 per cent glutaraldehyde (Sigma, UK), buffered in 0.1 M sodium cacodylate (Agar, UK; 4°C, 1 hour) after a three-week culture period to allow the viewing of any nascent bone nodules. The cells were then postfixed in 1 per cent osmium tetroxide (Agar), and 1 per cent tannic acid (Agar) was used as a mordant, dehydrated through a series of alcohol from 20 to 70 per cent, stained in 0.5 per cent uranyl acetate, followed by a further dehydration in 90, 96 and 100 per cent alcohol. The final dehydration was in hexamethyldisilazane (Sigma), followed by air-drying. Once dry, the samples were sputter coated with gold before examination with an Hitachi S800 field emission scanning electron microscopy (SEM).
2.5. Alizarin staining
Two per cent alizarin red stain (pH 4) was prepared by mixing 2 g of alizarin red S (Sigma) with 100 ml of water, and diluted ammonium hydroxide was added to adjust the pH. After three weeks of culture, in order to view nascent nodules, the osteoprogenitors were fixed in 4 per cent formaldehyde for 15 min at 37°C. Then, they were stained with 2 per cent alizarin red for 5 min before washing with water. The samples were viewed by brightfield optical microscopy. Pictures were taken with a greyscale digital camera (Scion Corporation Model CFW-1310M).
2.6. Protein extraction and protein precipitation
After five weeks' culture, to ensure sufficient levels of protein for analysis, the cell sheets were lysed in 1 ml of the DIGE lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS and 30 mM Tris base pH 8.0) with 1× final concentration of general purpose protease inhibitor cocktail (Sigma-Aldrich). The cell suspension was left at room temperature for 1 hour with vigorous mixing every 20 min. The suspension was then centrifuged at 2100g for 10 min to remove the insoluble material. The proteins were precipitated from the supernatant by addition of four volumes of 100 per cent cold acetone. After centrifugation, the protein pellets were washed with 80 per cent acetone and resuspended in the DIGE lysis buffer. The Bradford protein assay was used to determine the amount of protein extracted from each material. Briefly, varying concentrations of BSA (50, 25, 12.5, 6.25 and 3.125 μg ml−1) were prepared and used as a standard curve; 200 μl of protein assay reagent (Bio-Rad) was mixed with 10 μl of each standard and sample. The reaction was left to progress at room temperature for 5 min. Absorbance was measured at 595 nm. Protein concentrations of the protein extract from the test materials were determined from the standard curve.
2.7. Differential in-gel electrophoresis
2.7.1. Saturation labelling
Five micrograms of the extracted proteins were added into sterile microfuge tubes. The protein in each tube was reduced with 1 μl of 2 mM TCEP. The reactions were incubated at 37°C in the dark for 1 hour. The protein in each tube was labelled with the required volumes (2 μl) of Cy3 and Cy5 in the dark for 30 min (typically, 5 μg of protein requires 2 nmol TCEP and 4 nmol of CyDye). Equal volumes of 2× sample buffer (7 M urea, 2 M thiourea, 4% w/v CHAPS, 2% w/v IPG buffer pH 4–7 and 2% w/v DTT) were added to stop the reactions. The proteins labelled with Cy3 and Cy5 were mixed together. Two-dimensional gel electrophoresis was performed. Three pairs of tests and controls were used to compare with each other to meet the statistic criteria.
2.7.2. Two-dimensional gel electrophoresis
The first-dimension isoelectric focusing (IEF) was performed on IPG strips (24 cm; linear gradient pH 4–7) using an Ettan IPGphor system (GE-Healthcare). The IEF was performed using the following voltage programme: 30 V constant for 12 hours; 300 V constant for 1 hour; linear up to 600 V for over 1 hour; linear up to 1000 V for over 1 hour; linear up to 8000 V for over 3 hours; then, 8000 V constant for 8.5 hours. The current was limited to 50 μA per strip and the temperature was maintained at 20°C. After focusing, the strips were equilibrated for 15 min in 5 ml of reducing solution (6 M urea, 100 mM Tris–HCl pH 8, 30% v/v glycerol, 2% w/v SDS, 5 mg ml−1 DTT). For the second-dimension SDS–PAGE, IPG strips were placed on the top of 12 per cent acrylamide gels cast in low-fluorescence glass plates and then sealed by 0.5 per cent (w/v) agarose overlay solution. Gels were run at constant power 50 W/gel until the bromophenol blue tracking front had reached the base of the gel. Fluorescence images of the gels were obtained by scanning on a Typhoon 9400 scanner (GE Healthcare). Cy3 and Cy5 images were scanned at 532/580 nm and 633/670 nm excitation/emission wavelengths, respectively, at a pixel size of 100 μm resolution. Image analysis and statistical quantification of the relative protein expression was performed using DeCyder v. 5.1 software (GE Healthcare).
2.7.3. Preparative two-dimensional gel
Three hundred micrograms of protein extracted from human osteoprogenitors cultured in a tissue culture flask was reduced by 6 μl of 20 mM TCEP and then labelled with 20 μl of Cy3 DIGE flour. After this, two-dimensional gel electrophoresis was performed and the gel scanned as described earlier. The preparative gel image was matched with analytical DIGE gel images and the spots of interest were selected for further analysis. A pick list was generated, containing gel coordinates that were used to direct spot cutting for spots of interest. The gel spots were excised using an Ettan Spot Handling Workstation (Amersham Biosciences, UK), and each gel piece was placed in a separate well of a 96-well plate. The gel pieces were washed three times in 100 μl of 50 mM ammonium bicarbonate, 50 per cent v/v methanol and then twice in 100 μl 75 per cent v/v acetonitrile, before drying. The gel pieces were rehydrated with trypsin solution (20 μg trypsin ml−1 and 20 mM ammonium bicarbonate), and incubated for 4 hours at 37°C. Peptides were extracted from the gel pieces by washing twice in 100 μl of 50 per cent acetonitrile/0.1 per cent trifluoroacetic acid (v/v), before being transferred in solution to a fresh 96-well plate and dried before mass spectrometric (MS) analysis. The use of a preparative gel was required owing to the detection limits of the MS and the protein yield from our small samples.
2.7.4. MS/MS and database analysis
Trypsinized peptide solutions were mixed at a 1 : 1 ratio with a saturated α-cyano-4-hydroxycinnamic acid matrix in 0.3 per cent TFA, and spotted on stainless steel MALDI sample plates (Applied Biosystems, Framingham, MA). Peptide mixtures were then analysed using MALDI/TOF/TOF (4700 Proteomics Analyzer, Applied Biosystems). MALDI–TOF spectra were collected from m/z 800–2800 and up to 10 peaks were selected for the MS/MS analysis. Protein identification was performed using Global Proteome Server Explorer software (Applied Biosystems) using the NCBI human protein database. The identification was assigned to a protein spot feature if the protein score was calculated to be greater than 50, correlating with a 95 per cent confidence interval. The protein identifications were assigned using the Mascot search engine, which gives each protein a probability-based MOWSE score. In all cases, variable methionine oxidation was used for searches. An MS tolerance of 1.2 Da for MS and 0.4 Da for MS/MS analysis was used. Only proteins identified with a significant score (p<0.05) were included, corresponding to a MOWSE score greater than 100. Where multiple proteins were identified, only the highest scoring identification was included in the results.
3. Results
3.1. Cell morphology and histology
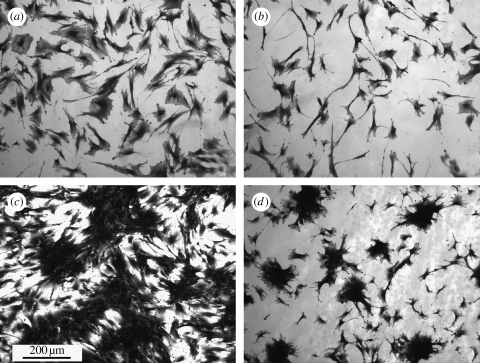
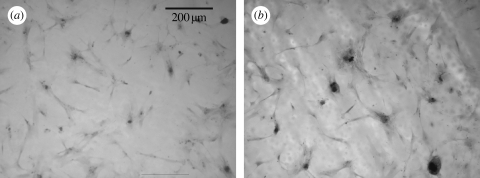
Coomassie blue staining after 5 days indicated that the OPGs on the flat control were observed to be well spread, while osteoprogenitors on the NSQ50 test surface appeared less well spread (figure 3). Higher magnification observation, however, showed that similar cell numbers could be observed in each image grab area, and that the cells on the NSQ50 topography were smaller (figure 4).
Figure 3.
OPGs cultured on PCL substrates in static culture: (a) 5 days on flat surface, (b) 5 days on nanopits, (c) three weeks on flat surface and (d) three weeks on nanopits. Note that the cells on the nanopits at day 5 were less spread than the flat surface. By three weeks, dense aggregates appear on nanopits, similar in appearance to bone nodules.
Figure 4.
A higher magnification of different areas on (a,c) flat surfaces and (b,d) nanopits showed that the numbers of osteoprogenitors after adhering to both surfaces were similar after 5 days' culture.
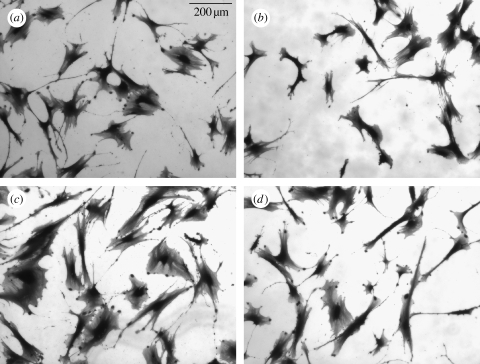
After three weeks of static culture, confluent cell layers (90%+) were observed on control. Dense clusters of cells were noted on the nanopit surface (figure 3) with the appearance of bone osteoid. Scanning electron microscopic evaluation confirmed these results with nodule formation only observed on NSQ50 (figure 5). Alizarin red staining of calcium was observed after three weeks' culture on both NSQ50 and control materials (figure 6). Dense aggregates with intense staining, however, were observed only on the NSQ50 test topography.
Figure 5.
Scanning electron micrographs of osteoprogenitor interaction with NSQ50 nanopits compared with planar control. On the NSQ50 topography (a–d) cells could adhere and differentiate to form a dense aggregate of cells similar in appearance to bone nodules (a,b). (c,d) Cells used their filopodia to sense nanopit topography surface. On the flat surface (e–h) cells were fully spread and proliferated to form a confluent tissue layer (e,f). Also, (g,h) cells using their filopodia to sense the substratum surface can be seen.
Figure 6.
(a,b) Alizarin red staining for calcium accumulation. Positive calcium staining could be seen three weeks after cell seeding. (b) Dense aggregates of cells, bone nodules, were observed on only NSQ50.
3.2. Differential in-gel electrophoresis
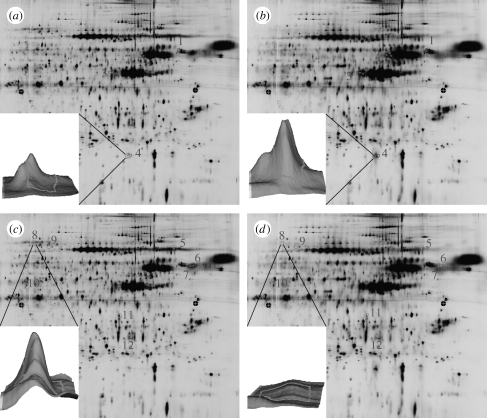
DIGE results showed that the expression of a number of proteins was significantly modulated following the culture of human OPGs on NSQ50 compared with those cultured on flat control (figure 7). The results, including name, direction and magnitude of regulation and at a glance function, of the identified proteins are shown in table 1.
Figure 7.
Typical DIGE images as analysed by DeCyder Image Analysis Software. (a,c) The images of proteins labelled by Cy3 and (b,d) the images of proteins labelled by Cy5. Peak volumes of each spot were compared with each other. A twofold difference in peak volume was used as a cut-off to determine the difference in protein expression between the test and the control. In all DIGE gels, there were areas showing results of up- and downregulation. These spots were used for protein identification.
Table 1.
Typical DIGE images as analysed by DeCyder Image Analysis Software. (Figure 7a,c shows the images of proteins labelled by Cy3. Figure 7b,d shows the images of proteins labelled by Cy5. Peak volumes of each spot were compared against each other; 2.0-fold difference in peak volume was used as a cut-off to determine the difference in protein expression between test and control. In all DIGE gels there were areas showing results of up- and downregulation. These spots were used for protein identification.)
| electrophoretic area | name | function summary | expression resulting from static culture |
|---|---|---|---|
| 1 | procollagen-proline, 2-oxoglutarate 4-dioxygenase and prolyl 4-hydroxylase | collagen formation | 2.50±1.13 |
| 2 | vimentin | cytoskeleton (intermediate filament) | 2.93±0.51 |
| 3 | actin | cytoskeleton (microfilament) | 3.55±1.57 |
| 4 | beta-galactoside-binding lectin precursor (galectin precursor) | regulation of cell adhesion | 3.63±0.82 |
| 5 | Hsp70 and GRP78 (BiP) | chaperone | −2.39±0.29 |
| 6 | GRASP55 | reassembling of Golgi apparatus | −2.81±0.82 |
| 7 | RNase inhibitor | RNA degradation inhibition | −2.77±0.77 |
| 8 | zyxin | focal adhesion forming | −2.83±0.89 |
| 9 | caldesmon | calmodulin and actin-binding protein | −5.03±0.99 |
| 10 | alpha-enolase | enzyme in glycolytic pathway | −2.58±0.31 |
| 11 | cathepsin D | lysosomal protease | −2.23±0.31 |
| 12 | heat shock protein 27 (Hsp27) | chaperone | −4.23±0.39 |
3.2.1. Proteins with increased expression
Significant upregulations of actin isoforms, beta-galectin1, vimentin and procollagen-proline, 2-oxoglutarate 4-dioxygenase and prolyl 4-hydroxylase were noted.
Electrophoretic area 1. Procollagen-proline, 2-oxoglutarate 4-dioxygenase and prolyl 4-hydroxylase are a group of enzymes that play critical roles in the maturation of collagen fibres (Pihlajaniemi et al. 1991). Hydroxyproline is involved in hydrogen bond formation, which is important for the stabilization of collagen fibres (Ramachandran et al. 1973). Prolyl 4-hydroxylase catalyses the formation of 4-hydroxyproline in collagens and the reaction which is catalysed by prolyl 4-hydroxylase requires Fe2+, 2-oxoglutarate, O2 and ascorbate and involves an oxidative decarboxylation of 2-oxoglutarate (Kivirikko et al. 1989). The upregulation of these enzymes suggests increasing collagen synthesis in cells cultured on nanopits.
Electrophoretic area 2. Vimentin is the major skeletal intermediate filament cytoskeletal protein and is involved in a wide range of cellular activities. Intermediate filaments play an important role in supporting the location of the organelles (Katsumoto et al. 1990). Furthermore, the intermediate filaments respond to mechanical stresses with great sensitivity (Thoumine et al. 1995) and have postulated roles in mechanosensitive signalling via, for example, cellular tensegrity (Wang et al. 1993; Ingber 1997). In a previous study, the osteoblast-like cell line MC3T3-E1 showed higher expression of vimentin during the differentiation and mineralization phase than in the proliferation phase, suggesting that vimentin may have a role in bone maturation (Kitching et al. 2002).
Electrophoretic area 3. Actin is a cytoskeletal protein known to be important in topographical contact guidance (Wojciak-Stothard et al. 1995). Changes in actin organization have also been reported in response to nanoscale topographies (Dalby et al. 2003). Increase in well-organized actin stress fibres is associated with enhanced osteogenic activity (Titushkin & Cho 2007). This may be through the generation of tension by actin stress fibres effecting the formation of focal adhesions that have critical roles in cell response to materials through transmembrane integrins linking the extracellular matrix to the cytoskeleton. Such changes to cellular adhesion will change signalling from associated proteins, such as FAK, extracellular receptor kinase (ERK) and Src family kinases, all of which interact during FAK autophosphorylation during adhesion maturation and stress fibre assembly (Chrzanowska-Wodnicka & Burridge 1996; Dalby et al. 2008).
Electrophoretic area 4. Upregulation of beta-galactoside-binding lectin precursor (galectin precursor) and galectin-1 was found. These results may indicate increased osteo-specific differentiation on NSQ50 compared with control (Aubin et al. 1996; Choi et al. 1998; Colnot et al. 1999), as the expression of galectin in skeletal tissue is controlled by Runx2 during the matrix maturation stage of osteoblastic development (Stock et al. 2003). Galectin-1 was found to be associated with the nuclear matrix in differentiated osteoblasts. It was detectable only in the nuclear matrix of differentiated osteoblasts (Choi et al. 1998).
3.2.2. Proteins with decreased expression
Significant downregulations for enolase, caldesmon, zyxin, GRASP55, Hsp70, (BiP/GRP78), RNH1, cathepsin D and Hsp27 were found.
Electrophoretic area 5. Previous studies have indicated that certain isoforms of Hsp70 were downregulated. Heat shock 70-kDa protein 8 isoform 1 (HSPA8) is expressed on the cell surface of the human embryonic stem cells and downregulated upon differentiation (Son et al. 2005). A decrease in mtHSP70 is important for the induced differentiation of HL-60 promyelocytic leukaemia cells (Xu et al. 1999). By contrast, other studies showed an increase in Hsp70 (GRP78/Bip) during differentiation (Nakai et al. 1995). In addition, Cotrupi & Maier (2004) have also suggested that cells in microgravity express Hsp70 in order to maintain their proliferative potential. While little is understood yet about this group of proteins, it seems likely that the heat shock proteins will be important in modulation, osteoblastic proliferation and differentiation. Some evidence suggests that mitogen-activated protein kinases (MAPKs) of the ERK family alter Hsp70 transcription. The ERK signalling pathway induces the expression of Hsp70 (Hung et al. 1998), and inhibition of ERK reduces the expression of Hsp70 in response to various stressors (Yang et al. 2004; Keller et al. 2008).
Electrophoretic area 6. Downregulation of RNase inhibitor was also noted in a previous study (Kantawong et al. in press) that considered progenitor cells on grooves. Activity of the RNase is inhibited by the formation of a complex with an inhibitor (such as RNH1). In vivo, more than 95 per cent of ribonuclease is complexed with an inhibitor, and the inhibitor/RNase ratio is elevated in proliferating tissues (Schneider et al. 1988). Thus, it may be that this result indicates the lower proliferative activity of the cells on the NSQ50 (compared with control) as a consequence of differentiation to matrix secreting cells.
Electrophoretic area 7. Downregulation of Golgi reassembly stacking protein 2 (GRASP55) might indicate the decrease of mitotic activity of the cultured cells on disordered nanopits. GRASP55 plays a mitogen-activated protein kinase kinase/extracellular-activated protein kinase (MEK/ERK)-regulated role in Golgi ribbon formation and cell cycle progression (Feinstein & Linstedt 2008). GRASP55 mediates assembly of Golgi stacks in an in vitro assay (Shorter et al. 1999).
Electrophoretic area 8. Zyxin is one of the LIM domain proteins at focal adhesion plaques (Wang & Gilmore 2003). Zyxin is a phosphoprotein localized at the sites of cell-substratum adhesion and also plays a role as an intracellular signal transducer. Zyxin and its partners have been implicated in the spatial control of actin filament assembly as well as in pathways important for cell differentiation (Beckerle 1997). This group of proteins also shuttle through the nucleus and may regulate gene transcription by interaction with transcription factors (Wang & Gilmore 2003).
Electrophoretic area 9. Downregulation of caldesmon can also be correlated with the previous proteomic study considering grooved topography (Kantawong et al. in press). Caldesmon is an actin-binding protein. In non-muscle cells, it influences contractility by interfering with focal adhesion and stress fibre assembly (Helfman et al. 1999; Li et al. 2004). Thus, this result appears to fit well with the previous observation of the increased actin isoform expression. This result is also in agreement with a further previous study (Inoue et al. 2000), which indicated that caldesmon is downregulated during osteoblastic differentiation.
Electrophoretic area 10. Downregulation of alpha-enolase provides further evidence informing us that culture cells on NSQ50 were differentiating (compared with control). Alpha-enolase was found upregulated in proliferating human keratinocytes and downregulated in differentiating cell types (Olsen et al. 1995).
Electrophoretic area 11. Cathepsin D is a well-characterized aspartic protease expressed ubiquitously in lysosomes. The enzyme has broad substrate specificity at acidic pH. Cathepsin D is important in bone degradation owing to a role in extracellular matrix degradation. Cathepsin D has also been implicated in cell growth and apoptosis, and expressed as a marker for osteoclast differentiation. Osteoclasts are macrophage-derived cells developed from the haemopoietic lineage. This suggests that the protein expression during cell differentiation depends on individual cell types.
Electrophoretic area 12. Heat shock protein 27 (Hsp27) is a downstream regulator of actin filament structure and dynamics (Landry & Huot 1995). Hsp27 is involved in the regulation of actin polymerization and stability (Benndorf et al. 1994; Huot et al. 1996), and participates in cell proliferation and differentiation during tissue development (Wang et al. 2003; Matalon et al. 2008; Takahashi-Horiuchi et al. 2008). By contrast, Hsp27 expression was decreased in this study. Hsp27 overexpression has been shown to increase FAK phosphorylation and focal adhesion formation, depending on integrin-mediated actin cytoskeleton polymerization (Lee et al. 2008).
4. Discussion
This study has examined the differential osteoblast proteomic profiles of the cells cultured on an osteogenic nanotopography, NSQ50, and planar control over a 35 day time frame and demonstrated significant regulation of a number of matrix, cytoskeletal, heat shock and molecular and biochemical players.
A previous study from our group considering DIGE of OPGs cultured on microtopography (grooves; Kantawong et al. in press) considered that the microgrooved topography altered the cell morphology and induced changes in the protein expression profiles via arrangement of the cytoskeletal network (Dalby et al. 2007a , b ). The cytoskeleton is thought to act as an integrated network transducing changes in cellular tension to the nucleus via the close relationship of the cytoskeletal intermediate filaments (e.g. vimentin) and the nucleoskeletal intermediate filaments (the lamins) (Ingber 2003). Furthermore, lamins have been described as being intimate with interphase chromatin (Bloom et al. 1996).
It has been reported that nanoscale topographies can induce changes in the cell adhesion complex formation (Biggs et al. 2007a , 2008), cell morphology (Biggs et al. 2007b ), cytoskeleton network and nuclear organization (Dalby et al. 2007b ). The present study was undertaken to investigate whether such changes translate to comparative changes in the protein expression on a highly novel nanotopography where the genome has been investigated; the protein expression profile providing definitive evidence of transcriptional protein changes as a consequence of alterations to the gene expression.
Here, we have studied the protein profiles in long-term culture (five weeks). As observed with cells on microgrooves, the cytoskeleton, and regulation of the cytoskeleton, is of central importance. The influence of microscale features on the cytoskeletal filaments and tubules is easier to appreciate owing to the scale of the features compared with the cell, i.e. they are of the same order of magnitude. However, nanoscale features are far smaller than the cell, corresponding in size to features such as filopodia, and thus it seems likely that nanotopography will change the cell behaviour through alteration in adhesion formation rather than mechanical constraint.
In this study, the novel osteogenic nanotopography, NSQ50, results in changes in the protein expression profiles of the cultured OPGs, confirming that nanotopography can direct cell differentiation with implications therein for the future design of scaffolds and biomaterials for reparative approaches.
In recent publications looking at nanoscale topographies (including NSQ50) and genomic regulations (Dalby et al. 2007c , 2008), we have shown that there are several canonical (well-defined) signalling pathways that appear to be specifically triggered by nanotopography. These include actin- and integrin-related signalling. The proteomic observation of the cytoskeletal and regulatory protein changes suggests translation of these pathways from the genome to the proteome. This is critical for the expression of phenotype from genomic regulations.
NSQ50 has previously been shown to modulate the osteoid formation in vitro in basal media, while OPGs grown on planar control appear to have a more fibroblastic morphology (Dalby et al. 2007c ). It is clear that NSQ50 attenuates cell adhesion as shown in the downregulation of zyxin and Hsp27, which will affect FAK signalling downstream. These changes will have effects on the cell cytoskeleton and subsequent cellular traffickings. Since cathepsin D is involved in the protein trafficking and protein degradation pathways, it might be postulated that the downregulation of this enzyme results from the changes in cellular trafficking activity. The maturation of cathepsin D depends on the clathrin-mediated trafficking of TGN-derived cargo, which is regulated via actin assembly (Carreno et al. 2004; Poupon et al. 2008).
The most important change influenced by NSQ50 is the attenuation of cell proliferation as shown in the downregulation of Hsp70 and Golgi reassembly stacking protein (GRASP55), the downstream regulator of the mitogen-activated protein kinase kinase (MEK)/extracellular-activated protein kinase (ERK) pathway (Jesch et al. 2001). Downregulation of RNH1 also shows agreement that the proliferation rate of cells on NSQ50 should be decreased. Changes in the expression of galectin and procollagen-proline, 2-oxoglutarate 4-dioxygenase and prolyl 4-hydroxylase all imply changes in the maturational state of the progenitor cells and thus this investigation suggests that the balance of these proteins could be important in topographically induced bone cell differentiation and bone formation.
The NSQ50 topography is of interest as it represents a previously untried middle route, that of controlled disorder. Previously, only highly ordered (sub-nanometre error) and totally random topographies have been tested. It would appear from these, and previous genomic and histological results, that this middle pathway provides a powerful stimulus for differentiation. It could be postulated that while nature can achieve high degrees of order, the potential for additional discrete levels of disorder to modulate the cell differentiation and activity offer exciting new avenues, as yet unexplored, in regenerative medicine.
5. Conclusions
Disordered nanopits imprinted onto a PCL substrate are a stimulator of human OPG differentiation. Here, we demonstrate a differential proteomic technique to study the cell response to nanotopography and show a number of significant changes in the protein expression. The changes implicate the modulation of focal adhesions and the cytoskeleton as having central roles in inducing cell differentiation on nanotopography.
The development of materials that can elicit desired responses in stem cell and progenitor cell populations will underpin tissue engineering, where a key driver is the formation of complex tissues in the laboratory. This requirement for complex nanostructures indicates the need to further elucidate the mechanisms and disordered structures implicated in stem cell differentiation and potential therein for developing innovative strategies to tissue repair.
Only tissue that would have been discarded was used with the approval of the Southampton and South West Hants Local Research Ethics Committee.
Acknowledgments
F.K. and M.J.D. thank the Royal Thai Government for the financial support for studentship and the BBSRC for the David Phillips Fellowship, respectively. Work in R.O.C.O.'s laboratory is supported by the BBSRC and EPSRC. The Sir Henry Wellcome Functional Genomics Facility (SHWFGF) was established with support from the Wellcome Trust. We would like to thank everyone in the Centre for Cell Engineering (CCE) and SHWFGF for their help, support and discussion. We thank Margaret K. Mullin for SEM sample preparation. Thanks to Andrew Hart, Anne McIntosh, Jay Jayawardena and Karl Burgess for their technical advice and discussion.
References
- Aubin J. E., Gupta A. K., Bhargava U., Turksen K. 1996. Expression and regulation of galectin 3 in rat osteoblastic cells. J. Cell Physiol. 169, 468–480. 10.1002/(SICI)1097-4652(199612)169:3%3C468::AID-JCP7%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Beckerle M. C. 1997. Zyxin: zinc fingers at sites of cell adhesion. Bioessays 19, 949–957. 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- Benndorf R., Hayess K., Ryazantsev S., Wieske M., Behlke J., Lutsch G. 1994. Phosphorylation and supramolecular organization of murine small heat shock protein HSP25 abolish its actin polymerization-inhibiting activity. J. Biol. Chem. 269, 20 780–20 784. [PubMed] [Google Scholar]
- Biggs M. J., Richards R. G., Gadegaard N., Wilkinson C. D., Dalby M. J. 2007a. The effects of nanoscale pits on primary human osteoblast adhesion formation and cellular spreading. J. Mater. Sci. Mater. Med. 18, 399–404. 10.1007/s10856-006-0705-6. [DOI] [PubMed] [Google Scholar]
- Biggs M. J., Richards R. G., Gadegaard N., Wilkinson C. D., Dalby M. J. 2007b. Regulation of implant surface cell adhesion: characterization and quantification of S-phase primary osteoblast adhesions on biomimetic nanoscale substrates. J. Orthop. Res. 25, 273–282. 10.1002/jor.20319. [DOI] [PubMed] [Google Scholar]
- Biggs M. J., Richards R. G., McFarlane S., Wilkinson C. D., Oreffo R. O., Dalby M. J. 2008. Adhesion formation of primary human osteoblasts and the functional response of mesenchymal stem cells to 330 nm deep microgrooves. J. R. Soc. Interface 5, 1231–1242. 10.1098/rsif.2008.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom S., Lockard V. G., Bloom M. 1996. Intermediate filament-mediated stretch-induced changes in chromatin: a hypothesis for growth initiation in cardiac myocytes. J. Mol. Cell Cardiol. 28, 2123–2127. 10.1006/jmcc.1996.0204. [DOI] [PubMed] [Google Scholar]
- Carreno S., Engqvist-Goldstein A. E., Zhang C. X., McDonald K. L., Drubin D. G. 2004. Actin dynamics coupled to clathrin-coated vesicle formation at the trans-Golgi network. J. Cell Biol. 165, 781–788. 10.1083/jcb.200403120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., van Wijnen A. J., Aslam F., Leszyk J. D., Stein J. L., Stein G. S., Lian J. B., Penman S. 1998. Developmental association of the beta-galactoside-binding protein galectin-1 with the nuclear matrix of rat calvarial osteoblasts. J. Cell Sci. 111, Pt 20, 3035–3043. [DOI] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., Burridge K. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133, 1403–1415. 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnot C., Sidhu S. S., Poirier F., Balmain N. 1999. Cellular and subcellular distribution of galectin-3 in the epiphyseal cartilage and bone of fetal and neonatal mice. Cell. Mol. Biol. (Noisy-le-grand) 45, 1191–1202. [PubMed] [Google Scholar]
- Cotrupi S., Maier J. A. 2004. Is HSP70 upregulation crucial for cellular proliferative response in simulated microgravity? J. Gravit. Physiol. 11, P173–P176. [PubMed] [Google Scholar]
- Dalby M. J., Riehle M. O., Johnstone H. J., Affrossman S., Curtis A. S. 2003. Nonadhesive nanotopography: fibroblast response to poly(n-butyl methacrylate)-poly(styrene) demixed surface features. J. Biomed. Mater. Res. A 67, 1025–1032. 10.1002/jbm.a.10139. [DOI] [PubMed] [Google Scholar]
- Dalby M. J., McCloy D., Robertson M., Agheli H., Sutherland D., Affrossman S., Oreffo R. O. 2006a. Osteoprogenitor response to semi-ordered and random nanotopographies. Biomaterials 27, 2980–2987. 10.1016/j.biomaterials.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Dalby M. J., McCloy D., Robertson M., Wilkinson C. D., Oreffo R. O. 2006b. Osteoprogenitor response to defined topographies with nanoscale depths. Biomaterials 27, 1306–1315. 10.1016/j.biomaterials.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Dalby M. J., Biggs M. J., Gadegaard N., Kalna G., Wilkinson C. D., Curtis A. S. 2007a. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. J. Cell Biochem. 100, 326–338. 10.1002/jcb.21058. [DOI] [PubMed] [Google Scholar]
- Dalby M. J., Gadegaard N., Herzyk P., Sutherland D., Agheli H., Wilkinson C. D., Curtis A. S. 2007b. Nanomechanotransduction and interphase nuclear organization influence on genomic control. J. Cell Biochem. 102, 1234–1244. 10.1002/jcb.21354. [DOI] [PubMed] [Google Scholar]
- Dalby M. J., Gadegaard N., Tare R., Andar A., Riehle M. O., Herzyk P., Wilkinson C. D., Oreffo R. O. 2007c. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 6, 997–1003. 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- Dalby M. J., Andar A., Nag A., Affrossman S., Tare R., McFarlane S., Oreffo R. O. 2008. Genomic expression of mesenchymal stem cells to altered nanoscale topographies. J. R. Soc. Interface 5, 1055–1065. 10.1098/rsif.2008.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T. N., Linstedt A. D. 2008. GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell 19, 2696–2707. 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Levy E. T., Berthier C., Shtutman M., Riveline D., Grosheva I., Lachish-Zalait A., Elbaum M., Bershadsky A. D. 1999. Caldesmon inhibits nonmuscle cell contractility and interferes with the formation of focal adhesions. Mol. Biol. Cell 10, 3097–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung J. J., Cheng T. J., Lai Y. K., Chang M. D. 1998. Differential activation of p38 mitogen-activated protein kinase and extracellular signal-regulated protein kinases confers cadmium-induced HSP70 expression in 9L rat brain tumor cells. J. Biol. Chem. 273, 31 924–31 931. 10.1074/jbc.273.48.31924. [DOI] [PubMed] [Google Scholar]
- Huot J., Houle F., Spitz D. R., Landry J. 1996. HSP27 phosphorylation-mediated resistance against actin fragmentation and cell death induced by oxidative stress. Cancer Res. 56, 273–279. [PubMed] [Google Scholar]
- Ingber D. E. 1997. Tensegrity: the architectural basis of cellular mechanotransduction. Annu. Rev. Physiol. 59, 575–599. 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- Ingber D. E. 2003. Tensegrity I. Cell structure and hierarchical systems biology. J. Cell Sci. 116, 1157–1173. 10.1242/jcs.00359. [DOI] [PubMed] [Google Scholar]
- Inoue A., Kamiya A., Ishiji A., Hiruma Y., Hirose S., Hagiwara H. 2000. Vasoactive peptide-regulated gene expression during osteoblastic differentiation. J. Cardiovasc. Pharmacol. 36, S286–S289. 10.1097/00005344-200036051-00084. [DOI] [PubMed] [Google Scholar]
- Jesch S. A., Lewis T. S., Ahn N. G., Linstedt A. D. 2001. Mitotic phosphorylation of Golgi reassembly stacking protein 55 by mitogen-activated protein kinase ERK2. Mol. Biol. Cell 12, 1811–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantawong F., Burchmore R., Wilkinson C. D. W., Oreffo R. O. C., Dalby M. J. In press Differential in gel electrophoresis (DIGE) analysis of human bone marrow stromal cell contact guidance. Acta Biomater. [DOI] [PubMed] [Google Scholar]
- Katsumoto T., Mitsushima A., Kurimura T. 1990. The role of the vimentin intermediate filaments in rat 3Y1 cells elucidated by immunoelectron microscopy and computer-graphic reconstruction. Biol. Cell 68, 139–146. 10.1016/0248-4900(90)90299-I. [DOI] [PubMed] [Google Scholar]
- Keller J. M., Escara-Wilke J. F., Keller E. T. 2008. Heat stress-induced heat shock protein 70 expression is dependent on ERK activation in zebrafish (Danio rerio) cells. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 150, 307–314. 10.1016/j.cbpa.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Kim P., Suh K., Kyu Choi S., Ho Lee S., Kim B. 2005. Modulation of adhesion and growth of cardiac myocytes by surface nanotopography. In Conf. Proc. IEEE Eng. Med. Biol. Soc., vol. 4, pp. 4091–4094. 10.1109/IEMBS.2005.1615362 [DOI] [PubMed] [Google Scholar]
- Kitching R., Qi S., Li V., Raouf A., Vary C. P., Seth A. 2002. Coordinate gene expression patterns during osteoblast maturation and retinoic acid treatment of MC3T3-E1 cells. J. Bone Miner. Metab. 20, 269–280. 10.1007/s007740200039. [DOI] [PubMed] [Google Scholar]
- Kivirikko K. I., Myllyla R., Pihlajaniemi T. 1989. Protein hydroxylation: prolyl 4-hydroxylase, an enzyme with four cosubstrates and a multifunctional subunit. FASEB J. 3, 1609–1617. [PubMed] [Google Scholar]
- Landry J., Huot J. 1995. Modulation of actin dynamics during stress and physiological stimulation by a signaling pathway involving p38 MAP kinase and heat-shock protein 27. Biochem. Cell Biol. 73, 703–707. [DOI] [PubMed] [Google Scholar]
- Lee J. W., et al. 2008. HSP27 regulates cell adhesion and invasion via modulation of focal adhesion kinase and MMP-2 expression. Eur. J. Cell Biol. 87, 377–387. 10.1016/j.ejcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Li Y., Lin J. L., Reiter R. S., Daniels K., Soll D. R., Lin J. J. 2004. Caldesmon mutant defective in Ca2+-calmodulin binding interferes with assembly of stress fibers and affects cell morphology, growth and motility. J. Cell Sci. 117, 3593–3604. 10.1242/jcs.01216. [DOI] [PubMed] [Google Scholar]
- Lim J. Y., Dreiss A. D., Zhou Z., Hansen J. C., Siedlecki C. A., Hengstebeck R. W., Cheng J., Winograd N., Donahue H. J. 2007. The regulation of integrin-mediated osteoblast focal adhesion and focal adhesion kinase expression by nanoscale topography. Biomaterials 28, 1787–1797. 10.1016/j.biomaterials.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Marouga R., David S., Hawkins E. 2005. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal. Bioanal. Chem. 382, 669–678. 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- Matalon S. T., Drucker L., Fishman A., Ornoy A., Lishner M. 2008. The role of heat shock protein 27 in extravillous trophoblast differentiation. J. Cell Biochem. 103, 719–729. 10.1002/jcb.21476. [DOI] [PubMed] [Google Scholar]
- Nakai A., et al. 1995. Expression and phosphorylation of BiP/GRP78, a molecular chaperone in the endoplasmic reticulum, during the differentiation of a mouse myeloblastic cell line. Cell Struct. Funct. 20, 33–39. [DOI] [PubMed] [Google Scholar]
- Nguyen K. T., Shukla K. P., Moctezuma M., Tang L. 2007. Cellular and molecular responses of smooth muscle cells to surface nanotopography. J. Nanosci. Nanotechnol. 7, 2823–2832. 10.1166/jnn.2007.610. [DOI] [PubMed] [Google Scholar]
- Olsen E., Rasmussen H. H., Celis J. E. 1995. Identification of proteins that are abnormally regulated in differentiated cultured human keratinocytes. Electrophoresis 16, 2241–2248. 10.1002/elps.11501601356. [DOI] [PubMed] [Google Scholar]
- Oreffo R. O., Cooper C., Mason C., Clements M. 2005. Mesenchymal stem cells: lineage, plasticity, and skeletal therapeutic potential. Stem Cell Rev. 1, 169–178. 10.1385/SCR:1:2:169. [DOI] [PubMed] [Google Scholar]
- Pihlajaniemi T., Myllyla R., Kivirikko K. I. 1991. Prolyl 4-hydroxylase and its role in collagen synthesis. J. Hepatol. 13, Suppl. 3, S2–S7. 10.1016/0168-8278(91)90002-S. [DOI] [PubMed] [Google Scholar]
- Poupon V., Girard M., Legendre-Guillemin V., Thomas S., Bourbonniere L., Philie J., Bright N. A., McPherson P. S. 2008. Clathrin light chains function in mannose phosphate receptor trafficking via regulation of actin assembly. Proc. Natl Acad. Sci. USA 105, 168–173. 10.1073/pnas.0707269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G. N., Bansal M., Bhatnagar R. S. 1973. A hypothesis on the role of hydroxyproline in stabilizing collagen structure. Biochim. Biophys. Acta 322, 166–171. 10.1016/0005-2795(73)90187-6. [DOI] [PubMed] [Google Scholar]
- Schneider R., Schneider-Scherzer E., Thurnher M., Auer B., Schweiger M. 1988. The primary structure of human ribonuclease/angiogenin inhibitor (RAI) discloses a novel highly diversified protein superfamily with a common repetitive module. EMBO J. 7, 4151–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Watson R., Giannakou M. E., Clarke M., Warren G., Barr F. A. 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18, 4949–4960. 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son Y. S., et al. 2005. Heat shock 70-kDa protein 8 isoform 1 is expressed on the surface of human embryonic stem cells and downregulated upon differentiation. Stem Cells 23, 1502–1513. 10.1634/stemcells.2004-0307. [DOI] [PubMed] [Google Scholar]
- Stock M., Schafer H., Stricker S., Gross G., Mundlos S., Otto F. 2003. Expression of galectin-3 in skeletal tissues is controlled by Runx2. J. Biol. Chem. 278, 17 360–17 367. 10.1074/jbc.M207631200. [DOI] [PubMed] [Google Scholar]
- Takahashi-Horiuchi Y., Sugiyama K., Sakashita H., Amano O. 2008. Expression of heat shock protein 27 with the transition from proliferation to differentiation of acinar precursor cell in regenerating submandibular gland of rats. Tohoku J. Exp. Med. 214, 221–230. 10.1620/tjem.214.221. [DOI] [PubMed] [Google Scholar]
- Tan J. L., Tien J., Pirone D. M., Gray D. S., Bhadriraju K., Chen C. S. 2003. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl Acad. Sci. USA 100, 1484–1489. 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoumine O., Ziegler T., Girard P. R., Nerem R. M. 1995. Elongation of confluent endothelial cells in culture: the importance of fields of force in the associated alterations of their cytoskeletal structure. Exp. Cell Res. 219, 427–441. 10.1006/excr.1995.1249. [DOI] [PubMed] [Google Scholar]
- Titushkin I., Cho M. 2007. Modulation of cellular mechanics during osteogenic differentiation of human mesenchymal stem cells. Biophys. J. 93, 3693–3702. 10.1529/biophysj.107.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triffitt J. T., Joyner C. J., Oreffo R. O., Virdi A. S. 1998. Osteogenesis: bone development from primitive progenitors. Biochem. Soc. Trans. 26, 21–27. [DOI] [PubMed] [Google Scholar]
- Wang Y., Gilmore T. D. 2003. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta 1593, 115–120. 10.1016/S0167-4889(02)00349-X. [DOI] [PubMed] [Google Scholar]
- Wang N., Butler J. P., Ingber D. E. 1993. Mechanotransduction across the cell surface and through the cytoskeleton. Science 260, 1124–1127. 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- Wang X., Tokuda H., Hatakeyama D., Hirade K., Niwa M., Ito H., Kato K., Kozawa O. 2003. Mechanism of simvastatin on induction of heat shock protein in osteoblasts. Arch. Biochem. Biophys. 415, 6–13. 10.1016/S0003-9861(03)00213-3. [DOI] [PubMed] [Google Scholar]
- Wojciak-Stothard B., Curtis A. S., Monaghan W., McGrath M., Sommer I., Wilkinson C. D. 1995. Role of the cytoskeleton in the reaction of fibroblasts to multiple grooved substrata. Cell Motil. Cytoskeleton 31, 147–158. 10.1002/cm.970310207. [DOI] [PubMed] [Google Scholar]
- Xu J., Xiao H. H., Sartorelli A. C. 1999. Attenuation of the induced differentiation of HL-60 leukemia cells by mitochondrial chaperone HSP70. Oncol. Res. 11, 429–435. [PubMed] [Google Scholar]
- Yang X., Tare R. S., Partridge K. A., Roach H. I., Clarke N. M., Howdle S. M., Shakesheff K. M., Oreffo R. O. 2003. Induction of human osteoprogenitor chemotaxis, proliferation, differentiation, and bone formation by osteoblast stimulating factor-1/pleiotrophin: osteoconductive biomimetic scaffolds for tissue engineering. J. Bone Miner. Res. 18, 47–57. 10.1359/jbmr.2003.18.1.47. [DOI] [PubMed] [Google Scholar]
- Yang J. B., Ke L., Jiang C. Z., Xu Q., He H. Z., Hu B. C., Wu T. C. 2004. Effects of mitogen activated protein kinase signal transduction pathways on heat shock protein 70 gene expression in endothelial cells exposed to benzo(a)pryene. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 22, 100–103. [PubMed] [Google Scholar]
- Yim E. K., Reano R. M., Pang S. W., Yee A. F., Chen C. S., Leong K. W. 2005. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials 26, 5405–5413. 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Zhang Q., Lu Q., Xu Y., Yin J., Hu J., Wang Z. 2004. Nanotopographical guidance of C6 glioma cell alignment and oriented growth. Biomaterials 25, 4215–4223. 10.1016/j.biomaterials.2003.11.020. [DOI] [PubMed] [Google Scholar]