Abstract
For many years, bacterial cells were considered primarily as selfish individuals, but, in recent years, it has become evident that, far from operating in isolation, they coordinate collective behaviour in response to environmental challenges using sophisticated intercellular communication networks. Cell-to-cell communication between bacteria is mediated by small diffusible signal molecules that trigger changes in gene expression in response to fluctuations in population density. This process, generally referred to as quorum sensing (QS), controls diverse phenotypes in numerous Gram-positive and Gram-negative bacteria. Recent advances have revealed that bacteria are not limited to communication within their own species but are capable of ‘listening in’ and ‘broadcasting to’ unrelated species to intercept messages and coerce cohabitants into behavioural modifications, either for the good of the population or for the benefit of one species over another. It is also evident that QS is not limited to the bacterial kingdom. The study of two-way intercellular signalling networks between bacteria and both uni- and multicellular eukaryotes as well as between eukaryotes is just beginning to unveil a rich diversity of communication pathways.
Keywords: quorum sensing, cell–cell communication, interspecies communication, inter-kingdom communication
1. Introduction
Over the last two decades, the notion that the individual cells in a bacterial population operate as autonomous units has been superseded with the realization that social interactions are common throughout the prokaryotic world. These confer on a bacterial population the ability to instigate a collective behavioural change to environmental challenges. Pivotal to such population-dependent adaptive behaviour is a change in gene expression in response to the perception and processing of chemical information in the form of diffusible signal molecules often referred to as autoinducers. This process is known as ‘quorum sensing’ (QS), and describes the phenomenon whereby an increase in bacterial cell population density is concomitant with an increase in the concentration of signal molecule(s) in the extracellular environment. Once the QS signal molecule concentration has reached a threshold level (at which point the population is considered to be ‘quorate’), activation of a signal transduction cascade will direct the expression or repression of target genes to bring about a collective behavioural adaptation (Cámara et al. 2002; Williams 2007b; Williams et al. 2007). In some instances, the genes responsible for QS signal production are positively regulated, facilitating a positive autoinductive loop driving autoinducer synthesis. The terms quorum diffusion sensing, compartment sensing and efficiency sensing have all been coined in an attempt to refine how information concerning the physiological state and spatial distribution of cells, their numbers and the specific environmental conditions being encountered will be conveyed (Redfield 2002; Winzer et al. 2002b; Hense et al. 2007). The term quorum diffusion sensing has recently been used to describe a threshold concentration of the QS signal eliciting a response that is achieved by a population rather than by a single cell. The number of individuals in a quorate population is therefore not fixed but will depend on the relative rates of synthesis and turnover/loss of QS signal molecule (Winzer et al. 2002b; Atkinson et al. 2007).
A range of chemically distinct compounds are employed as QS signal molecules, and in many bacterial species more than one compound class, along with their cognate synthase gene(s) and the associated signal transduction apparatus, form an interdependent network of regulatory units that govern how a bacterial population will respond to changes in the environment by regulating a diverse set of genes. Outside the laboratory, we must consider that in almost all environments a microbial community will contain multiple bacterial species and will normally be found in close association with eukaryotes. Cross-kingdom sensing of QS signal molecules may therefore constitute an adaptive survival strategy, enabling bacteria and eukaryotes to monitor their surroundings and adjust their behaviour in response to environmental challenge and population flux. This article will provide an overview of some of the key QS systems, followed by a consideration of the opportunities for crosstalk and eavesdropping between different bacterial species and between bacteria and eukaryotes.
2. Prokaryotic quorum-sensing systems
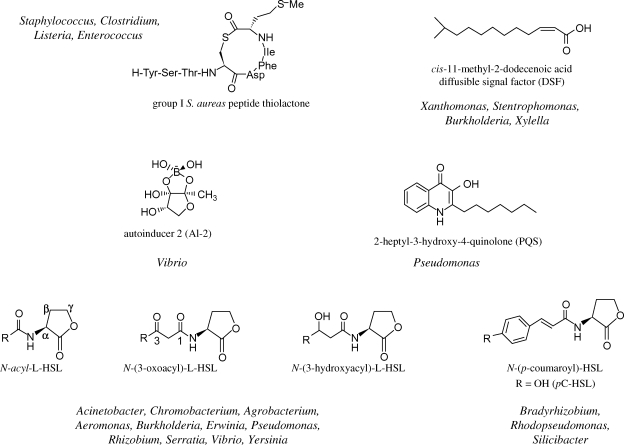
Several chemically distinct classes of QS signal molecules have been identified (figure 1), and in Gram-negative bacteria include the N-acylhomoserine lactones (AHLs), 2-alkyl-4-quinolones, γ-butyrolactones, furanones, long-chain fatty acid derivatives, fatty acid methyl esters, peptides, the 4,5-dihydroxy-2,3-pentandione (DPD) derivatives collectively referred to as autoinducer 2 (AI-2) and autoinducer 3 (AI-3, structure unknown) (Winzer et al. 2002b; Sperandio et al. 2003; Winzer & Williams 2003; Vendeville et al. 2005; Williams 2007b). The (AI-2)/LuxS QS system appears to be shared by both Gram-negative and Gram-positive bacteria (Winzer et al. 2002a; Winzer & Williams 2003; Vendeville et al. 2005) while autoinducing linear, modified or cyclic peptides and γ-butyrolactones (e.g. A-factor) are found in Gram-positive bacteria such as the streptococci, staphylococci and streptomycetes, respectively. Yim et al. (2006) suggested that any number of extracellular bacterial metabolites including compounds with antibiotic activity have the potential to function as signal molecules. However, it is important to differentiate between a true signal molecule involved in cell-to-cell communication and other metabolites. The features considered to define a molecule as a QS signal include: (i) accumulation in the extracellular milieu during a specific growth stage or under certain physiological conditions, or in response to specific environmental changes, (ii) recognition by a specific cell surface or cytoplasmic bacterial receptor, and (iii) the ability to induce a cellular response that extends beyond the physiological changes required to metabolize or detoxify the molecule (Winzer et al. 2002a).
Figure 1.
The structures of some common QS signal molecules.
2.1. Acylhomoserine lactone-mediated quorum-sensing systems
AHL biosynthesis is primarily dependent on members of the LuxI family of AHL synthases (over 100 of which currently appear in the bacterial genome databases) where they are responsible for the catalysis of an amide bond between the appropriately charged acyl–acyl carrier protein (acyl-CoA derivatives can also be substrates), the source of the fatty acyl side chain, and S-adenosyl-methionine, the source of the homoserine lactone ring (More et al. 1996; Jiang et al. 1998; Parsek & Greenberg 1999). Recently, Schaefer et al. (2008) identified an aroyl-CoA (p-coumaroyl-CoA) that served as an alternative source of the fatty acyl side chain and results in the formation of a novel AHL derivative, N-(p-coumaroyl)-homoserine lactone (pC-HSL) (figure 1) in the photosynthetic bacterium Rhodopseudomonas palustris. It is interesting to note that a LuxI homologue RpaI is responsible for catalysing this reaction rather than a novel enzyme specifically tailored to pC-HSL production. N-aroyl homoserine lactones may therefore be commonplace in situations where an aroyl-CoA is available as a precursor substrate to a LuxI homologue.
The response to an AHL usually takes place in the cytoplasm via the interaction between the QS signal and a transcription factor, which is usually a member of the LuxR family of transcriptional regulators (Fuqua et al. 1996; Swift et al. 2001; Cámara et al. 2002), although AHL signalling is sometimes transduced by a transmembrane histidine kinase sensor protein (Milton 2006). AHL–LuxR homologue complexes trigger target gene expression in response to the population size, its degree of compartmentalization and the QS signal concentration in the local environment (for reviews, see Lazdunski et al. 2004; Waters & Bassler 2005; Williams 2007b; Williams et al. 2007).
Two other classes of AHL synthase have been characterized. One of these, an enzyme found exclusively in Vibrio spp., is distinct in amino acid composition from LuxI homologues but is analogous in function and belongs to the LuxM protein family (Milton et al. 2001). These enzymes use the same substrates as, and coexist alongside, their LuxI counterparts in both Vibrio fischeri and Vibrio anguillarum (Hanzelka et al. 1999; Milton et al. 2001). HdtS represents a further putative class of AHL synthase that is distinct from LuxI and LuxM homologues. HdtS was originally described in Pseudomonas fluorescens (Laue et al. 2000) and more recently the homologue, Act, has been identified in Acidithiobacillus ferrooxidans (Rivas et al. 2007). When expressed in Escherichia coli HdtS directs the synthesis of [N-(3-hydroxy-7-cis-tetradecanoyl)homoserine lactone (3-hydroxy-C14:1HSL, which incorporates a double bond in the acyl side chain at position 7), N-decanoylhomoserine lactone and N-hexanoylhomoserine lactone (C6-HSL), the same AHLs as those produced in the P. fluorescens parent. Act also produces (3-hydroxy-C14:1HSL) in E. coli along with several shorter chain AHLs, which are apparently synthesized in small amounts (Rivas et al. 2007).
Often luxI and luxR orthologues are found at the same genetic locus, although a number of luxR ‘orphans’ have now been described (Fuqua 2006; Lee et al. 2006; Lequette et al. 2006; Duerkop et al. 2007; Ferluga et al. 2007). Many of these orphans are functionally similar to the luxR orthologues that are part of a luxR/I pair but may be different in size because of deletions or additions to their sequence. Alternatively, key conserved amino acid residues, which are known to be critical for successful LuxR function, may have been lost in these orphans altering or ablating their activity (Fuqua & Greenberg 2002; Fuqua 2006). There is the possibility that in some cases a LuxR orphan might form a heterodimer with another LuxR protein, the gene for which is genetically linked to a luxI orthologue. For example, dimeric TraR in Agrobacterium tumefaciens acts as a transcriptional activator in the presence of N-(3-oxooctanoyl)homoserine lactone (3-oxo-C8-HSL) (Fuqua et al. 1996; Zhu & Winans 1999), but its activity can be repressed when it forms heterodimers with either TrlR or TraM (Luo et al. 2000; Chai et al. 2001).
AHL-mediated QS systems have been extensively characterized in bacteria such as Vibrio (Milton 2006), Pseudomonas (Williams & Cámara 2009), Rhizobium (Sanchez-Contreras et al. 2007), Erwinia (Barnard et al. 2007) Agrobacterium (White & Winans 2007) and Yersinia (Atkinson et al. 2006b). Here we will consider the last.
2.1.1. Acylhomoserine lactone-mediated quorum sensing in Yersinia
The genus Yersinia comprises four species that are pathogenic to animals. Yersinia pseudotuberculosis and Yersinia enterocolitica are enteropathogens responsible for a self-limiting gastroenteritis presenting as fever, diarrhoea and abdominal pain (Cover & Aber 1989; Johnson 1992; Butler 1994). Yersinia pestis, which presents as pneumonic, bubonic or septicaemic plague, has a biphasic lifestyle alternating between flea and mammalian hosts. Yersinia pestis is a highly uniform clone of Y. pseudotuberculosis that arose shortly before the first recorded plague pandemic, which occurred between the sixth and the eighth century AD (Bercovier et al. 1980; Achtman et al. 1999). Yersinia ruckeri is the only member of the yersiniae that is not pathogenic to mammals but is a major cause of morbidity and mortality in salmonid fish (Ross et al. 1966).
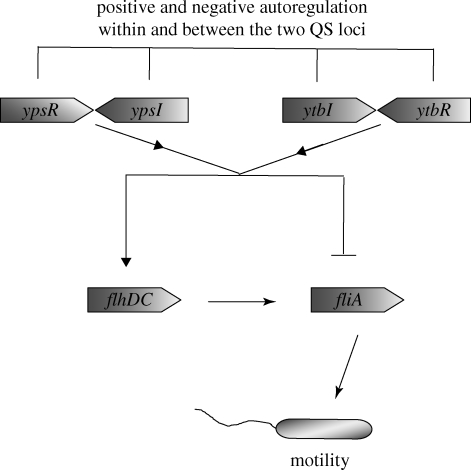
All four Yersinia species produce AHLs and both luxI and luxR orthologues have been identified (Throup et al. 1995; Atkinson et al. 1999; Swift et al. 1999; Temprano et al. 2001). Yersinia pseudotuberculosis possesses two LuxRI pairs termed YpsRI and YtbRI in which the corresponding I and R genes are convergently organized (figure 2). YpsI and YtbI are responsible for the production of at least 24 different AHLs, at least seven of which have been detected at levels considered to be capable of triggering a phenotypic change (Ortori et al. 2007). Both YpsI and YtbI are capable of synthesizing C6-HSL, N-(3-oxohexanoyl)homoserine lactone (3-oxo-C6-HSL) and N-3-(oxo-heptanoyl)homoserine lactone while YtbI is responsible for the synthesis of N-3-(hydroxy-octanonoyl)homoserine lactone, 3-oxo-C8-HSL, N-octanoylhomoserine lactone and N-3-(oxo-decanoyl)homoserine lactone (Ortori et al. 2007).
Figure 2.
The autoregulated QS circuits of Y. pseudotuberculosis, capable of producing up to eight AHLs at physiological concentrations, control flagella-mediated motility by regulating the expression of the motility master regulator flhDC and the alternative sigma factor fliA (adapted from Atkinson et al. 2008).
In Y. pseudotuberculosis, AHL-dependent QS controls cell-to-cell contact (aggregation) and flagellar-mediated motility (Atkinson et al. 1999). On soft-agar motility plates, ypsR and ypsI mutants, but not the parent strain, are motile within 24 h while, in liquid culture, motility is prematurely induced in the mutants but extensively delayed in the parent (Atkinson et al. 1999). The YpsRI and YtbRI systems in Y. pseudotuberculosis work in tandem such that YpsRI (Atkinson et al. 1999) negatively regulates whereas the YtbR/I system positively regulates swimming motility (Atkinson et al. 1999, 2008) (figure 2). In addition, the YpsRI system negatively autoregulates itself but positively regulates the expression of the ytbRI system whereas the ytbRI system is positively autoregulated.
It is perhaps not too surprising to find a similar range of AHLs and two pairs of luxRI homologues (ypeRI and yepRI) in Y. pestis given its close genetic relationship to Y. pseudotuberculosis (Swift et al. 1999; Isherwood 2001). However, Y. pestis is non-motile, and while a Y. pestis ypeR mutant exhibited reduced virulence in a mouse infection model (Swift et al. 1999), QS does not appear to be required for the transmission of plague by fleas because a triple mutant lacking ypeI, yepI and luxS (§2.5) retained the ability to form a biofilm in the flea foregut, a phenotype considered essential for transmissible infection (Jarrett et al. 2004).
Yersinia enterocolitica and Y. ruckeri each possess a single luxRI pair termed yenRI and yruRI, respectively, and exhibit similar AHL profiles to those of Y. pseudotuberculosis and Y. pestis. (Throup et al. 1995; Temprano et al. 2001; Bruhn et al. 2005; Atkinson et al. 2006a; Kirwan et al. 2006). Whether QS contributes to the virulence of these pathogens in their respective hosts is not known although tissues harvested from mice infected with Y. enterocolitica contain AHLs (Jacobi et al. 2003) and both swimming and swarming motility in these yersinia species are QS dependent (Atkinson et al. 2006a).
2.2. The Xanthomonas campestris diffusible signal factor
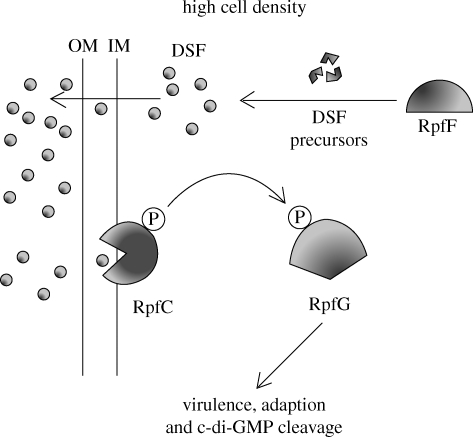
Cha et al. (1998) examined 106 plant-associated bacteria with representatives from seven different genera and found that all produced AHLs except for X. campestris. This organism, the cause of black rot disease in a variety of cruciferous plants such as cabbages (Onsando 1992), regulates the production of extracellular enzymes such as proteases, pectinases and cellulases (Dow & Daniels 1994) using a cell-to-cell communication system that does not employ AHLs. The expression of these exo-enzymes along with the control of biofilm dispersal, toxin resistance and survival (Dow et al. 2003; He et al. 2006) is driven by a small diffusible signal factor (DSF) (Barber et al. 1997; Slater et al. 2000) chemically characterized as cis-11-methyl-2-dodecenoic acid (figure 1) (Wang et al. 2004). The rpf genes are responsible for DSF signal synthesis (RpfF), detection (RpfC) and signal transduction (RpfG). Interestingly, RpfG is a phosphodiesterase that cleaves the intracellular second messenger, cyclic diguanosine monophosphate (c-di-GMP). RpfG contains an HD-GYP motif, the mutation of which abolishes c-di-GMP phosphodiesterase activity and virulence factor production (Ryan et al. 2006). DSF-mediated intercellular signalling is therefore coupled to intracellular signalling via c-di-GMP (Ryan et al. 2006) (figure 3).
Figure 3.
Xanthomonas campestris DSF autoinduction. RpfC inhibits the activity of the DSF synthase RpfF at low cell density. As the cell density increases and basal DSF production allows for signal accumulation, an RpfC conformational change mediated by DSF promotes the release of RpfF to synthesize more DSF and also to initiate phosphorelay from RpfC to RpfG, which will trigger the expression of target genes in the DSF regulon (adapted from He & Zhang 2008).
DSF is not unique to X. campestris and is also produced by Stenotrophomonas maltophilia together with seven other structurally related C12–C14 fatty acids (Fouhy et al. 2007; Boon et al. 2008). The S. maltophilia rpf locus is highly homologous to that of X. campestris, and the DSF signal regulates a number of virulence-associated phenotypes including swimming motility, extracellular protease and lipopolysaccharide along with antibiotic resistance. A further putative DSF signal has been identified as 12-methyltetradecanoic acid in Xylella fastidiosa (Colnaghi Simionato et al. 2007). An rpfF homologue has also been isolated from Burkholderia cenocepacia, which is highly conserved within this genus and is responsible for the synthesis of the DSF-related signal molecule, cis-2-dodecenoic acid. This compound is also able to restore the biofilm and extracellular polysaccharide production phenotypes of an X. campestris rpfF mutant. The conservation of Rpf proteins and the relatedness of DSF structures from different bacteria indicate that cross-species signalling between Xanthomonads and Burkholderia spp. may well occur in nature, particularly because many of these environmental organisms are associated with plants and soil. More recently, a novel fatty acid signalling molecule, cis-2-decenoic acid, produced by Pseudomonas aeruginosa was discovered to induce dispersion of biofilms not only in P. aeruginosa but also in a variety of other Gram-negative and Gram-positive bacteria and the yeast Candida albicans (Davies & Marques 2009). Pseudomonas aeruginosa does not appear to possess an rpf cluster homologue, although there are several enoyl CoA hydratases which share some similarities to RpfF which may be responsible for DSF signal synthesis.
2.3. Autoinducer 2, CAI-1 and Vibrio cholerae
The genus Vibrio contains over 50 species that are found in aquatic habitats and in association with coral, seagrass, fish and shrimp (Milton 2006). Depending on the species, they can associate with marine animal tissues as commensals, symbionts or pathogens. Bioluminescent vibrios such as V. fischeri and Vibrio harveyi as well as non-luminescent species such as V. anguillarum and V. cholerae are known to possess up to three parallel QS systems employing AHLs, AI-2 and a long-chain α-hydroxyketone (CAI-1) as QS signal molecules (Milton 2006). These QS pathways converge on the σ54-dependent activator, LuxO, to control the expression of a variety of target genes that, depending on the species, are involved in symbiosis, bioluminescence, biofilm development and virulence (Milton 2006).
Vibrio cholerae, the causative agent of the diarrhoeal disease cholera, infects humans usually through the ingestion of contaminated water. On reaching the intestine, V. cholerae expresses a number of virulence determinants, including the toxin-coregulated pilus, which promotes adherence to intestinal epithelium, and the ADP-ribosylating exotoxin, cholera toxin, which promotes the often fatal diarrhoea necessary for transmitting the organism back into the environment. Vibrio cholerae uses two QS signal molecules, AI-2 and CAI-1, to regulate synergistically virulence factor expression and biofilm development (Miller et al. 2002; Hammer & Bassler 2003; Zhu & Mekalanos 2003). While in V. harveyi AI-2 is the furanosyl borate diester (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate derived from S-ribosylhomocysteine via the action of LuxS, CAI-1 in V. cholerae is (S)-3-hydroxytridecan-4-one, a 13 carbon chain α-hydroxyketone derivative (Higgins et al. 2007). CAI-1 biosynthesis is dependent on CqsA, which is related to the pyridoxal phosphate-binding aminotransferases, which typically catalyse the condensation of amino acids and carboxylic thioesters, although the biochemical mechanism involved in CAI-1 production has yet to be elucidated.
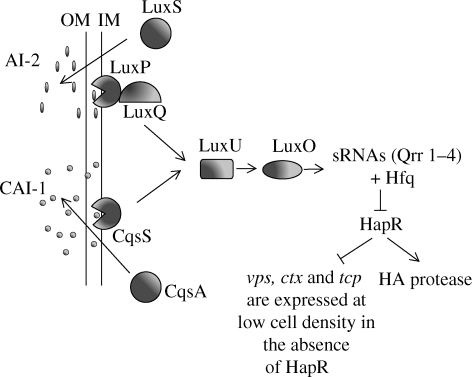
At low population densities in the absence of QS signals, the phosphorylated sensor kinases CqsS and LuxP/Q transfer their phosphates to the phosphotransfer protein, LuxU, which in turn phosphorylates the response regulator, LuxO (figure 4). In conjunction with σ54, LuxO activates the transcription of four small RNAs (sRNA), termed Qrr1–4, which destabilize the HapR transcript. At low cell population densities, this means that no HapR protein is available so that HapR-repressed but not HapR-activated genes are expressed. At high cell population densities where CAI-1 and AI-2 are present, the CqsS and LuxP/Q phosphorelay systems, respectively, are switched from kinases to phosphatases and, as a consequence, LuxO is dephosphorylated and HapR protein can now be produced in the absence of the sRNAs. In the absence of HapR, at low population densities, biofilm exopolysaccharide (vps), cholera toxin (ctx) and toxin-coregulated pilus (tcp) genes are expressed, whereas, at high population densities, they are repressed by HapR, which activates the expression of the HA protease. This response presumably promotes detachment from the intestinal epithelium during the latter stages of infection when V. cholerae prepares to exit the host and re-enter the environment.
Figure 4.
QS in V. cholerae. Two QS signals AI-2 and CAI-1 work synergistically to regulate virulence factor expression and biofilm development. At low population density in the absence of signal molecules, the absence of HapR triggers the expression of genes such as vps, ctx and tcp, which are normally repressed by HapR. As the cell density and therefore the signal molecule concentration increases HapR represses the expression of the genes responsible for colonization and promotes detachment from the intestinal epithelium to enable the organism to exit the host (adapted from Kaper & Sperandio 2005).
2.4. Agr-dependent quorum sensing and the staphylococci
In Staphylococcus aureus, the agr (accessory gene regulator) system is central to virulence gene regulation, intracellular survival in epithelial and endothelial cells as well as biofilm development (Chan et al. 2004; Cheung et al. 2004; Novick & Geisinger 2008). agr is a prototypic QS system shared by a number of different Gram-positive bacteria. Although there is some strain-dependent variation in the nature of the agr regulon in different S. aureus isolates, in experimental animal infection models agr mutants are attenuated, highlighting a key role for this regulatory locus in staphylococcal pathogenicity (Chan et al. 2004; Cheung et al. 2004; Novick & Geisinger 2008). Furthermore, agr regulates all three major exotoxin classes (α-toxin, PVL and the PSMs) implicated in invasive, community-acquired methicillin-resistant S. aureus infections as well as the methicillin antibiotic resistance gene, mecA (Novick & Geisinger 2008; Queck et al. 2008). A functional agr system is also required for endosomal escape, intracellular survival and replication (Qazi et al. 2001) and also contributes to biofilm dispersal and antimicrobial susceptibility (Boles & Horswill 2008).
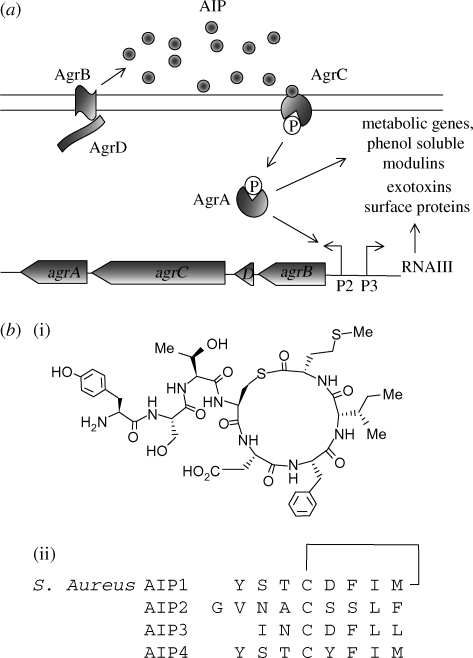
The agr locus consists of two adjacent but divergent transcriptional units (RNAII and RNAIII) controlled by the agrP2 and agrP3 promoters, respectively (Chan et al. 2004; Novick & Geisinger 2008) (figure 5a). The agrP2 operon consists of four genes, agrBDCA, which are required for the activation of transcription from the agrP2 and agrP3 promoters. The P3 transcript codes for both RNAIII, a 517 nucleotide regulatory RNA, and δ-toxin. AgrA and AgrC constitute a two-component system in which AgrA is a response regulator and AgrC is a transmembrane receptor histidine kinase that is activated upon binding to a cognate autoinducing peptide (AIP). This QS signal molecule is a peptide thiolactone consisting of seven to nine amino acid residues in which the central Cys residue is covalently linked to the C-terminal amino acid carboxylate forming a cyclic thioester (figure 5b(i)) and is generated by the action of AgrB on the AgrD pro-peptide (Chan et al. 2004; Novick & Geisinger 2008). Until recently, RNAIII was considered to be the primary effector of the agr response, although it is now clear that the agr regulon can be divided into RNAIII-dependent and RNAIII-independent, AgrA-dependent, genes (Queck et al. 2008).
Figure 5.
AIP-mediated QS in S. aureus. (a) The AIP activates the transmembrane receptor histidine kinase AgrC to trigger autophosphorylation and phosphotransfer to AgrA, which in turn drives the transcription of the agrP2 and agrP3 promoters. This triggers rightward and leftward transcription of the agr locus and RNAIII-dependent and -independent genes. (b)(i) Structures of AIP-1 and (ii) the linear sequences of the four S. aureus AIPs which constitute four different agr specificity groups.
Within the staphylococci, agr polymorphisms occur. Staphylococcus aureus has four different specificity groups with distinct AIPs (figure 5b(ii)) and AgrC sensor domain sequences (Novick & Geisinger 2008). Furthermore, there are at least 20 additional agr groups among the coagulase-negative staphylococci (Novick & Geisinger 2008). The agr system is also evolutionarily conserved across the Firmicutes, which include probiotics such as Lactobacillus plantarum as well as human pathogens such as Clostridium perfringens, Clostridium botulinum, Clostridium difficile, Listeria monocytogenes and Enterococcus faecalis (Wuster & Babu 2008). Apart from E. faecalis, which employs an 11 membered peptide lactone, the chemical identity of the AIPs in these bacteria is not yet known, although mutation of the agr locus in both Listeria (Riedel et al. 2009) and Enterococcus (Nakayama et al. 2006) results in the attenuation of virulence.
2.5. Autoinducer 2 ubiquity, metabolism and quorum sensing
When S-ribosyl-l-homocysteine (SRH) is cleaved to generate homocysteine and DPD, the latter spontaneously cyclizes to yield a number of furanones collectively known as AI-2 (Miller et al. 2004). The enzyme responsible for this conversion (LuxS) was originally discovered in V. harveyi where AI-2 is involved in the regulation of bioluminescence. Homologues of LuxS have been identified in over 50 different bacterial species (Schauder et al. 2001; Winzer et al. 2002b; Xavier & Bassler 2003; Vendeville et al. 2005), suggesting that AI-2-dependent QS is widespread and shared by both Gram-positive and Gram-negative bacteria. However, the LuxS-dependent cleavage of SRH is also an integral part of a metabolic pathway, the activated methyl pathway, that serves to recycle homocysteine from S-adenosyl methionine to maintain de novo methionine biosynthesis. The consequences of disrupting luxS may therefore lead to attenuation of either QS or metabolism or both depending on the organism (Winzer et al. 2002b; Xavier & Bassler 2003; Vendeville et al. 2005; Hardie & Heurlier 2008).
The LuxS/AI-2 system has been examined in detail in V. harveyi and V. cholerae (§2.3) where AI-2, in this case a furanosyl-borate diester, activates a sophisticated receptor-mediated sensor kinase/phosphorelay system (Lenz et al. 2004; Neiditch et al. 2005). Other autoinducers and their associated sensor kinases also feed into this pathway (§2.3; Miller et al. 2002; Henke & Bassler 2004a). In vibrios, the LuxS/AI-2 system functions as a QS system (Miller et al. 2002; Xavier & Bassler 2003; Henke & Bassler 2004a,b; Lenz et al. 2004). However, despite the ubiquity of LuxS, this is not the case for all bacteria. There are currently a limited number of reports (Vendeville et al. 2005; Hardie & Heurlier 2008) linking the expression of a specific gene, operon or phenotype to AI-2 apart from those described for vibrios, although Salmonella typhimurium and E. coli use AI-2 to regulate the uptake and phosphorylation of AI-2 (Taga et al. 2001, 2003; Xavier & Bassler 2005).
In S. typhimurium, the lsr operon encodes a periplasmic binding protein LsrB, which binds a chemically distinct form of AI-2 (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran and transports it into the cytoplasm whereupon it is phosphorylated by LsrK (Taga et al. 2003). It has been suggested that phosphorylated AI-2 relieves repression of the lsr operon by binding to and inhibiting the activity of the repressor LsrR, causing the subsequent upregulation of the lsr operon to increase the uptake of AI-2 from the environment (Taga et al. 2003). While provision of exogenous DPD (as a source of AI-2) restores the regulation of the lsr operon in a S. typhimurium luxS mutant, it was unable to restore a defect in biofilm formation in the same strain (De Keersmaecker et al. 2005). Similarly, mutation of luxS in S. typhimurium polarizes flagellar phase variation towards the more immunogenic phase 1 flagellin. While this phenotype could be complemented by the luxS gene supplied in trans it was not restored by provision of AI-2 and is, therefore, in common with the biofilm phenotype independent of QS (Karavolos et al. 2008). In contrast, Walters et al. (2006) reported that in enterohaemorrhagic E. coli (EHEC), the provision of exogenous AI-2 to a luxS mutant resulted in a gain of 62 phenotypes when compared with the luxS mutant using phenotype microarray analysis. The majority of these phenotypes were involved in carbon, nitrogen or phosphate metabolism. In other bacteria, LuxS and/or AI-2 have been implicated in biofilm development, flagellar-mediated motility, toxin production and virulence either as a consequence of the attenuation of QS or to a metabolic defect (for reviews, see Vendeville et al. 2005; Hardie & Heurlier 2008). Recently, a comparative study of genomic databases to try to establish an association between LuxS and the presence of AI-2 receptors such as LuxPQ and Lsr concluded that in most bacteria LuxS is primarily limited to a role in metabolism (Rezzonico & Duffy 2008), although the existence of as yet unidentified alternative AI-2 receptors could not be ruled out.
3. Quorum-sensing signal molecule-mediated polymicrobial interactions
In most environments, bacteria are found in mixed communities, and therefore populations of a given QS species that can sense a signal broadcast information both to themselves and to other species sharing the same ecological niche. Different bacterial species may ‘speak’ the same QS ‘language’, some may possess sensors for specific QS signals that facilitate eavesdropping while others may manipulate the QS activities of neighbouring bacteria by degrading the QS signal molecules within their locality. Although the impact of QS on microbial community parameters, such as species diversity and function, has not been extensively investigated, Valle et al. (2004) reported that the exogenous provision of AHLs to an industrial wastewater treatment bioreactor containing AHL-producing bacteria generated changes in both community function (phenol degradation) and composition. Furthermore, following AHL addition, one dominant functional member of the community was transiently supplanted by another, a finding which has important implications for ecosystem functionality.
3.1. Quorum-sensing-mediated interspecies cooperative interactions
Bacteria often exist in association with plants as members of polymicrobial biofilm communities, either in the rhizosphere or in planta. It has been reported that more AHL-producing bacteria live within the rhizosphere in association with plants than within the bulk soil population (Danhorn & Fuqua 2007). In mixed populations, the formation, development, maturation and dispersal of biofilms are likely to be tightly regulated through cooperative inter- and intraspecies signalling. Two different bacterial species that produce the same QS signal and coexist in the same environment will detect and respond to that signal. For example, Chromobacterium violaceum produces the purple pigment violacein in response to C6-HSL irrespective of whether the AHL is self-generated or supplied by another C6-HSL producer (McClean et al. 1997). While numerous studies have investigated the role of AHL-mediated signalling in single species biofilms (for review, see Atkinson et al. 2007), there have been relatively few studies of signalling in multi-species biofilms, which in turn have revealed cooperative behaviour. AHL-mediated communication between P. aeruginosa and B. cenocepacia in mixed biofilms appears to be unidirectional in that the latter responds to the P. aeruginosa AHLs but not vice versa (Riedel et al. 2001). In mixed Agrobacterium tumefaciens/P. aeruginosa coculture experiments, the A. tumefaciens biomass decreased in coculture with the wild-type but remained constant with a P. aeruginosa QS mutant (An et al. 2006), such findings are, however, more indicative of competitive rather than cooperative interactions.
Plant beneficial bacteria such as P. fluorescens produce a number of antibiotics and extracellular enzymes that suppress root diseases caused by fungal root pathogens (Dubuis & Haas 2007). These are regulated in a cell-density-dependent manner via the GacS/GacA two-component system that controls the expression of small regulatory RNAs, such as rsmZ and rsmY, which sequester RsmA/CsrA translational repressor family proteins and thereby relieve post-transcriptional repression of the exoproduct genes. Gac/Rsm signal transduction cascades occur in many different Gram-negative bacteria, and Dubuis & Haas (2007) reported that crude culture supernatant extracts taken from other plant beneficial and pathogenic pseudomonads could induce rsmY and rsmZ expression in the reciprocal host. Although the chemical identity of this putative cross-species signal molecule(s) has not yet been identified, the work suggests that it is possible for each species in a polymicrobial community to employ QS to monitor the local environment and to bring about a concerted change in behaviour that is beneficial to the community as a whole.
Sibley et al. (2008) developed a Drosophila melanogaster infection model to study polymicrobial interactions whereby a number of oropharyngeal isolates were coinoculated with P. aeruginosa. A number of the isolates which alone were avirulent or beneficial to the D. melanogaster synergistically exacerbated the pathogenicity of P. aeruginosa in a polymicrobial community. For example, when co-infected with P. aeruginosa, the oropharyngeal isolate C90 (a Streptococcus spp.), which alone appears to enhance D. melanogaster survival, significantly enhances fly killing. A number of P. aeruginosa QS-dependent genes, including the AHL synthase LasI, the AHL receptor RhlR, and the metalloprotease LasB, were apparently upregulated, and the authors suggested that AI-2, which is produced by C90 but not by P. aeruginosa, may be responsible for the change in gene expression (Sibley et al. 2008). As AI-2 is produced by many different bacteria, it has been suggested that AI-2 is a ‘universal language’ used by bacteria for interspecies communication (Bassler et al. 1997; Bassler 1999). However, not all AI-2 producers employ this metabolite as a QS signal molecule (§2.5) and not all bacteria synthesize AI-2 (Winzer et al. 2002b). As yet, there is little published data on AI-2-dependent interspecies signalling, which has been proposed to mediate interspecies communication between oral bacteria as well as biofilm community development within the human oral cavity (Kolenbrander et al. 2006).
3.2. Quorum-sensing-mediated signal interception and coercion
Most environments suitable for microbial growth will be populated by polymicrobial communities. While some of the interactions between species may be complementary, there is also the possibility for conflict through coercion, interception and chemical modification (reviewed by Keller & Surette 2006). LuxR homologues are most specific for their cognate AHL, but many can also be activated by other related compounds. A practical demonstration of this phenomenon is through the use of a range of bioluminescent biosensors based on the V. fischeri (luxI/R), P. aeruginosa (lasI/R) and Aeromonas hydrophila (ahyI/R) luxR/I homologues (pSB401, pSB1075 and pSB536, respectively) (Swift et al. 1997; Winson et al. 1998; Steindler & Venturi 2007). These plasmids contain the promoter region of the appropriate luxI homologue along with the response regulator fused to luxCDABE. While each reporter responds optimally to its cognate AHL to produce light, each also reports the presence of other signals that are structurally related to the cognate AHL (normally differing in the number of carbons in the fatty acyl chain) (Bainton et al. 1992a; Winson et al. 1998; Kirke et al. 2004).
Although these biosensors are engineered for in vitro use, they demonstrate an important point that the signal generated by one species can be detected by another species that does not synthesize the same signal molecule. This confers on one species the ability to intercept (Ahmer 2004) the signal from another, effectively using it as cue (Diggle et al. 2007a) to alter its behaviour as a direct response to the presence of the signal generator species. In vivo LuxR homologues may not be limited to recognizing AHLs. Riboflavin and its derivative lumichrome, produced from the single celled alga Chlamydomonas, is capable of stimulating P. aeruginosa LasR at the expense of the cognate AHL: N-(3-oxo-dodecanoylhomoserine) lactone (3-oxo-C12-HSL). Although there are no structural similarities between AHLs and riboflavin (and its derivative), amino acid substitutions in the AHL-binding domain of LasR altered responses to each class of molecule, which suggests that the AHL-binding domain of LasR is unable to discriminate between the two compounds. It is unclear from these initial observations whether this represents an example of cross-kingdom cooperation or coercion driven by the eukaryote (Rajamani et al. 2008).
While it is likely that cooperative interspecies interactions are beneficial in biofilms (as discussed above), there is also evidence that there may be non-cooperative interactions between species in these environments. Biofilms will develop when P. aeruginosa and Burkholderia cepacia are cocultured in flow chambers or on alginate beads in a mouse chronic lung infection model. Burkholderia cepacia is able to intercept and transduce the AHLs synthesized by P. aeruginosa but not vice versa. This was confirmed by analysing exoprotease production, which was restored to a B. cepacia cepI mutant when grown in the presence of some synthetic AHLs or supernatants from the AHL-producing strains P. aeruginosa PA01 or SH1. AHL-negative P. aeruginosa strains did not restore exoprotease activity. AHLs or supernatant extracts from the AHL-producing strain B. cepacia H111 did not restore exoprotease activity to a P. aeruginosa lasI/rhlI double mutant. However, the addition of spent PA01 culture supernatant or synthetic 3-oxo-C12-AHL did restore activity (Riedel et al. 2001). Pseudomonas aeruginosa may therefore have a mechanism for blocking the B. cepacia signals (possibly by detecting and reacting to an alternative B. cepacia-specific signal) to prevent B. cepacia-mediated coercion or alternatively is prevented from monitoring the activities of B. cepacia.
Escherichia coli and Salmonella spp. do not synthesize AHLs, but they are able to intercept and transduce AHL signals generated by other species because they possess the LuxR homologue SdiA (Ahmer 2004). Salmonella enterica SdiA responds to the presence of exogenous AHLs or bacterial species that produce AHLs. SdiA regulates the expression of srgE and the rck operon, so named because of its involvement in resistance to complement killing (Michael et al. 2001; Smith & Ahmer 2003; Ahmer 2004). In E. coli, several genes, including those involved in amino acid transport and metabolism and also in transcriptional regulation, have been shown to respond to C6-HSL in an SdiA-dependent manner (Van Houdt et al. 2006). This process of bacterial ‘eavesdropping’ confers on the organism the ability to intercept signals, which will provide important information about the local environment and the other species contained within. Sensing, but not synthesizing, a signal confers several advantages. For example, the organism may reduce the metabolic cost of QS signal biosynthesis and avoid conveying information about its whereabouts to other species in the immediate vicinity.
Some QS molecules exhibit biological functionalities beyond their role as signals. Pseudomonas aeruginosa employs 3-oxo-C12-HSL and 2-heptyl-3-hydroxy-4-quinolone (the pseudomonas quinolone signal (PQS)) as QS signals in a sophisticated gene regulatory network (Williams & Cámara 2009). However, apart from interacting with their cognate response regulator receptors (LasR and PqsR, respectively), 3-oxo-C12-HSL exhibits both antibacterial and quorum-quenching activity towards Gram-positive bacteria such as S. aureus (Qazi et al. 2006) while PQS functions as an iron chelator, a pro-oxidant and an inducer of an antioxidative stress response that shapes the population structure of P. aeruginosa (Diggle et al. 2007b; Haussler & Becker 2008). 3-oxo-C12-HSL and PQS also function as potent immune modulators (Hooi et al. 2004). Consequently, there is the possibility of crosstalk between the pathogen and host, which may, in return, elicit a counter-response from the host as it senses bacterial QS signal molecules. By intercepting but not synthesizing AHL signals, bacteria such as E. coli and Salmonella spp. are potentially capable of mounting a sustained stealth attack on the host while continually monitoring the local environment for potentially antagonistic AHL-producing bacteria.
In the staphylococci, the agr QS system has diverged such that an AIP produced by one strain may autoactivate but cross-inhibit other strains. For example, the four S. aureus agr groups can be distinguished on the basis of their ability to cross-activate or -inhibit agr expression, i.e. AIP-1 is an activator of AIP-1 producing group I strains but an inhibitor of agr groups II, III and IV. S. epidermidis group I strains cross-inhibit all four S. aureus agr groups but S. aureus AIPs have little inhibitory activity against non-group I S. epidermidis. In S. aureus, cross inhibition results in the downregulation of agr-dependent exotoxin synthesis. For example, exposure of an agr group I strain to AIP-2 or AIP-3 strains abolished production of TSST-1 and enterotoxin and attenuated infection in a mouse subcutaneous abscess model (Mayville et al. 1999; McDowell et al. 2001). Both intragroup activation and intergroup inhibition are mediated exclusively by the same group-specific AgrC receptor (Novick & Geisinger 2008). In addition, there is also some specificity in the processing of AgrD by AgrB; AgrB1 will process AgrD1, D3 and D4 but not AgrD2 or vice versa (Novick & Geisinger 2008). This raises some interesting questions with respect to the driving force behind the evolution of the agr system, its consequences for staphylococcal species divergence and the contribution of agr interference to competitive adaptation within a specific environmental niche. The natural causes of agr divergence within a species are, however, at present not known, although, in an insect infection model, competitive interference between different agr groups has been demonstrated (Fleming et al. 2006).
There is some evidence to suggest that bacteria, such as P. aeruginosa, that do not synthesize AI-2 can nevertheless sense AI-2, which may signify that AI-2 is prompting non-cooperative behaviour. Duan et al. (2003) examined the differential expression of several pathogenicity-associated genes from P. aeruginosa following exposure to exogenous AI-2 or to AI-2 produced by an uncharacterized avirulent, oropharyngeal bacterium recovered from the sputum of cystic fibrosis patients. The expression of several pseudomonas genes was altered in these assays, suggesting that P. aeruginosa is capable of intercepting AI-2 so using this information as a means of monitoring its environment and the composition of the local bacterial community. There is also the possibility that AI-2-producing oropharyngeal bacteria can coerce P. aeruginosa into altering gene expression to bring about a change in the environment that favours the former over the latter. Further support for the notion that P. aeruginosa will respond to AI-2 has recently been reported by Ganin et al. (2008), who have shown that two AI-2 synthetic analogues are capable of inhibiting QS as determined by the reduction in pyocyanin.
4. Quorum sensing in eukaryotes
Although QS has been the topic of intense study in prokaryotes, there is also evidence showing that QS systems are employed by eukaryotes. The dimorphic fungus C. albicans is a common human pathogen associated with high rates of mortality in cases of systemic candidiasis especially in immune-compromised individuals (Fridkin & Jarvis 1996; Viudes et al. 2002). Many fungi including C. albicans exhibit yeast–mycelial dimorphism and alternate between budding yeast or wheat-germ tubules and mycelia depending on environmental factors including temperature, pH, glucose levels, nitrogen source, carbon dioxide levels and cell population density (Nickerson et al. 2006; Romano 2008). In C. albicans a starting inoculum of less than 106 cells ml−1 favours mycelial growth, whereas budding yeast will be produced at a starting inoculum that is equal to or higher than 106 cells ml−1 (Hornby et al. 2001). This morphological shift, driven in part by cell population density, shows striking similarities to bacterial QS and prompted Hornby et al. (2001) to determine whether the process was signal molecule-mediated. Organic extracts of C. albicans-spent culture supernatants revealed that the active signal molecule, capable of repressing the yeast to mycelium conversion (without altering the growth rate), was a 15-carbon sesquiterpine known as farnesol (3,7,11-trimethyl-2,6,10-dodecatrien-1-ol). This molecule, which is secreted extracellularly, is produced throughout growth and is often considered to be the first QS signal molecule to be characterized in a eukaryotic microbe. Candida albicans also produces a second autoregulatory substance that represses the yeast to mycelium conversion (Hazen & Cutler 1983). This compound is chemically distinct from farnesol but its structure remains to be elucidated. The term ‘inoculum size effect’ has been use to describe the link between cell morphology and cell population density and has been observed in several different fungal species and apparently controls a number of different physiological processes (for review, see Nickerson et al. 2006). It is therefore likely that many other as yet undiscovered QS systems operate within the fungal world.
A fascinating extension to the concept of QS in eukaryotes was proposed by Hickson et al. (2008), who considered how single celled organisms behave in a community as compared with the behaviour of societies of cells integral within a multicellular organism. By way of example, tumorigenesis, resulting from the migration of metastatic ovarian cancer cells, was considered in relation to bacterial cell populations establishing a multicellular biofilm community during infection. Twenty years ago the idea that individual bacterial cells behaved cooperatively was greeted with scepticism in many quarters; today, cancer cells are still considered as autonomous, independent and non-cooperative entities. The challenge will now be to reconsider whether tumours are strictly a product of a few ‘unruly’ individuals or whether there is a population response to tumorigenesis, with division of labour and communication occurring within the developing tumour.
5. Inter-kingdom communication
Bacteria are the most abundant and adaptive form of life on Earth and, as such, have an impact on the habitats and lifestyles of all other living organisms. We would therefore be naive to think that, over the millennia, coevolution of pro- and eukaryotes has not stimulated the development of inter-kingdom signalling pathways promoting parasitic, symbiotic or commensal relationships; the members of each biological kingdom possess hormone-like molecules responsible for cell–cell communication, a fact that is fundamental to the concept that the members of any given kingdom will respond to the signals produced by another (Fox 2004; Rumbaugh 2007). Bacterially derived signal molecules are not limited to signalling between bacterial species but also modify the behaviour of eukaryotic organisms. This is not a one-way interaction, eukaryotes also manipulate bacterial signalling networks and therefore bacterial behaviour through molecular mimicry using cytokines, hormones and neurotransmitters. Bacterial signalling networks may also be disrupted following enzymatic degradation of the signal molecules or by the action of agonist/antagonist molecules that block QS (Bauer & Mathesius 2004; Pritchard 2006; Rasmussen & Givskov 2006; Kendall & Sperandio 2007; Williams 2007a). Earlier sections have outlined how signal interception and coercion play a key role in manipulating inter- and intraspecies communication among bacteria as does two-way signalling within and between species. Here we discuss inter-kingdom cooperative and non-cooperative behaviour.
5.1. Signal-mediated cooperative interactions between kingdoms
Symbioses across the pro- and eukaryotic divide are possible through the use of cooperative signalling systems, which undoubtedly modulate the behaviour and proliferation of the organisms involved in the relationship. There are more commensal bacterial cells in mammals than there are host cells and in humans, for example, the symbiosis that exists with the intestinal microbial flora is essential for nutrient assimilation and the development of the innate immune system (Hooper & Gordon 2001; Hughes & Sperandio 2008). Although, to date, there is little direct evidence of cooperative interactions between kingdoms, Fujiya et al. (2007) reported that, when the human colonic epithelial cell line Caco2 was exposed to a variety of cell-free supernatants derived from probiotic bacteria (bacteria used in food supplements which are thought to be beneficial in the treatment of a number of diseases; Hong et al. 2005) such as Bacillus subtilis, the cytoprotective heat shock protein Hsp27 was induced. When over-expressed, Hsps are capable of adhering to the intestinal epithelium and protecting it from oxidative injury (Tao et al. 2006), a contributing factor to the maintenance of a healthy gut. Upregulation of Hsp27 was a direct result of the presence of physiologically relevant concentrations of the QS signal peptide, CFS (also known as PhrC), the competence- and sporulation-stimulating factor. In B. subtilis CFS, ComX and other Phr homologues regulate multiple processes including the initiation of genetic competence, sporulation, antibiotic and exopolysaccharide synthesis and the production of degradative enzymes (Lazazzera 2001; Auchtung et al. 2006). These data suggest that a QS signal molecule-mediated symbiosis between B. subtilis and its host may exist whereby bacterially driven Hsp induction can confer protection to the host gut and in return provide the bacteria with a stable and nutrient-rich niche in which to reside. Mazmanian et al. (2008) similarly showed that the commensal human symbiont Bacteroides fragilis produces a molecule known as polysaccharide A, which apparently protects animals from Helicobacter hepaticus-induced colitis by suppressing pro-inflammatory interleukin-17 production from intestinal immune cells.
Recently, AHLs have been shown to mimic a eukaryotic signal causing profound effects on the growth of a cancer cell line. Thymidylate synthase (TS) plays a key role in DNA biosynthesis because it generates thymidine monophosphate which upon phosphorylation is converted into thymidine triphosphate. The TS inhibitor 5-fluorouracil (5-FU) has for decades been used to treat solid tumours (Curreri et al. 1958), but resistance to 5-FU is frequently associated with the over-expression of TS. The expression and/or function of TS is affected by rTS, which codes for an antisense TS RNA and two protein subunits rTSα and rTSβ (Dolnick 1993; Dolnick & Black 1996). Elevated levels of rTSβ are concomitant with an increase in the production of a novel signal molecule and a decrease in TS protein expression levels. The structure of this signal molecule is currently unknown but the rTSβ products are known to be metabolites of methionine biosynthesis formed directly from S-adenosylmethionine (and an uncharacterized substrate), which might indicate that this signal shares structural similarity with AHLs (Dolnick et al. 2003). The possible structural relationship between AHLs and the rTS signal prompted Dolnick et al. (2005) to test a range of AHLs for the ability to reduce TS protein levels. Western blot analysis showed that TS expression was reduced considerably in the presence of physiologically relevant concentrations of 3-oxo-C12-HSL, which also reduced the growth of human colorectal cancer cells (H630) by 50 per cent. The fact that N-dodecanoylhomoserine lactone showed no effect led the authors to speculate that the response was specific and receptor-mediated. Furthermore, 3-oxo-C12-HSL significantly enhanced the efficacy of 5-FU and Taxol (Dolnick et al. 2005), highlighting the therapeutic potential of this bacterial QS signal molecule. Indeed, comparative structure activity relationship studies have shown that cancer growth inhibitory activity in common with QS signalling in P. aeruginosa requires a long acyl chain with 3-oxo substitution for maximum activity (Oliver et al. 2009). Whether coevolution of these two systems has created a genuine biological relationship between rTS-signal-mediated TS expression and AHL action to trigger the death of malignant cells remains to be established. However, given that P. aeruginosa is an opportunistic human pathogen, it seems unlikely that such a cooperative relationship exists between the host and bacterium. Indeed, immune-compromised cancer patients often develop P. aeruginosa lung infections, and it is therefore feasible that the bacteria use AHLs to modulate the host response through the rTS-signal pathway. However, other commensal AHL producers might be involved in two-way signalling to communicate with the host via this pathway. Given the available information, it is not yet possible to determine whether the AHL–rTS signal relationship is combative or cooperative.
5.2. Interception and coercion
Both 3-oxo-C12-HSL and PQS modulate the host response in mammals such that these QS signals may facilitate P. aeruginosa infections both by regulating virulence factor expression and by modifying host inflammatory and immune responses (Hooi et al. 2004; Cooley et al. 2008). In cell culture-based assays, 3-oxo-C12-HSL, depending on the concentration used, is immunosuppressive at or below 10 µM but pro-inflammatory and pro-apoptotic at 20 µM and above. Investigation of the 3-oxo-C12-HSL structural requirements have revealed that, in common with QS activity, immune modulatory activity required an intact homoserine lactone ring, the L-configuration and an acyl chain of 11–13 carbons (Chhabra et al. 2003). Interestingly, 3-oxo-C12-HSL, but not the 3-oxo-C6 or 3-oxo-C13 analogues, prevented the invasion and destruction of pancreatic islet tissue by mononuclear cells normally associated with insulitis and cumulative diabetes in genetically predisposed non-obese diabetic (NOD) mice (Pritchard et al. 2005), suggesting that QS molecules such as 3-oxo-C12-HSL may represent a novel source of immune modulatory compounds for the treatment of the autoimmune disease type 1 diabetes, which afflicts more than two million individuals in Europe and North America. The signalling pathways by which 3-oxo-C12-HSL exerts its immune modulatory activity have been the subject of numerous studies (for reviews, see Cooley et al. 2008; Jahoor et al. 2008) and have shown that a potential target for the intracellular binding of 3-oxo-C12-HSL in mammalian cells may be the peroxisome proliferator-activated receptors (PPARs) that belong to the nuclear hormone receptor family (Jahoor et al. 2008). However, it is also possible that PPARs may be activated upon 3-oxo-C12-HSL binding to an as yet unknown protein target(s). Using an affinity chromatography strategy employing immobilized 3-oxo-C12-HSL, Seabra et al. (2008) isolated two 3-oxo-C12-HSL-binding proteins from mouse splenocyte and human granulocyte extracts, which were identified as S100A8 and S100A9 (calprotectin). Although these proteins clearly have the capacity to bind 3-oxo-C12-HSL, in murine proliferation assays using splenocytes from wild-type and S100A9 knockout mice, no differences in immune suppression were observed, suggesting that these proteins are unlikely to be the primary targets for the immune modulatory activity of 3-oxo-C12-HSL in lymphocytes.
Upon invasion by microbial pathogens, the innate immune response is triggered by activated NF-κB, and, recently, Kravchenko et al. (2008) reported that 3-oxo-C12-HSL may reduce the intensity of NF-κB signalling, which would reduce the expression of inflammatory cytokines and other related immune regulators in lipopolysaccharide-induced macrophages. These observations suggest a degree of coercion levied on the host by the signal molecules produced by the invading pathogens.
While there is clearly a considerable body of literature that shows that certain QS signals modify the host immune response, there are other examples where bacterial QS signals are intercepted by other organisms leading to behavioural modification in the latter. Conversely, QS signal molecules may coerce a eukaryote into modifying its behaviour in favour of the bacteria (for reviews, see Shiner et al. 2005; Rumbaugh 2007; Hughes & Sperandio 2008). A fascinating example of inter-kingdom signal interception can be seen in the relationship between Ulva intestinalis and biofilm-dwelling bacteria. Ulva intestinalis is a green microalga which occupies the inter-tidal zone and can reproduce asexually via the release of vast numbers of motile, asexual zoospores which seek and attach to a suitable surface. The zoospores discriminate between suitable and unsuitable surfaces by making a temporary attachment, which they leave rapidly if it proves to be an unsatisfactory environment for differentiation (Joint et al. 2007). Joint et al. (2000) noted that there was a positive correlation between zoospore settlement and attachment and the density of bacterial biofilms. Whether this was a property of the surface, the biofilm matrix or diffusible QS signals produced by the biofilm was unknown (Joint et al. 2002). Studies using parent and AHL-negative strains of Vibrio generated either by mutation of the AHL synthases or by introducing an AHL-degrading enzyme revealed that zoospores are preferentially attracted to AHL-producing bacterial biofilms (Joint et al. 2002; Tait et al. 2005). Zoospore swimming speeds decreased more rapidly over wild-type Vibrio biofilms when compared with those of an isogenic AHL-negative mutant, indicating that rather than a chemotactic response the zoospores appear to respond chemokinetically. The biological significance of AHL-mediated zoospore attachment to biofilms is unclear, although for many marine invertebrates a prerequisite to settlement on a surface is the presence of a microbial biofilm (Joint et al. 2007).
The nematode Caenorhabditis elegans is also capable of adapting its behaviour in the presence of AHLs (Beale et al. 2006). Some species of P. aeruginosa are natural pathogens of C. elegans, whereas others can serve as a food source for the nematodes. When exposed to P. aeruginosa or an isogenic AHL-negative mutant, C. elegans preferentially migrated to the AHL-producing strain on standard chemotaxis plate assays. When the assays were repeated using synthetic AHLs in the absence of bacteria, the worms were significantly more likely to migrate towards the AHL than the controls. Pseudomonas aeruginosa induces cyanide-mediated nematode paralysis after approximately 3.0 h exposure to the bacteria. To determine whether the nematodes were capable of sensing and ‘learning’ to avoid this pathogen, they were exposed to P. aeruginosa for 1.0 h, removed prior to paralysis and placed on AHL-containing chemotaxis plates. When compared with naive worms that had not been pre-exposed to P. aeruginosa, the pre-exposed worms were repelled rather than attracted to the AHLs. Conversely, worms that had been pre-exposed to a non-pathogenic food source in the presence of an AHL were more likely to be attracted to synthetic AHLs (Beale et al. 2006). In this context, C. elegans appears to be capable of intercepting AHL signals and altering its behaviour to respond appropriately to the presence of what it may perceive as a pathogen or a food source. This example of cross-kingdom signal interception illustrates that this type of attractive and aversive behaviour represents an essential strategy for nematode survival given that at any time in its natural environment C. elegans will be exposed to a variety of potential bacterial food sources, some of which may be harmful.
AI-3, a QS signal of an as yet unknown structure, is produced by bacteria such as EHEC, which causes severe gastric infections and haemolytic–uraemic syndrome. AI-3 is involved in the regulation of virulence motility and a type III secretion system encoded by the LEE locus (Locus of Enterocyte Effacement). The mammalian neuroendocrine stress hormones adrenaline and noradrenaline can substitute for AI-3 in activating EHEC virulence gene expression and the action of all three signals can be blocked by mammalian α- and β-adrenergic receptor antagonists (Sperandio et al. 2003). AI-3, adrenaline and noradrenaline can all bind to the histidine sensor kinase, QseC. Upon sensing any of these signalling molecules, QseC autophosphorylates and subsequently transfers the phosphate to the response regulator, QseB, which activates a sophisticated regulatory cascade, resulting in the transcription of key virulence genes (Clarke & Sperandio 2005; Hughes & Sperandio 2008). As it is likely that AI-3, adrenaline and noradrenaline interact with the same bacterial receptor, AI-3 is likely to share some common structural features with the two catecholamines. With this in mind, it is possible that during EHEC infection the host and pathogen are involved in a signal-mediated battle involving interception and coercion. However, while bacteria clearly respond to the catecholamines, the effect of AI-3 on the host, if any, has not yet been reported.
The sesquiterpine farnesol is a QS signal molecule that represses C. albicans mycelial development in a density-dependent manner. It is possible that the molecule can act as an interspecies signal for morphogenesis because it will inhibit mycelial development in Candida dubliniensis (Henriques et al. 2006). There also appears to be an inter-kingdom signalling relationship between C. albicans and P. aeruginosa such that C. albicans-derived farnesol reduces PQS and pyocyanin production in P. aeruginosa. Farnesol appears to affect pqsA (the first gene in the PQS biosynthetic operon) transcription in the parent strain and in recombinant E. coli via a non-productive interaction with the LysR-type regulatory protein PqsR (Cugini et al. 2007). This interaction is not one-way because P. aeruginosa 3-oxo-C12-HSL can also inhibit C. albicans mycelial development (Hogan et al. 2004). This is a good example of how two organisms apparently coerce each other into phenotypic change using a QS signal molecule. By reducing the amount of P. aeruginosa pyocyanin, C. albicans can improve the growth conditions in its immediate environment while P. aeruginosa may prevent the fungus from differentiating into mycelia from the free-living yeast form, so preventing the fungus from establishing a foothold in the micro-environment in which the two organisms reside.
6. Conclusions and future perspectives
QS has been extensively used as a term to describe density-dependent bacterial cell–cell communication since the early 1990s. However, the concept of individual bacteria coordinating a population-wide response to a signal or diffusible factor was first recognized over 50 years ago, initially by Lev (1954), by Jennings (1961) and by McVittie et al. (1964) while investigating fruiting body formation in Myxococcus xanthus. Genetic competence in Streptococcus pneumoniae and streptomycin biosynthesis and aerial mycelium formation in Streptomyces griseus followed as examples of other species of bacteria exhibiting multicellular behaviour in a population of individuals (Tomasz 1965; Khoklov et al. 1967). However, it was perhaps the discovery that bioluminescence in V. fischeri (Nealson et al. 1970) was mediated by an AHL that was subsequently shown to be employed as a QS signal by numerous other bacterial species (Bainton et al. 1992a,b) that resulted in the widespread recognition that bacteria, in general, were capable of coordinating their behaviour collectively. QS is now a well-established concept in bacteriology, is increasingly being discovered in unicellular eukaryotic microbes and has even been used to describe the behaviour of ants (Pratt et al. 2002), metastatic cells and even iron-impregnated particles (Taylor et al. 2009). While many studies have focused on unravelling the QS network operating within a single bacterial species, there has been the general realization that this approach is limited because most environments harbour multiple bacterial species and higher organisms. When we consider QS signalling networks in the context of these multi-species/multi-kingdom communities, there are several ways in which each representative group can interact. Within and between species and kingdoms there may be signal molecule-mediated cooperation or coercion. Alternatively, individuals from one group may intercept signals generated by another. There is also the intriguing possibility that different species from one kingdom may coordinate their behaviour against the individual/s from another kingdom. Irrespective of how the signals are processed, the results will either be a collective change in behaviour for the good of the community as a whole or will favour one or more species over others. These relationships are complex and the finer details of signal-mediated cross-species/kingdom communication are still to be investigated in depth, and this is reflected in the limited number of reports that have addressed the issue to date. This may be due in part to technical difficulties because by their very nature these communities are complex and difficult to standardize in the laboratory, but there is also a need for a broader cross-disciplinary approach involving collaborations between pro- and eukaryotic biologists.
Although we are only just beginning to investigate the possible benefits of exploiting QS the mechanisms underpinning bacterially driven cell–cell and cell–host communication systems constitute potential targets for novel anti-bacterials. Given that QS is not essential for the metabolic processes required for bacterial growth also means that using QS as a target for novel anti-bacterials will not impose on the bacteria the severe selective pressures that would normally lead to the development of resistance similar to that observed with conventional bactericidal agents (Rasmussen & Givskov 2006). One area that may be exploited is that of enzyme-mediated signal molecule inactivation. With respect to AHL QS signals, many bacteria produce acylase or lactonase enzymes that either hydrolyse the homoserine lactone ring or cleave the amide bond (Zhang & Dong 2004; Dong et al. 2007). In mammalian cells, the paraoxanase (PON) enzymes, which are a common component of serum and airway epithelia, exhibit a variety of hydrolytic activities. Both PON1 and PON2 have lactonase activity, while PON1 also has arylesterase and organophosphatase activities (Dong et al. 2007). Alternatively, antibodies against QS signals have been developed and shown to be capable of attenuating staphylococcal infections in experimental animal models (Park et al. 2007). Analogues of QS molecules such as 3-oxo-C12-HSL clearly have potential as immune modulators for treating autoimmune and cardiovascular diseases (Gardiner et al. 2001; Chhabra et al. 2003; Pritchard et al. 2005). Several reports have shown that plant, animal and algal cells all produce compounds that are either antagonistic or agonistic towards Gram-negative and Gram-positive bacterial QS systems (for reviews, see Bauer & Mathesius 2004; Dudler & Eberl 2006; Rasmussen & Givskov 2006; Williams 2007b). Both natural products and the structure-based design offer multiple opportunities (Suga & Smith 2003; Raffa et al. 2005) for discovery of novel inhibitors of QS. To date, a number of compounds have been synthesized which inhibit QS systems in different bacteria and include AHL and AIP analogues as well as compounds that are structurally unrelated to QS signals (Scott et al. 2003; Suga & Smith 2003; Glansdorp et al. 2004; Persson et al. 2005; Kaufmann et al. 2006; Muh et al. 2006; Singh et al. 2006; Geske et al. 2007; Pomianek & Semmelhack 2007; Ni et al. 2009).
It is now clear that both prokaryotes and eukaryotes respond to their own and each other's signals, but, at present, we can only hypothesize how significant our understanding of these relationships and their potential for exploitation might be. In the coming years, it is likely that detailed knowledge of intra- and inter-kingdom signalling systems will emerge. This will undoubtedly inform our understanding of host–pathogen relationships, the mechanisms that underlie infectious and perhaps malignant diseases and aid the design and discovery of novel therapeutic strategies.
References
- Achtman M., Zurth K., Morelli C., Torrea G., Guiyoule A., Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA 96, 14 043–14 048. ( 10.1073/pnas.96.24.14043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmer B. M. M. 2004. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol 52, 933–945. ( 10.1111/j.1365-2958.2004.04054.x) [DOI] [PubMed] [Google Scholar]
- An D. D., Danhorn T., Fuqua C., Parsek M. R. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl Acad. Sci. USA 103, 3828–3833. ( 10.1073/pnas.0511323103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S., Throup J. P., Stewart G. S. A. B., Williams P. 1999. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol. Microbiol. 33, 1267–1277. ( 10.1046/j.1365-2958.1999.01578.x) [DOI] [PubMed] [Google Scholar]
- Atkinson S., Chang C. Y., Sockett R. E., Cámara M., Williams P. 2006a. Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility. J. Bacteriol. 188, 1451–1461. ( 10.1128/JB.188.4.1451-1461.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S., Sockett R. E., Cámara M., Williams P. 2006b. Quorum sensing and the lifestyle of Yersinia. Curr. Issues Mol. Biol. 8, 1–10. [PubMed] [Google Scholar]
- Atkinson S., Cámara M., Williams P. 2007. N-acylhomoserine lactones, quorum sensing and biofilm development in Gram-negative bacteria. In The biofilm mode of life. Mechanisms and adaptations (eds Kjellberg S., Givskov M.), pp. 95–122. Wymondham, UK: Horizon Bioscience. [Google Scholar]
- Atkinson S., Chang C. Y., Patrick H. L., Buckley C. M. F., Wang Y., Sockett R. E., Cámara M., Williams P. 2008. Functional interplay between the Yersinia pseudotuberculosis YpsRI and YtbRI quorum sensing systems modulates swimming motility by controlling expression of flhDC and fliA. Mol. Microbiol. 69, 137–151. ( 10.1111/j.1365-2958.2008.06268.x) [DOI] [PubMed] [Google Scholar]
- Auchtung J. M., Lee C. A., Grossman A. D. 2006. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J. Bacteriol. 188, 5273–5285. ( 10.1128/JB.00300-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton N. J., et al. 1992a. A general role for the lux autoinducer in bacterial-cell signaling-control of antibiotic biosynthesis in Erwinia. Gene 116, 87–91. ( 10.1016/0378-1119(92)90633-Z) [DOI] [PubMed] [Google Scholar]
- Bainton N. J., Stead P., Chhabra S. R., Bycroft B., Salmond G., Stewart G. S. A. B., Williams P. 1992b. N-(3-oxohexanoyl)-l-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. J. Biochem. 288, 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber C. E., Tang J. L., Feng J. X., Pan M. Q., Wilson T. J. G., Slater H., Dow J. M., Williams P., Daniels M. J. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24, 555–566. ( 10.1046/j.1365-2958.1997.3721736.x) [DOI] [PubMed] [Google Scholar]
- Barnard A. M. L., Bowden S. D., Burr T., Coulthurst S. J., Monson R. E., Salmond G. P. C. 2007. Quorum sensing, virulence and secondary metabolite production in plant soft-rotting bacteria. Phil. Trans. R. Soc. B 362, 1165–1183. ( 10.1098/rstb.2007.2042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2, 582–587. ( 10.1016/S1369-5274(99)00025-9) [DOI] [PubMed] [Google Scholar]
- Bassler B. L., Greenberg E. P., Stevens A. M. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer W. D., Mathesius U. 2004. Plant responses to bacterial quorum sensing signals. Curr. Opin. Plant Biol. 7, 429–433. ( 10.1016/j.pbi.2004.05.008) [DOI] [PubMed] [Google Scholar]
- Beale E., Li G., Tan M. W., Rumbaugh K. P. 2006. Caenorhabditis elegans senses bacterial autoinducers. Appl. Environ. Microbiol. 72, 5135–5137. ( 10.1128/AEM.00611-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercovier H., Mollaret H. H., Alonso J. M., Brault J., Fanning G. R., Steigerwalt A. G., Brenner D. J. 1980. Intra- and interspecies relatedness of Yersinia pestis by DNA hybridisation and its relationship to Yersinia pseudotuberculosis. Curr. Microbiol. 4, 225–229. ( 10.1007/BF02605861) [DOI] [Google Scholar]
- Boles B. R., Horswill A. R. 2008. agr-Mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052 ( 10.1371/journal.ppat.1000052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon C., Deng Y. Y., Wang L. H., He Y. W., Xu J. L., Fan Y., Pan S. Q., Zhang L. H. 2008. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2, 27–36. ( 10.1038/ismej.2007.76) [DOI] [PubMed] [Google Scholar]
- Bruhn J. B., Dalsgaard I., Nielsen K. F., Buchholtz C., Larsen J. L., Gram L. 2005. Quorum sensing signal molecules (acylated homoserine lactones) in Gram-negative fish pathogenic bacteria. Dis. Aquat. Organ. 65, 43–52. ( 10.3354/dao065043) [DOI] [PubMed] [Google Scholar]
- Butler T. 1994. Yersinia infections: centennial of the discovery of the plague bacillus. Clin. Infect. Dis. 19, 655–663. [DOI] [PubMed] [Google Scholar]
- Cámara M., Williams P., Hardman A. 2002. Controlling infection by tuning in and turning down the volume of bacterial small-talk. Lancet Infect. Dis. 2, 667–676. ( 10.1016/S1473-3099(02)00447-4) [DOI] [PubMed] [Google Scholar]
- Cha C., Gao P., Chen Y. C., Shaw P. D., Farrand S. K. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant. Microbe Interact. 11, 1119–1129. ( 10.1094/MPMI.1998.11.11.1119) [DOI] [PubMed] [Google Scholar]
- Chai Y., Zhu J., Winans S. C. 2001. TrIR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrIR:TraR dimers. Mol. Microbiol. 40, 414–421. ( 10.1046/j.1365-2958.2001.02385.x) [DOI] [PubMed] [Google Scholar]
- Chan W. C., Coyle B. J., Williams P. 2004. Virulence regulation and quorum sensing in Staphylococcal infections: competitive AgrC antagonists as quorum sensing inhibitors. J. Med. Chem. 47, 4633–4641. ( 10.1021/jm0400754) [DOI] [PubMed] [Google Scholar]
- Cheung A. L., Bayer A. S., Zhang G. Y., Gresham H., Xiong Y. Q. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40, 1–9. ( 10.1016/S0928-8244(03)00309-2) [DOI] [PubMed] [Google Scholar]
- Chhabra S. R., Harty C., Hooi D. S. W., Daykin M., Williams P., Telford G., Pritchard D. I., Bycroft B. W. 2003. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-l-homoserine lactone as immune modulators. J. Med. Chem. 46, 97–104. ( 10.1021/jm020909n) [DOI] [PubMed] [Google Scholar]
- Clarke M. B., Sperandio V. 2005. Transcriptional regulation of flhDC by QseBC and sigma(28) (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57, 1734–1749. ( 10.1111/j.1365-2958.2005.04792.x) [DOI] [PubMed] [Google Scholar]
- Colnaghi Simionato A. V., da Silva D. S., Lambais M. R., Carrilho E. 2007. Characterization of a putative Xylella fastidiosa diffusible signial factor by HRGC-EI-MS. J. Mass Spectrom. 42, 490–496. ( 10.1002/jms.1181) [DOI] [PubMed] [Google Scholar]
- Cooley M., Chhabra S. R., Williams P. 2008. N-acylhomoserine lactone-mediated quorum sensing: A twist in the tail and a blow for host immunity. Chem. Biol. 15, 1141–1147. ( 10.1016/j.chembiol.2008.10.010) [DOI] [PubMed] [Google Scholar]
- Cover T. L., Aber R. C. 1989. Yersinia enterocolitica. N. Engl. J. Med. 321, 16–24. [DOI] [PubMed] [Google Scholar]
- Cugini C., Calfee M. W., Farrow J. M., Morales D. K., Pesci E. C., Hogan D. A. 2007. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 65, 896–906. ( 10.1111/j.1365-2958.2007.05840.x) [DOI] [PubMed] [Google Scholar]
- Curreri A. R., Ansfield F. J., Mciver F. A., Waisman H. A., Heidelberger C. 1958. Clinical studies with 5-fluorouracil. Cancer Res. 18, 478–484. [PubMed] [Google Scholar]
- Danhorn T., Fuqua C. 2007. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61, 401–422. ( 10.1146/annurev.micro.61.080706.093316) [DOI] [PubMed] [Google Scholar]
- Davies D. G., Marques C. N. H. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403. ( 10.1128/JB.01214-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker S. C. J., Varszegi C., van Boxel N., Habel L. W., Metzger K., Daniels R., Marchal K., De Vos D., Vanderleyden J. 2005. Chemical synthesis of (S)-4,5-dihydroxy-2,3-pentanedione, a bacterial signal molecule precursor, and validation of its activity in Salmonella typhimurium. J. Biol. Chem. 280, 19 563–19 568. ( 10.1074/jbc.M412660200) [DOI] [PubMed] [Google Scholar]
- Diggle S. P., Gardner A., West S. A., Griffin A. S. 2007a. Evolutionary theory of bacterial quorum sensing: when is a signal not a signal? Phil. Trans. R. Soc. B 362, 1241–1249. ( 10.1098/rstb.2007.2049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle S. P., et al. 2007b. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 14, 87–94. ( 10.1016/j.chembiol.2006.11.014) [DOI] [PubMed] [Google Scholar]
- Dolnick B. J. 1993. Cloning and characterization of a naturally occurring antisense RNA to human thymidylate synthase messenger RNA. Nucleic Acids Res. 21, 1747–1752. ( 10.1093/nar/21.8.1747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnick B. J., Black A. R. 1996. Alternate splicing of the rTS gene product and its overexpression in a 5-fluorouracil-resistant cell line. Cancer Res. 56, 3207–3210. [PubMed] [Google Scholar]
- Dolnick B. J., Angelino N. J., Dolnick R., Sufrin J. R. 2003. A novel function for the rTS gene. Cancer Biol. Ther. 2, 364–369. [DOI] [PubMed] [Google Scholar]
- Dolnick R., Wu Q., Angelino N. J., Stephanie L. V., Chow K. C., Sufrin J. R., Dolnick B. J. 2005. Enhancement of 5-fluorouracil sensitivity by an rTS signaling mimic in H630 colon cancer cells. Cancer Res. 65, 5917–5924. ( 10.1158/0008-5472.CAN-05-0431) [DOI] [PubMed] [Google Scholar]
- Dong Y. H., Wang L. H., Zhang L. H. 2007. Quorum-quenching microbial infections: mechanisms and implications. Phil. Trans. R. Soc. B 362, 1201–1211. ( 10.1098/rstb.2007.2045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow J. M., Daniels M. J. 1994. Pathogenicity determinants and global regulation of pathogenicity of Xanthomonas campestris pv. campestris. In Molecular and cellular mechanisms in bacterial pathogenesis of plants and animals (ed. Dangl J. L.), pp. 29–41. Berlin, Germany: Springer-Verlag. [DOI] [PubMed] [Google Scholar]
- Dow J. M., Crossman L., Findlay K., He Y. Q., Feng J. X., Tang J. L. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell–cell signaling and is required for full virulence to plants. Proc. Natl Acad. Sci. USA 100, 10 995–11 000. ( 10.1073/pnas.1833360100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K. M., Dammel C., Stein J., Rabin H., Surette M. G. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50, 1477–1491. ( 10.1046/j.1365-2958.2003.03803.x) [DOI] [PubMed] [Google Scholar]
- Dubuis C., Haas D. 2007. Cross-species GacA-controlled induction of antibiosis in pseudomonads. Appl. Environ. Microbiol. 73, 650–654. ( 10.1128/AEM.01681-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudler R., Eberl L. 2006. Interactions between bacteria and eukaryotes via small molecules. Curr. Opin. Biotechnol. 17, 268–273. ( 10.1016/j.copbio.2006.04.004) [DOI] [PubMed] [Google Scholar]
- Duerkop B. A., Ulrich R. L., Greenberg E. P. 2007. Octanoyl-homoserine lactone is the cognate signal for Burkholderia mallei BmaR1-BmaI1 quorum sensing. J. Bacteriol. 189, 5034–5040. ( 10.1128/JB.00317-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferluga S., Bigirimana J., Hofte M., Venturi V. 2007. A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol. Plant Pathol. 8, 529–538. ( 10.1111/j.1364-3703.2007.00415.x) [DOI] [PubMed] [Google Scholar]
- Fleming V., Feil E., Sewell A. K., Day N., Buckling A., Massey R. C. 2006. Agr interference between clinical Staphylococcus aureus strains in an insect model of virulence. J. Bacteriol. 188, 7686–7688. ( 10.1128/JB.00700-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouhy Y., Scanlon K., Schouest K., Spillane C., Crossman L., Avison M. B., Ryan R. P., Dow J. M. 2007. Diffusible signal factor-dependent cell–cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J. Bacteriol. 189, 4964–4968. ( 10.1128/JB.00310-07) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fox J. E. 2004. Chemical communication threatened by endocrine-disrupting chemicals. Environ. Health Perspect. 112, 648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin S. K., Jarvis W. R. 1996. Epidemiology of nosocomial fungal infections. Clin. Microbiol. Rev. 9, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiya M., Musch M. W., Nakagawa Y., Hu S., Alverdy J., Kohgo Y., Schneewind O., Jabri B., Chang E. B. 2007. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 1, 299–308. ( 10.1016/j.chom.2007.05.004) [DOI] [PubMed] [Google Scholar]
- Fuqua C. 2006. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J. Bacteriol. 188, 3169–3171. ( 10.1128/JB.188.9.3169-3171.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C., Greenberg E. P. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3, 685–695. ( 10.1038/nrm907) [DOI] [PubMed] [Google Scholar]
- Fuqua C., Winans S. C., Greenberg E. P. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50, 727–751. ( 10.1146/annurev.micro.50.1.727) [DOI] [PubMed] [Google Scholar]
- Ganin H., Tang X., Meijler M. M. 2008. Inhibition of Pseudomonas aeruginosa quorum sensing by AI-2 analogs. Bioorg. Med. Chem. Lett. 19, 3941–3944. ( 10.1016/j.bmcl.2009.03.163) [DOI] [PubMed] [Google Scholar]
- Gardiner S. M., Chhabra S. R., Harty C., Williams P., Pritchard D. I., Bycroft B. W., Bennett T. 2001. Haemodynamic effects of the bacterial quorum sensing signal molecule, N-(3-oxododecanoyl)-l-homoserine lactone, in conscious, normal and endotoxaemic rats. Br. J. Pharmacol. 133, 1047–1054. ( 10.1038/sj.bjp.0704174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske G. D., O'Neill J. C., Blackwell H. E. 2007. N-phenylacetanoyl-l-homoserine lactones can strongly antagonize or superagonize quorum sensing in Vibrio fischeri. ACS Chem. Biol. 2, 315–319. ( 10.1021/cb700036x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glansdorp F. G., Thomas G. L., Lee J. J. K., Dutton J. M., Salmond G. P. C., Welch M., Spring D. R. 2004. Synthesis and stability of small molecule probes for Pseudomonas aeruginosa quorum sensing modulation. Org. Biomol. Chem. 2, 3329–3336. ( 10.1039/b412802h) [DOI] [PubMed] [Google Scholar]
- Hammer B. K., Bassler B. L. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50, 101–114. ( 10.1046/j.1365-2958.2003.03688.x) [DOI] [PubMed] [Google Scholar]
- Hanzelka B. L., Parsek M. R., Val D. L., Dunlap P. V., Cronan J. E., Greenberg E. P. 1999. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J. Bacteriol. 181, 5766–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie K. R., Heurlier K. 2008. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer-2 in biofilm development. Nat. Rev. Microbiol. 6, 635–643. ( 10.1038/nrmicro1916) [DOI] [PubMed] [Google Scholar]
- Haussler S., Becker T. 2008. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 4, e1000166 ( 10.1371/journal.ppat.1000166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen K. C., Cutler J. E. 1983. Isolation and purification of morphogenic autoregulatory substance produced by Candida albicans. J. Biochem. (Tokyo) 94, 777–783. [DOI] [PubMed] [Google Scholar]
- He Y. W., Zhang L. H. 2008. Quorum sensing and virulence regulation in Xanthomonas campestris. FEMS Microbiol. Rev. 32, 842–857. ( 10.1111/j.1574-6976.2008.00120.x) [DOI] [PubMed] [Google Scholar]
- He Y. W., et al. 2006. Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell–cell communication-dependent genes and functions. Mol. Microbiol. 59, 610–622. ( 10.1111/j.1365-2958.2005.04961.x) [DOI] [PubMed] [Google Scholar]
- Henke J. M., Bassler B. L. 2004a. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186, 3794–3805. ( 10.1128/JB.186.12.3794-3805.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke J. M., Bassler B. L. 2004b. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186, 6902–6914. ( 10.1128/JB.186.20.6902-6914.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques M., Azeredo J., Oliveira R. 2006. Candida albicans and Candida dubliniensis: comparison of biofilm formation in terms of biomass and activity. Br. J. Biomed. Sci. 63, 5–11. [DOI] [PubMed] [Google Scholar]
- Hense B. A., Kuttler C., Mueller J., Rothballer M., Hartmann A., Kreft J. U. 2007. Opinion—does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5, 230–239. ( 10.1038/nrmicro1600) [DOI] [PubMed] [Google Scholar]
- Hickson J., Yamada S. D., Berger J., Alverdy J., O'Keefe J., Bassler B. L., Rinker-Schaeffer C. 2008. Societal interactions in ovarian cancer metastasis: a quorum-sensing hypothesis. Clin. Exp. Metastasis 26, 67–76. ( 10.1007/s10585-008-9177-z) [DOI] [PubMed] [Google Scholar]
- Higgins D. A., Pomianek M. E., Kraml C. M., Taylor R. K., Semmelhack M. F., Bassler B. L. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450, 883–886. ( 10.1038/nature06284) [DOI] [PubMed] [Google Scholar]
- Hogan D. A., Vik A., Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54, 1212–1223. ( 10.1111/j.1365-2958.2004.04349.x) [DOI] [PubMed] [Google Scholar]
- Hong H. A., Duc L. H., Cutting S. M. 2005. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 29, 813–835. ( 10.1016/j.femsre.2004.12.001) [DOI] [PubMed] [Google Scholar]
- Hooi D. S. W., Bycroft B. W., Chhabra S. R., Williams P., Pritchard D. I. 2004. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect. Immun. 72, 6463–6470. ( 10.1128/IAI.72.11.6463-6470.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L. V., Gordon J. I. 2001. Commensal host-bacterial relationships in the gut. Science 292, 1115–1118. ( 10.1126/science.1058709) [DOI] [PubMed] [Google Scholar]
- Hornby J. M., Jensen E. C., Lisec A. D., Tasto J. J., Jahnke B., Shoemaker R., Dussault P., Nickerson K. W. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67, 2982–2992. ( 10.1128/AEM.67.7.2982-2992.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. T., Sperandio V. 2008. Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6, 111–120. ( 10.1038/nrmicro1836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isherwood K. E. 2001. Quorum sensing in Yersinia pestis. PhD thesis, The University of Nottingham, Nottingham, UK. [Google Scholar]
- Jacobi C. A., Bach A., Eberl L., Steidle A., Heesemann J. 2003. Detection of N-(3-oxohexanoyl)-l-homoserine lactone in mice infected with Yersinia enterocolitica serotype O8. Infect. Immun. 71, 6624–6626. ( 10.1128/IAI.71.11.6624-6626.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahoor A., et al. 2008. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J. Bacteriol. 190, 4408–4415. ( 10.1128/JB.01444-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett C. O., Deak E., Isherwood K. E., Oyston P. C., Fischer E. R., Whitney A. R., Kobayashi S. D., DeLeo F. R., Hinnebusch B. J. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190, 783–792. ( 10.1086/422695) [DOI] [PubMed] [Google Scholar]
- Jennings J. 1961. Association of a steroid and a pigment with a diffusible fruiting factor in Myxococcus virescens. Nature 190, 190 ( 10.1038/190190b0) [DOI] [PubMed] [Google Scholar]
- Jiang Y., Cámara M., Chhabra S. R., Hardie K. R., Bycroft B. W., Lazdunski A., Salmond G. P. C., Stewart G. S. A. B., Williams P. 1998. In vitro biosynthesis of the Pseudomonas aeruginosa quorum-sensing signal molecule N-butanoyl-l-homoserine lactone. Mol. Microbiol. 28, 193–203. ( 10.1046/j.1365-2958.1998.00789.x) [DOI] [PubMed] [Google Scholar]
- Johnson R. H. 1992. Yersinia infections. Curr. Opin. Infect. Dis. 5, 654–658. ( 10.1097/00001432-199210000-00005) [DOI] [Google Scholar]
- Joint I., Callow M. E., Callow J. A., Clarke K. R. 2000. The attachment of Enteromorpha zoospores to a bacterial biofilm assemblage. Biofouling 16, 151–158. ( 10.1080/08927010009378440) [DOI] [Google Scholar]
- Joint I., Tait K., Callow M. E., Callow J. A., Milton D., Williams P., Cámara M. 2002. Cell-to-cell communication across the prokaryote–eukaryote boundary. Science 298, 1207 ( 10.1126/science.1077075) [DOI] [PubMed] [Google Scholar]
- Joint I., Tait K., Wheeler G. 2007. Cross-kingdom signalling: exploitation of bacterial quorum sensing molecules by the green seaweed Ulva. Phil. Trans. R. Soc. B 362, 1223–1233. ( 10.1098/rstb.2007.2047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. B., Sperandio V. 2005. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect. Immun. 73, 3197–3209. ( 10.1128/IAI.73.6.3197-3209.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavolos M. H., Bulmer D. M., Winzer K., Wilson M., Mastroeni P., Williams P., Khan C. M. A. 2008. LuxS affects flagellar phase variation independently of quorum sensing in Salmonella enterica serovar typhimurium. J. Bacteriol. 190, 769–771. ( 10.1128/JB.01253-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann G. F., et al. 2006. Antibody interference with N-acyl homoserine lactone-mediated bacterial quorum sensing. J. Am. Chem. Soc. 128, 2802–2803. ( 10.1021/ja0578698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L., Surette M. G. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4, 249–258. ( 10.1038/nrmicro1383) [DOI] [PubMed] [Google Scholar]
- Kendall M. M., Sperandio V. 2007. Quorum sensing by enteric pathogens. Curr. Opin. Gastroenterol. 23, 10–15. ( 10.1097/MOG.0b013e3280118289) [DOI] [PubMed] [Google Scholar]
- Khoklov A. S., Tovarova I. I., Borisova L. N., Pliner S. A., Shevchenko L. A., Kornitskaya E. Y., Ivkina N. S., Rapoport I. A. 1967. A-factor assuring the biosynthesis of streptomycin by a mutant strain of Actinomyces steptomycini. Dokl. Akad. Nauk SSSR 1777, 232–235. [PubMed] [Google Scholar]
- Kirke D. F., Swift S., Lynch M. J., Williams P. 2004. The Aeromonas hydrophila LuxR homologue AhyR regulates the N-acyl homoserine lactone synthase, AhyI positively and negatively in a growth phase-dependent manner. FEMS Microbiol. Lett. 241, 109–117. ( 10.1016/j.femsle.2004.10.011) [DOI] [PubMed] [Google Scholar]
- Kirwan J. P., Gould T. A., Schweizer H. P., Bearden S. W., Murphy R. C., Churchill M. E. A. 2006. Quorum-sensing signal synthesis by the Yersinia pestis acyl-homoserine lactone synthase YspI. J. Bacteriol. 188, 784–788. ( 10.1128/JB.188.2.784-788.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P. E., Palmer R. J., Rickard A. H., Jakubovics N. S., Chalmers N. I., Diaz P. I. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42, 47–79. ( 10.1111/j.1600-0757.2006.00187.x) [DOI] [PubMed] [Google Scholar]
- Kravchenko V. V., et al. 2008. Modulation of gene expression via disruption of NF-kappa B signaling by a bacterial small molecule. Science 321, 259–263. ( 10.1126/science.1156499) [DOI] [PubMed] [Google Scholar]
- Laue R. E., Jiang Y., Chhabra S. R., Jacob S., Stewart G. S. A. B., Hardman A., Downie J. A., O'Gara F., Williams P. 2000. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology-UK 146, 2469–2480. [DOI] [PubMed] [Google Scholar]
- Lazazzera B. A. 2001. The intracellular function of extracellular signaling peptides. Peptides 22, 1519–1527. ( 10.1016/S0196-9781(01)00488-0) [DOI] [PubMed] [Google Scholar]
- Lazdunski A. M., Ventre I., Sturgis J. N. 2004. Regulatory circuits and communication in gram-negative bacteria. Nat. Rev. Microbiol. 2, 581–592. ( 10.1038/nrmicro924) [DOI] [PubMed] [Google Scholar]
- Lee J. H., Lequette Y., Greenberg E. P. 2006. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59, 602–609. ( 10.1111/j.1365-2958.2005.04960.x) [DOI] [PubMed] [Google Scholar]
- Lenz D. H., Mok K. C., Lilley B. N., Kulkarni R. V., Wingreen N. S., Bassler B. L. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118, 69–82. ( 10.1016/j.cell.2004.06.009) [DOI] [PubMed] [Google Scholar]
- Lequette Y., Lee J. H., Ledgham F., Lazdunski A., Greenberg E. P. 2006. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 188, 3365–3370. ( 10.1128/JB.188.9.3365-3370.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev M. 1954. Demonstration of a diffusible fruiting factor in Myxobacteria. Nature 173, 501 ( 10.1038/173501a0)13144794 [DOI] [Google Scholar]
- Luo Z. Q., Qin Y. P., Farrand S. K. 2000. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J. Biol. Chem. 275, 7713–7722. ( 10.1074/jbc.275.11.7713) [DOI] [PubMed] [Google Scholar]
- Mayville P., Ji G. Y., Beavis R., Yang H. M., Goger M., Novick R. P., Muir T. W. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl Acad. Sci. USA 96, 1218–1223. ( 10.1073/pnas.96.4.1218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L., Kasper D. L. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. ( 10.1038/nature07008) [DOI] [PubMed] [Google Scholar]
- McClean K. H., et al. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology UK 143, 3703–3711. ( 10.1099/00221287-143-12-3703) [DOI] [PubMed] [Google Scholar]
- McDowell P., et al. 2001. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol. Microbiol. 41, 503–512. ( 10.1046/j.1365-2958.2001.02539.x) [DOI] [PubMed] [Google Scholar]
- McVittie A., Zahler S. A., Messik F. 1964. Developmental biology of Myxococcus. J. Bacteriol. 84, 546–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael B., Smith J. N., Swift S., Heffron F., Ahmer B. M. M. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183, 5733–5742. ( 10.1128/JB.183.19.5733-5742.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. B., Skorupski K., Lenz D. H., Taylor R. K., Bassler B. L. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110, 303–314. ( 10.1016/S0092-8674(02)00829-2) [DOI] [PubMed] [Google Scholar]
- Miller S. T., Xavier K. B., Campagna S. R., Taga M. E., Semmelhack M. F., Bassler B. L., Hughson F. M. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal Al-2. Mol. Cell 15, 677–687. ( 10.1016/j.molcel.2004.07.020) [DOI] [PubMed] [Google Scholar]
- Milton D. L. 2006. Quorum sensing in vibrios: complexity for diversification. Int. J. Med. Microbiol. 296, 61–71. ( 10.1016/j.ijmm.2006.01.044) [DOI] [PubMed] [Google Scholar]
- Milton D. L., Chalker V. J., Kirke D., Hardman A., Cámara M., Williams P. 2001. The luxM homologue vanM from Vibrio anguillanrum directs the synthesis of N-(3-hydroxyhexanoyl)homoserine lactone and N-hexanoylhomoserine lactone. J. Bacteriol. 183, 3537–3547. ( 10.1128/JB.183.12.3537-3547.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- More M. I., Finger L. D., Stryker J. L., Fuqua C., Eberhard A., Winans S. C. 1996. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science 272, 1655–1658. ( 10.1126/science.272.5268.1655) [DOI] [PubMed] [Google Scholar]
- Muh U., Hare B. J., Duerkop B. A., Schuster M., Hanzelka B. L., Heim R., Olson E. R., Greenberg E. P. 2006. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc. Natl Acad. Sci. USA 103, 16 948–16 952. ( 10.1073/pnas.0608348103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Chen S. M., Oyama N., Nishiguchi K., Azab E. A., Tanaka E., Kariyama R., Sonomoto K. 2006. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal AgrD. J. Bacteriol. 188, 8321–8326. ( 10.1128/JB.00865-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson K. H., Platt T., Hastings J. W. 1970. Cellular control of synthesis and activity of bacterial luminescent system. J. Bacteriol. 104, 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch M. B., Federle M. J., Miller S. T., Bassler B. L., Hughson F. M. 2005. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell 18, 507–518. ( 10.1016/j.molcel.2005.04.020) [DOI] [PubMed] [Google Scholar]
- Ni N. T., Li M. Y., Wang J. F., Wang B. H. 2009. Inhibitors and antagonists of bacterial quorum sensing. Med. Res. Rev. 29, 65–124. ( 10.1002/med.20145) [DOI] [PubMed] [Google Scholar]
- Nickerson K. W., Atkin A. L., Hornby J. M. 2006. Quorum sensing in dimorphic fungi: farnesol and beyond. Appl. Environ. Microbiol. 72, 3805–3813. ( 10.1128/AEM.02765-05) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Geisinger E. 2008. Quorum sensing in Staphylococci. Annu. Rev. Genet. 42, 541–564. ( 10.1146/annurev.genet.42.110807.091640) [DOI] [PubMed] [Google Scholar]
- Oliver C. M., Schaefer A. L., Greenberg E. P., Sufrin J. R. 2009. Microwave synthesis and evaluation of phenacylhomoserine lactones as anticancer compounds that minimally activate quorum sensing pathways in Pseudomonas aeruginosa. J. Med. Chem. 52, 1569–1575. ( 10.1021/jm8015377) [DOI] [PubMed] [Google Scholar]
- Onsando J. M. 1992. Black rot of crucifers. In Diseases of vegetables and oil seed crops (eds Chaube H. S., Kumar J., Mukhopadhyay A. N., Singh U. S.), pp. 243–252. Englewood Cliffs, NJ: Prentice Hall. [Google Scholar]
- Ortori C. A., Atkinson S., Chhabra S. R., Cámara M., Williams P., Barrett D. A. 2007. Comprehensive profiling of N-acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadruple–linear ion trap mass spectrometry. Anal. Bioanal. Chem. 387, 497–511. ( 10.1007/s00216-006-0710-0) [DOI] [PubMed] [Google Scholar]
- Park J., Jagasia R., Kaufmann G. F., Mathison J. C., Ruiz D. I., Moss J. A., Meijler M. M., Ulevitch R. J., Janda K. D. 2007. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 14, 1119–1127. ( 10.1016/j.chembiol.2007.08.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M. R., Greenberg E. P. 1999. Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Biofilms 310, 43–55. ( 10.1016/S0076-6879(99)10005-3) [DOI] [PubMed] [Google Scholar]
- Persson T., Hansen T. H., Rasmussen T. B., Skinderso M. E., Givskov M., Nielsen J. 2005. Rational design and synthesis of new quorum-sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org. Biomol. Chem. 3, 253–262. ( 10.1039/b415761c) [DOI] [PubMed] [Google Scholar]
- Pomianek M. E., Semmelhack M. F. 2007. Making bacteria behave: new agonists and antagonists of quorum sensing. ACS Chem. Biol. 2, 293–295. ( 10.1021/cb700098c) [DOI] [PubMed] [Google Scholar]
- Pratt S. C., Mallon E. B., Sumpter D. J. T., Franks N. R. 2002. Quorum sensing, recruitment, and collective decision-making during colony emigration by the ant Leptothorax albipennis. Behav. Ecol. Sociobiol. 52, 117–127. ( 10.1007/s00265-002-0487-x) [DOI] [Google Scholar]
- Pritchard D. I. 2006. Immune modulation by Pseudomonas aeruginosa quorum-sensing signal molecules. Int. J. Med. Microbiol. 296, 111–116. ( 10.1016/j.ijmm.2006.01.037) [DOI] [PubMed] [Google Scholar]
- Pritchard D. I., Todd I., Brown A., Bycroft B. W., Chhabra S. R., Williams P., Wood P. 2005. Alleviation of insulitis and moderation of diabetes in NOD mice following treatment with a synthetic Pseudomonas aeruginosa signal molecule, N-(3-oxododecanoyl)-l-homoserine lactone. Acta Diabetol. 42, 119–122. ( 10.1007/s00592-005-0190-2) [DOI] [PubMed] [Google Scholar]
- Qazi S. N. A., Counil E., Morrissey J., Rees C. E. D., Cockayne A., Winzer K., Chan W. C., Williams P., Hill P. J. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69, 7074–7082. ( 10.1128/IAI.69.11.7074-7082.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi S. N. A., Middleton B., Muharram S. H., Cockayne A., Hill P., O'Shea P., Chhabra S. R., Cámara M., Williams P. 2006. N-acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 74, 910–919. ( 10.1128/IAI.74.2.910-919.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T. H. L., Khan B. A., Sturdevant D. E., Ricklefs S. M., Li M., Otto M. 2008. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158. ( 10.1016/j.molcel.2008.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa R. B., Iannuzzo J. R., Levine D. R., Saeid K. K., Schwartz R. C., Sucic N. T., Terleckyj O. D., Young J. M. 2005. Bacterial communication (‘Quorum sensing’) via ligands and receptors: a novel pharmacologic target for the design of antibiotic drugs. J. Pharmacol. Exp. Ther. 312, 417–423. ( 10.1124/jpet.104.075150) [DOI] [PubMed] [Google Scholar]
- Rajamani S., Bauer W. D., Robinson J. B., Farrow J. M., Pesci E. C., Teplitski M., Gao M., Sayre R. T., Phillips D. A. 2008. The vitamin riboflavin and its derivative lumichrome activate the LasR bacterial quorum-sensing receptor. Mol. Plant. Microbe Interact. 21, 1184–1192. ( 10.1094/MPMI-21-9-1184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. B., Givskov M. 2006. Quorum sensing inhibitors: a bargain of effects. Microbiology-Sgm 152, 895–904. ( 10.1099/mic.0.28601-0) [DOI] [PubMed] [Google Scholar]
- Redfield R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10, 365–370. ( 10.1016/S0966-842X(02)02400-9) [DOI] [PubMed] [Google Scholar]
- Rezzonico F., Duffy B. 2008. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 8, 154 ( 10.1186/1471-2180-8-154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel K., et al. 2001. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology-Sgm 147, 3249–3262. [DOI] [PubMed] [Google Scholar]
- Riedel C. U., Monk I. R., Casey P. G., Waidmann M. S., Gahan C. G. M., Hill C. 2009. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 71, 1177–1189. ( 10.1111/j.1365-2958.2008.06589.x) [DOI] [PubMed] [Google Scholar]
- Rivas M., Seeger M., Jedlicki E., Holmes D. S. 2007. Second acyl homoserine lactone production system in the extreme acidophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 73, 3225–3231. ( 10.1128/AEM.02948-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A. 2008. Dimorphism. In The fungi, vol. 2 (eds Ainsworth G. C., Sussman A. S.), pp. 181–209. New York, NY: Academic Press. [Google Scholar]
- Ross A. J., Rucker R. R., Ewing W. H. 1966. Description of a bacterium associated with redmouth disease of rainbow trout (Salmo Gairdneri). Can. J. Microbiol. 12, 763–770. [DOI] [PubMed] [Google Scholar]
- Rumbaugh K. P. 2007. Convergence of hormones and autoinducers at the host/pathogen interface. Anal. Bioanal. Chem. 387, 425–435. ( 10.1007/s00216-006-0694-9) [DOI] [PubMed] [Google Scholar]
- Ryan R. P., et al. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl Acad. Sci. USA 103, 6712–6717. ( 10.1073/pnas.0600345103) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sanchez-Contreras M., Bauer W. D., Gao M. S., Robinson J. B., Downie J. A. 2007. Quorum-sensing regulation in Rhizobia and its role in symbiotic interactions with legumes. Phil. Trans. R. Soc. B 362, 1149–1163. ( 10.1098/rstb.2007.2041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. L., et al. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454, 595–599. ( 10.1038/nature07088) [DOI] [PubMed] [Google Scholar]
- Schauder S., Shokat K., Surette M. G., Bassler B. L. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476. ( 10.1046/j.1365-2958.2001.02532.x) [DOI] [PubMed] [Google Scholar]
- Scott R. J., Lian L. Y., Muharram S. H., Cockayne A., Wood S. J., Bycroft B. W., Williams P., Chan W. C. 2003. Side-chain-to-tail thiolactone peptide inhibitors of the staphylococcal quorum-sensing system. Bioorg. Med. Chem. Lett. 13, 2449–2453. ( 10.1016/S0960-894X(03)00497-9) [DOI] [PubMed] [Google Scholar]
- Seabra R., Brown A., Hooi D., Kerkhoff C., Chhabra S. R., Harty C., Williams P., Pritchard D. I. 2008. A eukaryotic binding protein for the immune modulatory bacterial quorum sensing molecule N-(3-oxododecanoyl)-l-homoserine lactone. Calcium Binding Proteins 3, 31–37. [Google Scholar]
- Shiner E. K., Rumbaugh K. P., Williams S. C. 2005. Interkingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol. Rev. 29, 935–947. ( 10.1016/j.femsre.2005.03.001) [DOI] [PubMed] [Google Scholar]
- Sibley C. D., Duan K. M., Fischer C., Parkins M. D., Storey D. G., Rabin H. R., Surette M. G. 2008. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4, e1000184 ( 10.1371/journal.ppat.1000184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., et al. 2006. Structure and inhibition of a quorum sensing target from Streptococcus pneumoniae. Biochemistry (Mosc.). 45, 12 929–12 941. ( 10.1021/bi061184i) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater H., Varez-Morales A., Barber C. E., Daniels M. J., Dow J. M. 2000. A two-component system involving an HD-GYP domain protein links cell–cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38, 986–1003. ( 10.1046/j.1365-2958.2000.02196.x) [DOI] [PubMed] [Google Scholar]
- Smith J. N., Ahmer B. M. M. 2003. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 185, 1357–1366. ( 10.1128/JB.185.4.1357-1366.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V., Torres A. G., Jarvis B., Nataro J. P., Kaper J. B. 2003. Bacteria-host communication: the language of hormones. Proc. Natl Acad. Sci. USA 100, 8951–8956. ( 10.1073/pnas.1537100100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler L., Venturi V. 2007. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 266, 1–9. ( 10.1111/j.1574-6968.2006.00501.x) [DOI] [PubMed] [Google Scholar]
- Suga H., Smith K. M. 2003. Molecular mechanisms of bacterial quorum sensing as a new drug target. Curr. Opin. Chem. Biol. 7, 586–591. ( 10.1016/j.cbpa.2003.08.001) [DOI] [PubMed] [Google Scholar]
- Swift S., Karlyshev A. V., Fish L., Durant E. L., Winson M. K., Chhabra S. R., Williams P., MacIntyre S., Stewart G. S. A. B. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179, 5271–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift S., Isherwood K. E., Atkinson S., Oyston P., Stewart G. S. A. B. 1999. Quorum sensing in Aeromonas and Yersinia. In Microbial signalling and communication (eds England R., Hobbs G., Bainton N. J., Roberts D. M.), pp. 85–104. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Swift S., Downie J. A., Whitehead N. A., Barnard A. M. L., Salmond G. P. C., Williams P. 2001. Quorum sensing as a population-density-dependent determinant of bacterial physiology. Adv. Microb. Physiol. 45, 199–270. ( 10.1016/S0065-2911(01)45005-3) [DOI] [PubMed] [Google Scholar]
- Taga M. E., Semmelhack J. L., Bassler B. L. 2001. The LuxS-dependent autoinducer Al-2 controls the expression of an ABC transporter that functions in Al-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42, 777–793. ( 10.1046/j.1365-2958.2001.02669.x) [DOI] [PubMed] [Google Scholar]
- Taga M. E., Miller S. T., Bassler B. L. 2003. Lsr-mediated transport and processing of Al-2 in Salmonella typhimurium. Mol. Microbiol. 50, 1411–1427. ( 10.1046/j.1365-2958.2003.03781.x) [DOI] [PubMed] [Google Scholar]
- Tait K., Joint I., Daykin M., Milton D. L., Williams P., Cámara M. 2005. Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ. Microbiol. 7, 229–240. ( 10.1111/j.1462-2920.2004.00706.x) [DOI] [PubMed] [Google Scholar]
- Tao Y., Drabik K. A., Waypa T. S., Musch M. W., Alverdy J. C., Schneewind O., Chang E. B., Petrof E. O. 2006. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 290, C1018–C1030. ( 10.1152/ajpcell.00131.2005) [DOI] [PubMed] [Google Scholar]
- Taylor A. F., Tinsley M. R., Wang F., Huang Z. Y., Showalter K. 2009. Dynamical quorum sensing and synchronization in large populations of chemical oscillators. Science 323, 614–617. ( 10.1126/science.1166253) [DOI] [PubMed] [Google Scholar]
- Temprano A., Yugueros J., Hernanz C., Sanchez M., Berzal B., Luengo J. M., Naharro G. 2001. Rapid identification of Yersinia ruckeri by PCR amplification of yrul-yruR quorum sensing. J. Fish Dis. 24, 253–261. ( 10.1046/j.1365-2761.2001.00261.x) [DOI] [Google Scholar]
- Throup J. P., Cámara M., Bainton N. J., Briggs G. S., Chhabra S. R., Bycroft B. W., Williams P., Stewart G. S. A. B. 1995. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acyl homoserine lactone signal molecules. Mol. Microbiol. 17, 345–356. ( 10.1111/j.1365-2958.1995.mmi_17020345.x) [DOI] [PubMed] [Google Scholar]
- Tomasz A. 1965. Control of competent state in Pneumococcus by a hormone-like cell product—an example for a new type of regulatory mechanism in bacteria. Nature 208, 155–159. ( 10.1038/208155a0) [DOI] [PubMed] [Google Scholar]
- Valle A., Bailey M. J., Whiteley A. S., Manefield M. 2004. N-acyl-l-homoserine lactones (AHLs) affect microbial community composition and function in activated sludge. Environ. Microbiol. 6, 424–433. ( 10.1111/j.1462-2920.2004.00581.x) [DOI] [PubMed] [Google Scholar]
- Van Houdt R., Aertsen A., Moons P., Vanoirbeek K., Michiels C. W. 2006. N-acyl-l-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett. 256, 83–89. ( 10.1111/j.1574-6968.2006.00103.x) [DOI] [PubMed] [Google Scholar]
- Vendeville A., Winzer K., Heurlier K., Tang C. M., Hardie K. R. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3, 383–396. ( 10.1038/nrmicro1146) [DOI] [PubMed] [Google Scholar]
- Viudes A., Peman J., Canton E., Ubeda P., Lopez-Ribot J. L., Gobernado M. 2002. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur. J.Clin. Microbiol. Infect. Dis. 21, 767–774. ( 10.1007/s10096-002-0822-1) [DOI] [PubMed] [Google Scholar]
- Walters M., Sircili M. P., Sperandio V. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188, 5668–5681. ( 10.1128/JB.00648-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., et al. 2004. A bacterial cell–cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51, 903–912. ( 10.1046/j.1365-2958.2003.03883.x) [DOI] [PubMed] [Google Scholar]
- Waters C. M., Bassler B. L. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. ( 10.1146/annurev.cellbio.21.012704.131001) [DOI] [PubMed] [Google Scholar]
- White C. E., Winans S. C. 2007. Cell–cell communication in the plant pathogen Agrobacterium tumefaciens. Phil. Trans. R. Soc. B 362, 1135–1148. ( 10.1098/rstb.2007.2040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. 2007a. Bacillus subtilis: a shocking message from a probiotic. Cell Host Microbe 1, 248–249. ( 10.1016/j.chom.2007.05.010) [DOI] [PubMed] [Google Scholar]
- Williams P. 2007b. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 153, 3923–3938. ( 10.1099/mic.0.2007/012856-0) [DOI] [PubMed] [Google Scholar]
- Williams P., Cámara M. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12, 182–191. ( 10.1016/j.mib.2009.01.005) [DOI] [PubMed] [Google Scholar]
- Williams P., Winzer K., Chan W. C., Cámara M. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Phil. Trans. R. Soc. B 362, 1119–1134. ( 10.1098/rstb.2007.2039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson M. K., Swift S., Fish L., Throup J. P., Jorgensen F., Chhabra S. R., Bycroft B. W., Williams P., Stewart G. S. A. B. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163, 185–192. ( 10.1111/j.1574-6968.1998.tb13044.x) [DOI] [PubMed] [Google Scholar]
- Winzer K., Williams P. 2003. Escherichia coli gets the message. Nat. Med. 9, 1118–1119. ( 10.1038/nm0903-1118) [DOI] [PubMed] [Google Scholar]
- Winzer K., et al. 2002a. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology-Sgm 148, 909–922. [DOI] [PubMed] [Google Scholar]
- Winzer K., Hardie K. R., Williams P. 2002b. Bacterial cell-to-cell communication: sorry, can't talk now—gone to lunch! Curr. Opin. Microbiol. 5, 216–222. ( 10.1016/S1369-5274(02)00304-1) [DOI] [PubMed] [Google Scholar]
- Wuster A., Babu M. M. 2008. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J. Bacteriol. 190, 743–746. ( 10.1128/JB.01135-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier K. B., Bassler B. L. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6, 191–197. ( 10.1016/S1369-5274(03)00028-6) [DOI] [PubMed] [Google Scholar]
- Xavier K. B., Bassler B. L. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187, 238–248. ( 10.1128/JB.187.1.238-248.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim G., Wang H. H. M., Davies J. 2006. The truth about antibiotics. Int. J. Med. Microbiol. 296, 163–170. ( 10.1016/j.ijmm.2006.01.039) [DOI] [PubMed] [Google Scholar]
- Zhang L. H., Dong Y. H. 2004. Quorum sensing and signal interference: diverse implications. Mol. Microbiol. 53, 1563–1571. ( 10.1111/j.1365-2958.2004.04234.x) [DOI] [PubMed] [Google Scholar]
- Zhu J., Mekalanos J. J. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5, 647–656. ( 10.1016/S1534-5807(03)00295-8) [DOI] [PubMed] [Google Scholar]
- Zhu J., Winans S. C. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl Acad. Sci. USA 96, 4832–4837. ( 10.1073/pnas.96.9.4832) [DOI] [PMC free article] [PubMed] [Google Scholar]