Abstract
Two prehistoric migrations peopled the Pacific. One reached New Guinea and Australia, and a second, more recent, migration extended through Melanesia and from there to the Polynesian islands. These migrations were accompanied by two distinct populations of the specific human pathogen Helicobacter pylori, called hpSahul and hspMaori, respectively. hpSahul split from Asian populations of H. pylori 31,000 to 37,000 years ago, in concordance with archaeological history. The hpSahul populations in New Guinea and Australia have diverged sufficiently to indicate that they have remained isolated for the past 23,000 to 32,000 years. The second human expansion from Taiwan 5000 years ago dispersed one of several subgroups of the Austronesian language family along with one of several hspMaori clades into Melanesia and Polynesia, where both language and parasite have continued to diverge.
After modern humans dispersed “out of Africa” about 60,000 years ago (60 ka) (1), they reached Asia via a southern coastal route (2). That route extended along the Pleistocene landmass, known as Sundaland (i.e., the Malay peninsula, Sumatra, Java, Borneo, and Bali), that was joined to the Asian mainland as a result of low sea levels during the last ice age (12 to 43 ka) (3). Low sea levels also meant that Australia, New Guinea, and Tasmania were connected in a continent called Sahul, separated from Sundaland by a few narrow deep-sea channels. It seems Sahul was colonized only once, ~40 to 50 ka (3, 4), although backed-blade stone tool technology and the dingo appear to have been introduced from India at a later date (5, 6).
Human genetic data are compatible with these interpretations, but have not provided the details. Redd and Stoneking identified multiple mitochondrial DNA (mtDNA) lineages among New Guinea peoples with coalescence times of 80,000 to 122,000 years (80 to 122 ky), predating the out-of-Africa migrations (5). In subsequent analyses, Australian aboriginals and Melanesians fell into multiple, distinct mtDNA haplogroups inter-dispersed among lineages from East Asia and India (4), with one exception: haplogroup Q, which had a coalescent estimate of 32 ka and contained both Australian and Melanesian lineages. Y-chromosome markers yielded one lineage for Australians and a second one for Melanesians (4). Australia and New Guinea remained connected by a land bridge until sea levels rose ~8 to 12 ka, and it is surprising that the native inhabitants of Sahul are not genetically associated except for haplogroup Q.
Subsequent prehistoric migrations to island East Asia and the Pacific have been designated differently depending on whether they were traced by language, archaeological remains, or genetic studies. Most of the native Pacific languages from near the African coast (Madagascar) through to Polynesia are Malayo-Polynesian, a subgroup of the Austronesian language family (7). The nine other subgroups of Austronesian are only spoken in Taiwan, suggesting that Taiwan is the origin of Austronesian (7). In support of this interpretation, agriculturists spread from Taiwan via insular and coastal Melanesia into the Pacific, as marked by the Lapita cultural complex, including red-slipped pottery, Neolithic tools, chickens, pigs, and farming (8). A human genetic marker of this route of spread is the “Polynesian” mtDNA HV1 motif of lineage B4a1a, which is found at high frequency among native Taiwanese (9), Melanesians, and Polynesians (10, 11).
We attempted to trace human prehistory in the Pacific by analyzing the distribution of a bacterial parasite of humans, Helicobacter pylori. H. pylori accompanied modern humans during their migrations out of Africa (12). Subsequent founder effects, plus geographic separation, have resulted in populations of bacterial strains specific for large continental areas. Thus, Africans are infected by the H. pylori populations hpAfrica1 and hpAfrica2, Asians are infected by hpAsia2 and hpEastAsia, and Europeans are infected by hpEurope (12, 13). It seemed possible that the distribution of H. pylori genotypes among native inhabitants might provide insights into migrations throughout the Pacific. We cultivated 212 bacterial isolates from gastric biopsies or mucus obtained from aboriginals in Taiwan and Australia, highlanders in New Guinea, as well as Melanesians and Polynesians in New Caledonia (table S1). Concatenated sequences of seven gene fragments (3406 base pairs, of which half are polymorphic) from these isolates yielded 196 unique haplotypes. These were compared with 99 unique haplotypes from 100 Europeans in Australia and 222 other unique haplotypes from Asia and the Pacific, including 15 haplotypes from Chinese inhabitants of Taiwan, as well as ~1700 haplotypes from other sources.
According to Bayesian assignment analysis, our samples from native inhabitants yielded 50 unique haplotypes that formed a distinct bio-geographic group called hpSahul (14). Twenty-eight percent (26 of 92) of the haplotypes from aboriginals in Australia and 89% (24 of 27) of the haplotypes from highlanders in New Guinea were hpSahul (Fig. 1A). One hpSahul haplotype was found among 99 haplotypes from Europeans in Australia and none among the other haplotypes from elsewhere.
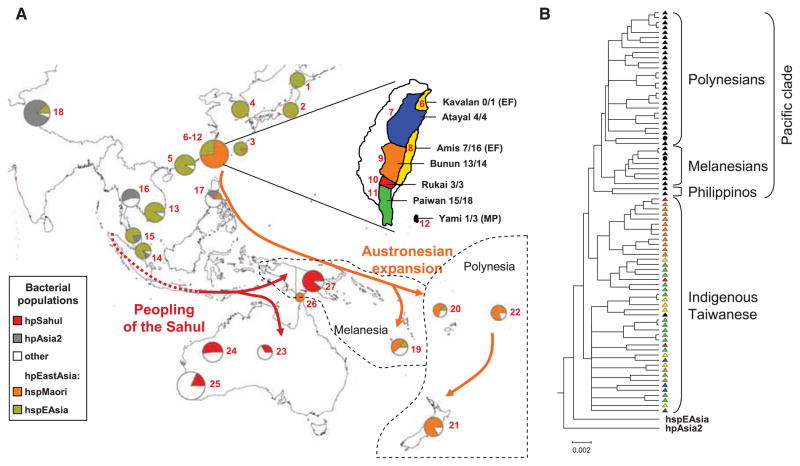
Fig. 1.
(A) The distribution of H. pylori populations in Asia and the Pacific. The proportions of haplotypes at each sampling location (red numbers; table S1) that were assigned to different bacterial populations are displayed as pie charts whose sizes indicate the numbers of haplotypes. The geographic location of Melanesia and Polynesia is depicted. The term “Austronesia” refers to the entire region inhabited by Austronesian-speaking people from Madagascar through to the Easter Islands. (Inset) A detailed map of Taiwan showing the distribution of aboriginal tribes. The names of the tribes plus the proportion of hspMaori haplotypes among all haplotypes are shown in black at the right. The language-family designations are the same as the tribal names except where indicated by parentheses (EF, East Formosan; MP, Malayo-Polynesian). (B) Phylogenetic relationships among hspMaori strains (80% consensus of 100 ClonalFrame analyses). One haplotype each of hpAsia2 and hspEAsia was used to root the tree. Strains are color-coded according to Austronesian language family in (A). Two black circles within the Pacific clade indicate haplotypes isolated from the Torres Strait islands, and a black triangle among indigenous Taiwanese indicates an hspMaori haplotype from Yami.
hspMaori is a subpopulation of hpEastAsia, isolated from Polynesians (Maoris, Tongans, and Samoans) in New Zealand (13) and three individuals in the Philippines and Japan. hspMaori isolates have not previously been isolated from other individuals, including the 15 Chinese inhabitants of Taiwan (12). Fifty-four of the 196 unique haplotypes from native inhabitants were hspMaori (14), and all came from Austronesian sources. These included native Taiwanese (43 of 59, 73%), Melanesians (6 of 13, 46%), and Polynesians (3 of 5, 60%) in New Caledonia, and two inhabitants of the Torres Straits islands that lie between Australia and New Guinea and which have been visited extensively by Polynesians (Fig. 1A and table S1). These observations suggest that hspMaori is a marker for the entire Austronesian expansions rather than only for Polynesians. The remaining unique haplotypes from native inhabitants were hpEurope, hspEAsia, and hpAfrica1, which can be attributed to very recent human travels.
If Taiwan were the source of the Austronesian expansions, hspMaori haplotypes would be expected to be widespread among aboriginal Taiwanese tribes. Indeed, hspMaori was isolated frequently (44 to 100%) from five of the six tribes sampled (Fig. 1A). Taiwan should also harbor the greatest diversity, and the branching order within a phylogenetic tree should reflect the direction of subsequent migrations. The phylogenetic analyses showed that genetic diversity was significantly higher in Taiwanese hspMaori (Π95 = 1.79 to 1.82%) than in non-Taiwanese hspMaori (Π95 = 1.58 to 1.62%). All non-Taiwanese hspMaori haplotypes form a single clade, the Pacific clade, which originates from one of several clades among indigenous Taiwanese haplotypes (Fig. 1B). The sequence of branching events within the Pacific clade is consistent with sequential migrations from Taiwan via the Philippines and island Melanesia to Polynesia (Fig. 1B). These results also support an association between language and haplotype group. The indigenous Taiwanese haplotypes were isolated from tribes that speak 5 of the 10 subgroups of the Austronesian family of languages, whereas the Pacific clade was isolated from individuals that speak variants of Malayo-Polynesian. The sole exception to these generalizations was one haplotype from the Yami of Lanyu, a small island off the coast of Taiwan, where the language is a variant of Malayo-Polynesian but the haplotype clustered with the indigenous Taiwanese haplotypes. Together, these observations provide support for a Taiwanese source of the Austronesian expansions.
Using the isolation with migration model (IMa), we calculated the magnitude of migrations in both directions after the initial split between the Taiwan and Pacific clades of hspMaori (15). IMa uses sequence data within a probabilistic framework to simulate a model of initial geographic separation between two populations followed by occasional migration in both directions. Because homologous recombination is frequent within H. pylori (13, 16), we excluded blocks of sequences that had a high likelihood of recombination (14). The calculations indicated that migrations subsequent to the initial split were unidirectional, from Taiwan to the Pacific (Fig. 2A).
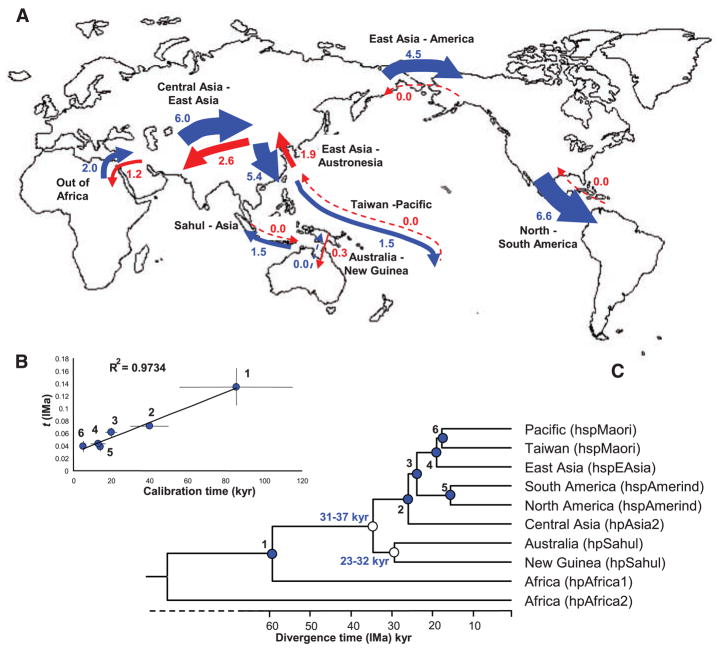
Fig. 2.
Global patterns of migration between eight pairs of H. pylori populations as calculated by the isolation with migration model (IMa). (A) Map. The magnitudes of migration are denoted by numbers and arrow thickness and their direction is indicated in blue or red. (B) Graph showing a linear relation between the calibration time (table S2) of six events (filled blue circles) that are dated by archaeological estimates and the estimated time (t). (C) Population tree reconstructed from a consensus of 1000 bootstrap samples from the range of calculated t values to determine the ages of nodes (thousands of years, kyr) associated with the peopling of the Sahul (unfilled circles). Ages (in light blue) are the 95% confidence limits of estimated coalescence times obtained by applying global rate minimum deformation (GRMD) rate-smoothing, as implemented in Treefinder, to the range of t values within the limits of calibration dates.
Other splits between pairs of H. pylori populations were also unidirectional: for example, the Amerind colonization over the Bering Strait and the subsequent colonization of South America from North America. However, migrations out of Africa, from Central to East Asia, and from East Asia to Taiwan were followed by appreciable levels of return migration (Fig. 2A).
Molecular mutation rates are unknown for most bacteria, so we cannot directly use IMa data to calculate the dates of initial splits. Instead, we calibrated against known dates for splits among human populations. The archaeologically attributed split between Taiwan and the Pacific Clade is 5 ka (8). Five other calibration dates are presented in table S2. The time when populations split (t) calculated by IMa varied linearly with the calibration dates (Fig. 2B). We used random values within the range of five t values that were calculated for each split between all pairs of populations (table S2) to construct 1000 bootstrap trees using Treefinder (17). These trees were then used to calculate the age of the Sahulian migration by rate-smoothing within the limits of the six calibration dates (14).
The dates and numbers of migrations to the Sahul are controversial. According to our IMa calculations, the population split leading to hpSahul postdated the out-of-Africa migrations but predated the splits that resulted in hpAsia2 (found in Central Asia) and hpEastAsia [East Asia (hspEAsia); the Pacific (hspMaori); the Americas (hspAmerind)]. The 95% confidence limits of the date of the split between hpSahul and the Asian populations were estimated as 31 to 37 ka and the split between hpSahul in New Guinea and Australia as 23 to 32 ka. The combined data presented here indicate that hpSahul migrated only once from Asia toward Sahul, and once between New Guinea and Australia, and subsequent migration did not occur from Australia to New Guinea (Fig. 2A).
To verify the use of IMa for dating of population splits in a bacterial species like H. pylori, we also used a haplotype-based coalescent approach, which accounts for recombination with unrelated sources of DNA, as implemented in the program ClonalFrame (18). ClonalFrame generated a haplotype tree whose branch order agreed with the population tree generated by IMa (Fig. 3A). It also assigned individual haplotypes to clades that are congruent with the population assignments, including the separation between hpSahul and other populations. The observation that all hpSahul strains clustered in a monophyletic clade verifies a single colonization event and confirms that modern Asians and the inhabitants of the Sahul have undergone independent evolutionary trajectories since they first split. The two hpSahul clades in New Guinea and Australia are also distinct, confirming a lack of migration between the two areas.
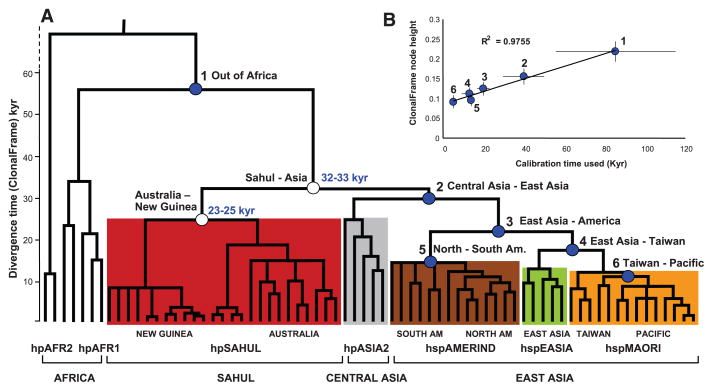
Fig. 3.
Global phylogeny of H. pylori as calculated by a haplotype approach based on the 80% consensus of 100 ClonalFrame analyses. (A) Phylogenetic tree of divergence time, as indicated by node height versus geographic sources (bottom line) and population assignments (second line). Detailed sources of clades within populations are indicated in the third line from the bottom. Node heights were used to date the two hpSahul nodes (unfilled circles) based on six calibration times (filled blue circles, table S2). Age ranges (light blue numbers) are the 95% confidence limits of estimated coalescence times obtained with GRMD rate-smoothing over the range of node height values and calibration time limits. hpAFR2, hpAfrica2; hpAFR1, hpAfrica1; AM, America. (B) Graph showing a linear relation of calibration time with the range of heights for each node.
Similarly to the IMa analyses, we observed a linear relation between the calibration dates and time of splitting calculated by ClonalFrame as node heights (Fig. 3B). Applying the same rate-smoothing calibration method as above, we estimated that hpSahul split from the Asian population 32 to 33 ka. Subsequently, hpSahul from New Guinea and Australia split 23 to 25 ka. Both estimates overlap with the range of IMa estimates (31 to 37 ka and 23 to 32 ka, respectively). The date of origin of hpSahul is comparable to the estimated age of 32 ka for the Q mtDNA haplogroup (4), but less than the 40 to 50 ky associated with the oldest archaeological finding of human artefacts in Australia (3).
Our results lend support for two distinct waves of migrations into the Pacific. First, early migrations to New Guinea and Australia accompanied by hpSahul and second, a much later dispersal of hspMaori from Taiwan through the Pacific by the Malayo-Polynesian–speaking Lapita culture. Each sampling area yielded either hpSahul or hspMaori haplotypes, but not both. The lack of overlap between these populations may reflect differential fitness of the parasite, as has been inferred for the modern replacement of hspAmerind haplotypes by European and African H. pylori in South America (19, 20). Alternatively, hpSahul and hspMaori may still coexist in unsampled islands of East Asia, Melanesia, and coastal New Guinea, where their identification might help to unravel the details of human history in those areas.
Supplementary Material
Acknowledgments
We gratefully acknowledge C. Stamer for technical assistance, F. Balloux and D. Falush for helpful discussions, and J. Hey for advice on IMa. Support was provided by grants from the ERA-NET PathoGenoMics (project HELDIVNET) to M.A. and S.B., the Science Foundation of Ireland (05/FE1/B882) to M.A., the NIH (grant R01 DK62813) to Y.Y., and the Institut Pasteur and the Institut de Veille Sanitaire to J.-M.T. This publication made use of the Helicobacter pylori Multi Locus Sequence Typing Web site (http://pubmlst.org/helicobacter/) developed by K. Jolley and sited at the University of Oxford. Each strain has an ID number, and the strains newly isolated here have the continuous block of IDs from 930 to 1242. The development of this site has been funded by the Wellcome Trust and European Union.
Footnotes
References and Notes
- 1.Liu H, Prugnolle F, Manica A, Balloux F. Am J Hum Genet. 2006;79:230. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macaulay V, et al. Science. 2005;308:1034. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 3.Pope KO, Terrell JE. J Biogeography. 2008;35:1. [Google Scholar]
- 4.Hudjashov G, et al. Proc Natl Acad Sci USA. 2007;104:8726. doi: 10.1073/pnas.0702928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redd AJ, Stoneking M. Am J Hum Genet. 1999;65:808. doi: 10.1086/302533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savolainen P, Leitner T, Wilton AN, Matisoo-Smith E, Lundeberg J. Proc Natl Acad Sci USA. 2004;101:12387. doi: 10.1073/pnas.0401814101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray RD, Jordan FM. Nature. 2000;405:1052. doi: 10.1038/35016575. [DOI] [PubMed] [Google Scholar]
- 8.Spriggs MJT. In: Prehistoric Mongoloid Dispersals. Akazawa T, Szathmary E, editors. Oxford Univ. Press; Oxford: 1996. pp. 322–346. [Google Scholar]
- 9.Trejaut JA, et al. PLoS Biol. 2005;3:e247. doi: 10.1371/journal.pbio.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melton T, et al. Am J Hum Genet. 1995;57:403. [PMC free article] [PubMed] [Google Scholar]
- 11.Sykes B, Leiboff A, Low-Beer J, Tetzner S, Richards M. Am J Hum Genet. 1995;57:1463. [PMC free article] [PubMed] [Google Scholar]
- 12.Linz B, et al. Nature. 2007;445:915. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falush D, et al. Science. 2003;299:1582. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 14.Materials and methods and supplementary tables, figures, and scripts are available on Science Online.
- 15.Hey J, Nielsen R. Proc Natl Acad Sci USA. 2007;104:2785. doi: 10.1073/pnas.0611164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falush D, et al. Proc Natl Acad Sci USA. 2001;98:15056. doi: 10.1073/pnas.251396098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jobb G, von Haeseler A, Strimmer K. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Didelot X, Falush D. Genetics. 2007;175:1251. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaoka Y, et al. FEBS Lett. 2002;517:180. doi: 10.1016/s0014-5793(02)02617-0. [DOI] [PubMed] [Google Scholar]
- 20.Ghose C, et al. Proc Natl Acad Sci USA. 2002;99:15107. doi: 10.1073/pnas.242574599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.