Abstract
Periodontitis and caries are infectious diseases of the oral cavity in which oral biofilms play a causative role. Moreover, oral biofilms are widely studied as model systems for bacterial adhesion, biofilm development, and biofilm resistance to antibiotics, due to their widespread presence and accessibility. Despite descriptions of initial plaque formation on the tooth surface, studies on mature plaque and plaque structure below the gum are limited to landmark studies from the 1970s, without appreciating the breadth of microbial diversity in the plaque. We used fluorescent in situ hybridization to localize in vivo the most abundant species from different phyla and species associated with periodontitis on seven embedded teeth obtained from four different subjects. The data showed convincingly the dominance of Actinomyces sp., Tannerella forsythia, Fusobacterium nucleatum, Spirochaetes, and Synergistetes in subgingival plaque. The latter proved to be new with a possibly important role in host-pathogen interaction due to its localization in close proximity to immune cells. The present study identified for the first time in vivo that Lactobacillus sp. are the central cells of bacterial aggregates in subgingival plaque, and that Streptococcus sp. and the yeast Candida albicans form corncob structures in supragingival plaque. Finally, periodontal pathogens colonize already formed biofilms and form microcolonies therein. These in vivo observations on oral biofilms provide a clear vision on biofilm architecture and the spatial distribution of predominant species.
Introduction
Oral microbial biofilms are three-dimensional structured bacterial communities [1] attached to a solid surface like the enamel of the teeth, the surface of the root or dental implants [2] and are embedded in an exo-polysaccharide matrix [3]. Oral biofilms are exemplary and served as a model system for bacterial adhesion [4], [5] and antibiotic resistance [6].
The appreciation of the complex nature of oral biofilms was highlighted decades ago by the work of Listgarten and co-workers who described the architecture of biofilms by light and electron microscopy on epoxy resin crowns and extracted teeth [7], [8]. Supragingivally, on the enamel, they observed the formation of columnar micro-colonies with their long axis perpendicular to the crown surface. Gram-positive cocci dominated these columns and occasionally, some isolated branching filaments were found after one day of growth. After one week filaments appeared on top of the columns. After three weeks, the biofilm was predominantly filamentous without any sign of cocci left. Filaments seemed to have colonized and subsequently replaced the predominantly coccoid population. A loose layer of so-called corncobs covered the three-week-old biofilm. Corncobs were thought to be bacterial aggregates with a central filamentous cell surrounded by cocci attached to it. After two months, the general features of the biofilm resembled those found at the three weeks time point. Most noticeably was the gingival area, where a fuzzy layer of spirochetes covered the biofilm. This fuzzy layer contained bacterial aggregates resembling test-tube brushes. There were rough and fine types of these brushes. In a study examining biofilm structure at varying degrees of periodontal health, the gingivitis and periodontitis associated biofilms resembled largely the two months old plaque on epoxy resin crowns. Filamentous bacteria were predominant in the biofilm. Between the adhered biofilm and the soft tissue of the pocket, a layer without a well-defined extracellular matrix was observed. This layer consisted of spirochetes, flagellated bacteria and test-tube brushes [8]. The major hindrance of these electron microscopy studies was the inability to identify the species in the biofilm, corncobs or test-tube brushes.
Using fluorescent in situ hybridization (FISH), it was shown for the first time in vivo that initial biofilm formation was the result of co aggregation and adhesion between Streptococcus spp. and Actinomyces spp. [9]. In a later study with the same technique, it was shown in vivo, that after seven days the proportion of streptococci decreased and the proportion of Fusobacterium nucleatum increased [10]. Subgingival biofilms formed on expanded polytetrafluoroethylene carriers that had been inserted into the depth of periodontal pockets have been studied with FISH with only two probes, one with specificity for a large group of oral treponemes and the other recognizing all oral bacteria [11]. The bacterial diversity in the oral cavity is estimated to be more than 700 different species and phylotypes, belonging to nine phyla; Deferribacteres, Spirochaetes, Fusobacteria, Actinobacteria, Firmicutes, Bacteroidetes, Proteobacteria and two phyla without cultiviable members; OP11 and TM7, which is summarized in Figure 1. Little is known about the spatial distribution of these taxa in oral biofilms. The aim of the present study therefore was to reveal the in vivo architecture of supra and subgingival plaque with a panel of 16S or 18S rRNA targeted FISH-probes covering the most important groups of oral microorganisms, and to provide an essential step from oral microbial diversity to oral biofilm function.
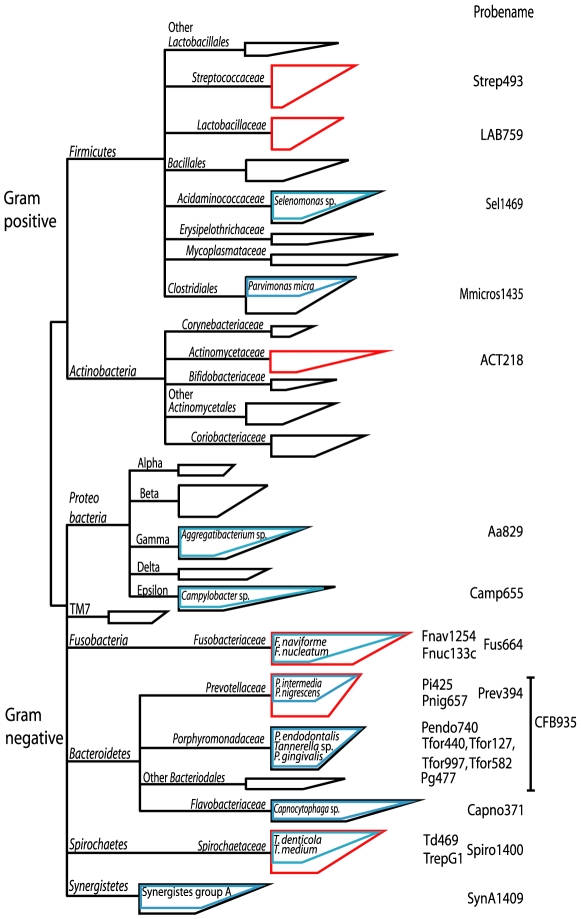
Figure 1. Phylogenetic tree representing oral microbial diversity.
The tree is based on >1500 sequences derived from oral-cavity studies and shows the schematic coverage of the diversity by our probe set. The branching of the tree was simplified for clarity. The boxes represent groups of bacteria. The vertical size of the box reflect the number of sequences and the angular side the genetic diversity. The blue boxes indicate the part that is covered by the species or subgroup-specific probes and the red box indicate genus-specific probes.
Results
From 10 examined teeth, seven showed a positive signal after hybridization with fluorescently labeled probes. Macroscopic analysis of Gram-stained sections revealed the localization of the plaque in relation to the cemento-enamel junction and gingival tissue. A phylogenetic tree based on approximately 1500 nearly complete (>1300 bp) sequences was constructed. Sequences were derived from molecular studies of oral microbial diversity and a manual search through the SILVA database [12]. The coverage of the applied probes is represented in Figure 1. It shows that a representative part of the oral microbial diversity is covered.
Subgingival Biofilm Architecture
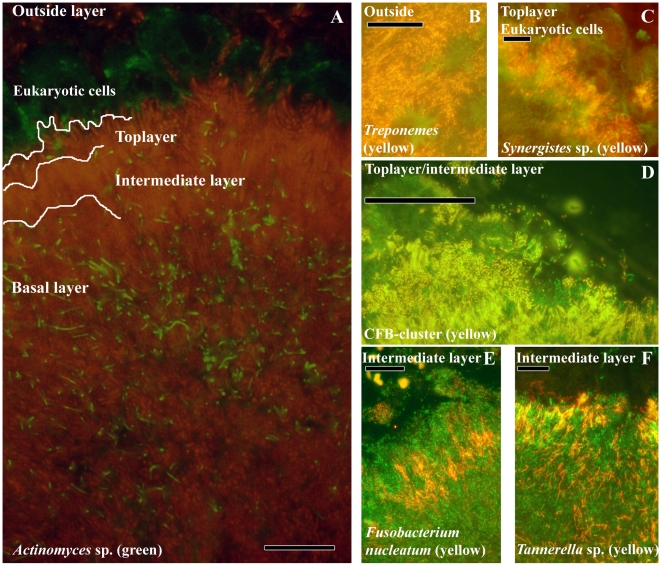
The localization of the most abundant subgingival bacteria is summarized in Figure 2. Panel A shows a typical subgingival biofilm with increasing fluorescent intensity from the tooth side towards the epithelium side. Based on differences in bacterial morphologies and fluorescence intensities, four different layers were distinguished. The first layer of the biofilm is composed of cells displaying little fluorescence relative to cells in the top of the biofilm. Of all the probes tested, only Actinomyces sp. gave a positive signal in this layer. The intermediate layer is composed of many spindle-shaped cells of which F. nucleatum, T. forsythia and possibly other Tannerella sp. positive with probe Tfor127 are visible as the red/yellow band of bacteria in panels E and F of Figure 2. The top layer of the biofilm and part of the intermediate layer contain mainly bacteria belonging to the Cytophaga-Flavobacterium-Bacteroides cluster (CFB-cluster) as detected with probe CFB935 and shown in panel D. CFB935 positive cells are filamentous, rod-shaped or even coccoid. Samples double stained with CFB935 and Tannerella-specific probes showed that most filamentous bacteria are Tannerella sp., while many of the rod-shaped bacteria are Prevotella sp. and Bacteroidetes species as detected with the group-specific probes PREV392 and BAC303, respectively. Besides the presence of bacteria from the CFB-cluster, large cigar-like bacteria were in the top layer. These cells belong to the Synergistetes group A of bacteria and form a ‘palisade’-like lining. They were in close contact to eukaryotic cells resembling polymorphonuclear leukocytes (PMN's) according to the presence of polymorph nuclei (panel C). Outside the biofilm, a fourth layer without clear organization was observed. Spirochaetes were primary localized in the fourth layer where they are the most dominant species. Bacterial aggregates, called rough and fine test-tube brushes were detected between the Spirochaetes (Panel B).
Figure 2. Localization of the most abundant species in subgingival biofilms.
(A) Overview of the subgingival biofilm with Actinomyces sp. (green bacteria), bacteria (red) and eukaryotic cells (large green cells on top). (B) Spirochaetes (yellow) outside the biofilm. (C) Detail of Synergistetes (yellow) in the top layer in close proximity to eukaryotic cells (green). (D) CFB-cluster (yellow) in the top and intermediate layer. (E) F. nucleatum in the intermediate layer. (F) Tannerella sp. (yellow) in the intermediate layer. Each panel is double-stained with probe EUB338 labeled with FITC or Cy3. The yellow color results from the simultaneous staning with FITC and Cy3 labeled probes. Bars are 10 µm.
Supragingival Biofilm Architecture
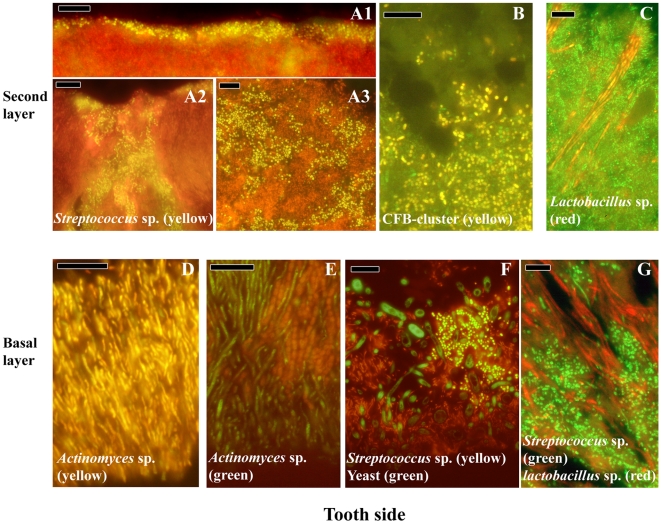
Supragingival biofilms are more heterogeneous in architecture compared to subgingival biofilms. In general, two different layers could be observed. The basal layer adheres to the tooth surface and four different biofilm types were observed (Figure 3). First, a biofilm composed of only rod shaped Actinomyces cells perpendicularly orientated to the tooth surface (panel D). The second type is a mixture of Actinomyces sp. and chains of cocci, not identified as streptococci, perpendicularly orientated to the tooth surface (panel E). The third type shows a biofilm with filamentous bacteria, streptococci and yeasts, where streptococci form a distinct colony around yeast cells (panel F). The fourth type is a biofilm composed of mainly streptococci growing in close proximity to Lactobacillus sp. that are orientated perpendicularly to the tooth surface (panel G).
Figure 3. Localization of the most abundant species in supragingival biofilms.
Streptococcus sp. (yellow) form a thin band on top of the biofilm (A1), almost engulfing in the biofilm (A2) or present as small cells scattered through the top layer of the biofilm (A3). (B) Cells from the CFB-cluster of bacteria in the top layer of the biofilm, without defined structure. (C) Lactobacillus sp. (red) forming long strings through the top layer. (D) Actinomyces sp. (yellow) plaque attached to the tooth. (E) Actinomyces sp. (green) and cocci forming initial plaque.(F) Multispecies initial plaque composed of Streptococcus sp. (yellow), yeast cells (green) and bacteria unidentified (red). (G) Streptococcus sp. (green) and Lactobacillus sp. (red) forming initial plaque. Black holes might be channels through the biofilm. Panels A, B, C, E, F are double stained with probe EUB338 labeled with FITC or Cy3. Bars are 10 µm.
The second layer can be found on top of any biofilm type of the basal layer. Streptococcus sp. can be present as heterogeneously scattered cells through the second layer of the biofilm without any apparent organization (panel A3), or they can be aligned on top of the second biofilm layer as a thin coat (panel A1). In addition, they colonize cracks in the biofilm (panel A2). Also, there is a heterogeneous scattering of bacterial cells belonging to the CFB-cluster (panel B). Finally, Lactobacillus sp. that are orientated away from the tooth surface are surrounded by cells with different morphologies (panel C).
Subgingival Localization of Periodontal Pathogens
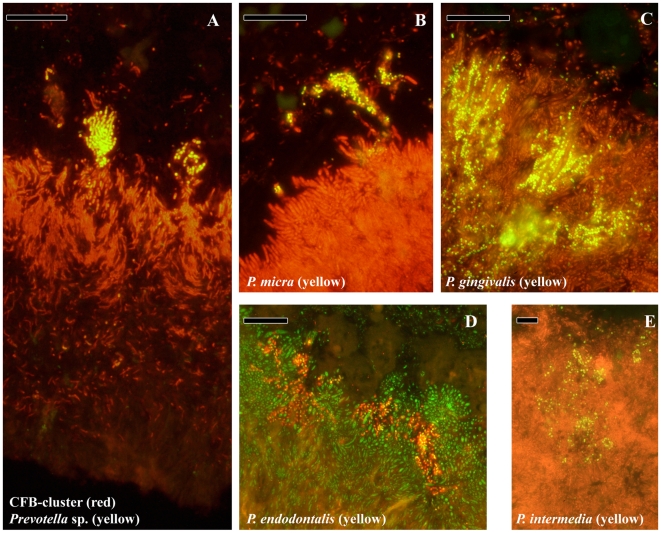
The localization of presumptive periodontal pathogens in the subgingival biofilm is shown in Figure 4. Most of the periodontal pathogens belong to the gram-negative group of bacteria united in the CFB-cluster like P. gingivalis, P. intermedia, P. endodontalis or P.nigrescens. Most CFB-cluster cells are evenly distributed in the top and intermediate layer of the biofilm. Prevotella sp. however, colonize the biofilm in micro-colonies (panel A) which sometimes are located on top of the biofilm, but, as is shown for P. intermedia, also reside within the top layer (panel E). Porphyromonas gingivalis and Porphyromonas endodontalis appear mainly as micro-colonies within the top layer (panel C and D). Parvimonas micra, an example of a gram-positive species that is associated with periodontitis, is also found in micro-colonies in the top layer (panel B). Apparently, the microorganisms considered pathogens are mostly present in micro-colonies in the top layer and in the fourth layer of the subgingival biofilm or can be part of bacterial aggregates.
Figure 4. Localization of species associated with periodontitis.
(A) Overview of the subgingival biofilm with CFB-cluster species (red) and Prevotella sp. (yellow). Since Prevotella sp. are part of the CFB-cluster of bacteria, cells appear in yellow. (B) Top of the biofilm with a micro-colony of P. micra (yellow). (C) Micro-colonies of P. gingivalis (yellow) in the top layer. (D) Micro-colonies of P. endodontalis (yellow) in the top layer. (E) Micro-colonies of P. intermedia in the top layer. Panels B, C, D and E are double stained with probe EUB338 labeled with FITC or Cy3. Bars are 10 µm.
Bacterial Aggregates or Structures in Dental Plaque
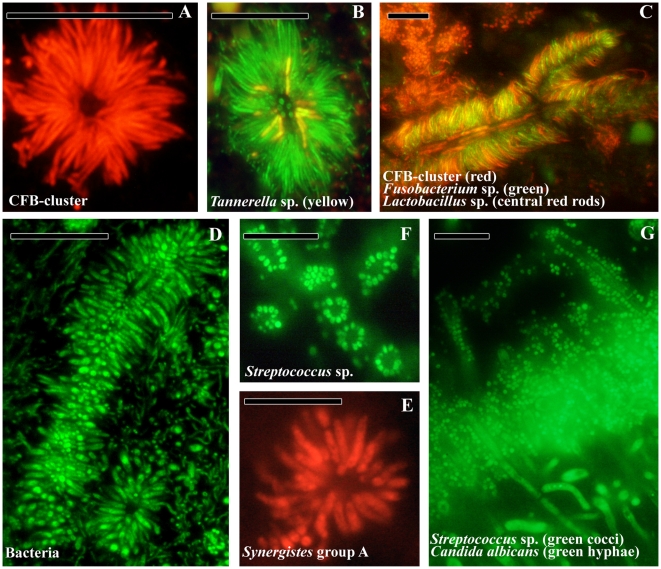
Aggregates of microorganisms have been detected in both sub and supragingival plaque (Figure 5). In line with previous reports, different aggregate morphologies were observed in the fourth layer of subgingival plaque. Filaments from the CFB-cluster, morphologically like T. forsythia, and F. nucleatum are arranged perpendicularly around lactobacilli, forming fine test-tube brushes (panel A–C). There have also been observations of test-tube brushes composed of a complex mixture of cells with T. forsythia, Campylobacter sp., P. micra, Fusobacteria and Synergistetes group A, among others. Synergistetes cells may also form aggregates exclusively with themselves (panel D and E). In supragingival plaque, corncob structures consist of streptococci adhering to a central axis of yeast cells or hyphae.
Figure 5. Bacterial aggregates in oral plaque.
(A) Transversal view of a test-tube brush found in subgingival plaque composed of filamentous cells from the CFB-cluster. (B) Tannerella sp. (yellow) in a test-tube brush. (C) Longitudinal view of a test-tube brush with Lactobacillus sp. (red rods) as central structures. F. nucleatum (green) and CFB-cluster filaments radiating from the central structures. (D) Longitudinal and transversal view of a test-tube brush stained with the eubacterial probe. (E) Transversal view of the test-tube brush in panel D, composed of Synergistetes group A species. (F) Transversal view of Streptococcus sp. (green) aggregation around a central cell (not stained) in supragingival plaque. (G) Transversal view of supragingival plaque with Streptococcus sp. (green cocci) and Candida albicans (green hyphae) in the top layer of the biofilm and forming corn cob structures growing outwards. Bars are 10 µm.
Discussion
The aim of the present study was to unravel tooth attached biofilm architecture. For the first time, bacteria in these biofilms are identified and localized in vivo using FISH. The results present new insights into the architecture of tooth-attached biofilms and visualize the interaction of the biofilm with the human immune system. FISH offers the opportunity to obtain positional information of bacteria in intact biofilms, its application overcomes the limits of culturability and can relatively easily be extended to new identified species and phylotypes. Synergistetes sp. for example, only recently gained attention [13] but may account for 3–11% of the bacterial population [14]. In addition, they form a palisade-like lining along the outer length of the biofilm and were in direct contact with host immune cells suggesting an important role in host-biofilm interactions.
In our current experimental design, group-level probes and species-specific probes were applied to efficiently identify cells that might be of interest due to their location in the biofilm, this was seen with Lactobacillus sp. as the central axis of test-tube brushes and streptococci and Candida as species that form corncobs in supragingival plaque.
Understanding the role of microorganisms in oral diseases needs a unifying concept incorporating biofilm diversity, structure and function. A first oral biofilm model was based on co-aggregation experiments [15]. The model presents a final composition of the biofilm, without taking into consideration the spatio-temporal dynamics of biofilm formation. The figures of the present study are composed of consistent observations from hundreds of thin sections from seven different teeth of four different persons. Each observation provides only a “snap shot” of plaque architecture without quantitative measurements or dynamic time lap observations. Combining multiple observations of supra- and subgingival biofilms reflects to some degree the dynamics of biofilm formation, which leads us to the following view of plaque formation.
Initial plaque formation starts with the deposition of a salivary pellicle on the tooth surface. Planktonic cells or aggregates of cells adhere to this pellicle via specialized adhesins on the bacterial cell surface that recognize pellicle proteins [16] and by non-specific physico-chemical interactions [5]. These phenomena may result in a scattered pattern of bacterial deposits [9], [17] composed of initial colonizers like Actinomyces sp., Streptococcus sp., Lactobacillus sp. and Candida sp. [18]–[20] and is reflected in the different biofilm types of the first layer of supragingival plaque (Figure 3). Maturation of the biofilm proceeds via co-aggregation of planktonic bacteria to the already adhered biofilm [21] and bacterial growth, as has been shown for Streptococcus sanguinis [22]. The second layer may be the result of both processes. The presence of either Streptococcus sp. or bacteria from the CFB-cluster in the second layer of Figure 3 may reflect a crucial transition in supragingival plaque from a predominantly gram-positive saccharolytic plaque to a gram-negative proteolytic plaque that might be the result of the availability of nutrients e.g. dietary sugars or proteins from saliva and crevicular fluids. It was noticed that after three weeks, undisturbed supragingival plaque morphologically resembled subgingival plaque [7]. In our observations of subgingival plaque, the fluorescence intensity of the bacterial cells stained with the eubacterial probe increased from the tooth side towards the epithelium. This reflects differences in physiological activity of the cells [23]. In the basal layer of Figure 2, only Actinomyces sp. showed positive of all the probes tested. The unidentified cells may belong to new species for which no probes have been developed. Another explanation might be that the basal layer constitutes previous stages of the biofilm that have become secluded from nutrients and contain dead or physiologically inactive cells with lower fluorescence intensity as has been shown in vitro [24], [25]. Of the initial colonizers, only Actinomyces sp. might survive, maybe due to their capacity to store intracellular glycogen [26] or their capacity to scavenge on biofilm material like extracellular polymeric substances and on compounds from dead bacterial cells. These are the first in vivo observations of graduated differences in the physiological activity of cells within the biofilm, and support the idea that bacterial growth is an important determent of oral biofilm development [27].
In the intermediate layer, T. forsythia may benefit from its close proximity to dead cells in the basal layer. These may serve as a source of exogenous N-acetyl-muramic acid, a bacterial cell wall sugar on which T. forsythia is dependent [28]. In the presence of F. nucleatum, T. forsythia synergistically forms robust biofilms via cell-cell contacts in vitro [29] as is reflected in their prominent and abundant co-localization of both species along the entire length of the biofilm. The presence of F. nucleatum in the intermediate layer, as proposed by Kolenbrander and London [15], is confirmed for the first time in vivo in the present study. These structural observations on the dominance of Actinomyces sp., Fusobacteria and T. forsythia are supported by dot-blot analysis of subgingival plaque [30]. In contrast, P. gingivalis, P. intermedia, P. endodontalis and P. micra are mainly located in the top layer of the biofilm in micro-colonies [31], [32]. The presence of micro-colonies proposes a distinction between species that are structurally present, probably forming the framework of the biofilm, and transient species that can colonize the already established biofilm forming micro-colonies.
Summarizing, the present study on oral biofilms links early studies on biofilm structure and recent molecular insights in oral bacterial diversity. This resulted in important new observations on oral biofilms, in architecture and in dynamics. First, the species that form test-tube brushes and corncobs are identified for the first time in vivo. Second, the localization of T. forsythia in the intermediate layer of oral biofilms should be incorporated in the biofilm model, as well as a fourth layer of unattached plaque consisting of mainly Spirochaetes. Third, the observation of bacteria that are either structural members of the subgingival biofilm, e.g. Actinomyces, Fusobacteria, Tannerella sp., or species that colonize an already formed biofilm, e.g. P. intermedia, P. gingivalis and P. micra. Fourth, the biofilm model based on co-aggregation should include bacterial growth and appreciate the dynamics of biofilm maturation. Finally, the finding of a palisade lining of Synergistetes sp. in the close proximity to host defence cells suggests a major role in host biofilm interactions. These results provide an oral biofilm model and show that in vivo observations on biofilm architecture are an essential link between molecular diversity and bacterial function in relation to oral diseases.
Materials and Methods
Ethics Statement
All protocols were approved by the Medical Ethical board of the University Medical Center Groningen. Extracted teeth were collected as anonymous by-products of regular treatment. As such, the Medical Ethical board stated that the performed research was not conducted under the regulations of the Act on Medical Research Involving Human Subjects (METc 2009.305). A written informed consent was therefore not compulsory. Nevertheless, patients were informed about the research purposes and gave verbal informed consent, which was not recorded to keep the procedure anonymously.
Sample Collection and Handling
Ten teeth from four patients were used in this study. The patients were referred to the Dept. of Oral Surgery and Periodontology for extraction of their remaining teeth and the fabrication of complete dentures. Teeth were diagnosed with advanced generalized periodontitis based on pocket depth recordings of >6 mm and x-rays indicating more than 30% bone loss. Subjects had not taken antibiotics within the last three months and did not suffer from systemic diseases. An experienced dentist carefully extracted the teeth without the use of elevators, not to disturb the subgingival plaque. Immediately after extraction, the teeth were placed in 3% (wt/vol) paraformaldehyde (PFA) in phosphate-buffered saline (PBS) (8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4 and 0.24 g of KH2PO4 per liter; pH 7.2) and fixed at 4°C for 16 h. Fixed teeth were dehydrated in 50%, 60%, 70%, 80%, 90% and 100% (vol/vol) ethanol-PBS in subsequent sessions of 1 hour. Teeth were either stored in 60% (vol/vol) ethanol-PBS at −20°C until further use or processed immediately.
Sample Processing
Fixed and dyhydrated teeth were carefully embedded in Technovit 8100 (Heraeus Kultzer GmbH) at 4°C. The embedded tissue was either cut transversal or a combination of longitudinal and transversal (50/50) in cross-sections of 1 mm with a water-cooled rotary saw. The embedded cross-sections were decalcified with a 17% ethylene-diamine-tetraacetic acid decalcifying solution (pH 7.0), which was renewed regularly during the course of decalcification, varying in duration from 16 to 22 days depending on the size of the specimens. Regular x-ray analysis confirmed completion of decalcification. Decalcified cross-sections were re-embedded in Technovit 8100 and stored at 4°C. Sections of two micron were obtained with a Tungsten carbide knife in a rotary microtome (Reichart-Jung) and stretched on water. Stretched sections were mounted to polysine precoated glass slides (Thermo Scientific) for FISH analysis.
FISH Analysis
Oligonucleotide probes, labeled at the 5′- and 3′ end with fluorescein (FITC) or at the 5′- end with Cy3 were purchased from Eurogentec (Eurogentec, Maastricht, the Netherlands). A set of 29 FISH probes, specific at the domain or group level were used together with species-specific probes (Table 1). The target bacteria of probe LAB759 needed pre-treatment with Labmix (25 mM Tris-HCl pH 7.5, 10 mM EDTA, 585 mM sucrose, 5 mM CaCl2, 0.3 mg/ml sodiumtaurocholate, 0.1 mg/ml lipase and 2 mg/ml lysozyme) for 1 h at 37°C. To enable probe penetration, other gram-positive targets needed lysozyme pre-treatment for 15 min at room temperature with lysozyme buffer (2 mg/ml lysozyme, 100 mM Tris-HCl, pH 8.0) as indicated in Table 1. Standard FISH procedures were followed with a hybridization time of three hours and formamide concentration and hybridization temperature (46°C or 50°C) according to the references (Table 1), with the aim of achieving optimal stringency and specificity. The biofilms were examined using a Leica DM RXA microscope (Leica Mikroskopie). Filters were set to 500–540 nm for FITC and 570–630 nm for Cy3. Images were obtained using 63× (numeric aperture 1.0) oil immersion objectives. Color micrographs were taken with a digital Canon EOS400 Camera, transferred to an HP personal computer and processed using Photoshop 6.0 (Adobe) without any qualitative changes to the raw images.
Table 1. Oligonucleotide probe sequences, their targets and the hybridization conditions used in this study.
| Target | Probename | Label | Sequence 5′→3′ | Tm (°C) | Pretreatment | Formamide (%) | Hyb. Temp. (°C) | Reference |
| most Bacteria | EUB338 | FITC/Cy3 | GCT GCC TCC CGT AGG AGT | 55 | No | 0–50 | 50 | [33] |
| Eukarya | EUK502 | FITC | ACC AGA CTT GCC CTC C | 52 | No | 15–45 | 50 | [33] |
| Candida albicans | CAAL | Cy3 | GCC AAG GCT TAT ACT CGC T | 51 | No | 30 | 50 | [34] |
| most Streptococcus spp. and some Lactococcus spp. | STR493 | FITC | GTT AGC CGT CCC TTT CTG G | 53 | 15 min 37°C lysis buffer | 0 | 50 | [35] |
| Lactobacillus sp., Ruminococcaceae sp. and Pediococcus sp. | LAB759 | Cy3 | CTA CCC ATR CTT TCG AGC C | 59 | 60 min 37°C labmix | 35 | 46 | This study |
| Parvimonas micra | Mmicros1435 | FITC | GCC GCC GAT CTA ACC GCA | 64 | No | 0 | 50 | [36] |
| Selenomonas sp.but not S. sputigena | Sel1469 | Cy3 | CCA GTC ACC TTC CCC ACC | 58 | No | 30–50 | 46 | This study |
| Synergistetes group A | SynA1409 | Cy3 | ACA CCC GGC TCG GGT GGT | 62 | No | 40–50 | 46 | This study |
| Actinomyces sp. | ACT218 | FITC | CGA GCC CAT CCC CCA CCA | 57 | 15 min 37°C lysis buffer | 0 | 50 | This study |
| most Fusobacterium sp. | Fus664 | FITC | CTT GTA GTT CCG CYT ACC TC | 52 | No | 40 | 46 | [37] |
| F. naviforme, F. nucleatum subsp. fusiforme | Fnav1254 | FITC | CTT CAC AGC TTT GCG ACT C | 58 | No | 30 | 46 | This study |
| F. nucleatum, F. periodonticum | Fnuc133c | FITC | GTT GTC CCT ANC TGT GAG GC | 46 | No | 30 | 46 | [37] |
| subgroup of the Spirochaetaceae | SPIRO1400 | FITC | CTC GGA TGG TGT GAC GGG CG | 60 | No | ND | 50 | [38] |
| T. medium and T. denticola | TrepG1 | Cy3 | GAT TCC ACC CCT ACA CTT | 58 | No | 20–50 | 46 | This study |
| T. denticola | Td469 | FITC | CAT GAC TAC CGT CAT CAA AGA AGC | 56 | No | ND | 50 | [39] |
| A. actinomycetemcomitans | Aa829 | FITC | GGG CTA AAC CCC AAT CCC | 53 | No | ND | 50 | This study |
| Campylobacter sp. | Camp655 | FITC | CAT CTG CCT CTC CCT YAC | 57 | No | 30 | 46 | [40] |
| Cytophaga-Flavobacterium-Bacteroides cluster | CFB935 | Cy3 | CCA CAT GTT CCT CCG CTT GT | 54 | No | >40 | 50 | [38] |
| Bacteroidaceae and Prevotellaceae | BAC303 | Cy3 | CCA ATG TGG GGG ACC TT | 49 | No | 0 | 50 | [41] |
| Prevotella sp. | Prev394 | FITC | GCA CGC TAC TTG GCT GG | 56 | No | 25 | 46 | [17] |
| P. intermedia | Pi425 | FITC | CTT TAC TCC CCA ACA AAA GCA GTT TAC AA | 57 | No | 20 | 46 | [42] |
| P. nigrescens | Pnig657 | Cy3 | TCC GCC TGC GCT GCG TGT A | 64 | No | ≤40 | 46 | [43] |
| T. forsythia | Tfor582 | Cy3 | GCG GAC TTA ACA GCC CAC CT | 64 | No | 40 | 46 | [44] |
| T. forsythia | Tfor440 | Cy3 | CGT ATC TCA TTT TAT TCC CCT GTA | 52 | No | 20 | 46 | [42] |
| T. forsythia | Tfor127 | Cy3 | CTC TGT TGC GGG CAG GTT AC | 65 | No | 40 | 46 | [44] |
| T. forsythia | Tfor997 | Cy3 | TCA CTC TCC GTC GTC TAC | 56 | No | 40 | 46 | [44] |
| P.gingivalis | Pg477 | FITC | CAA TAC TCG TAT CGC CCG TTA TTC | 56 | No | 20 | 46 | [42] |
| P. endodontalis | Pendo740 | Cy3 | CAG TGT CAG ACG GAG CCT | 58 | No | 30–40 | 46 | This study |
| Capnocytophaga genus | Capno371 | FITC | TCA GTC TTC CGA CCA TTG | 54 | No | 0 | 50 | [45] |
New probes developed in the present study have been designed with the ARB software package [46] and tested against a panel of reference strains for specificity (ACT218, Aa829, LAB759, Sel1469 and Fnav1254) or tested on subgingival plaque samples at increasing formamide concentrations to define assay conditions for maximum stringency and optimal specificity (TrepG1, Pendo740 and SynA1409). Probe LAB759 showed cross-reactivity with Eikenella corrodens and not identified cocci. In the present study, only rods gave a positive signal with probe LAB759.
Acknowledgments
We thank Helga Lüthi-Schaller for laboratory assistance and Rudi Tonk for the microscope set-up. Linda Wildeboer-Veloo is gratefully acknowledged for validation of probe ACT218.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Junior Scientific Master class program of the University Medical Center of the University of Groningen. RG and TT were supported by the University of Zürich. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wood SR, Kirkham J, Marsh PD, Shore RC, Nattress B, et al. Architecture Of Intact Natural Human Plaque Biofilms Studied By Confocal Laser Scanning Microscopy. J Dent Res. 2000;79:21–27. doi: 10.1177/00220345000790010201. [DOI] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD. Dental Biofilms: Difficult Therapeutic Targets. Periodontol 2000. 2002;28:12–55. doi: 10.1034/j.1600-0757.2002.280102.x. [DOI] [PubMed] [Google Scholar]
- 3.Reese S, Guggenheim B. A Novel TEM Contrasting Technique For Extracellular Polysaccharides In In Vitro Biofilms. Microsc Res Tech. 2007;70:816–822. doi: 10.1002/jemt.20471. [DOI] [PubMed] [Google Scholar]
- 4.Bos R, Van Der Mei HC, Busscher HJ. Physico-Chemistry Of Initial Microbial Adhesive Interactions–Its Mechanisms And Methods For Study. FEMS Microbiol Rev. 1999;23:179–230. doi: 10.1111/j.1574-6976.1999.tb00396.x. [DOI] [PubMed] [Google Scholar]
- 5.Busscher HJ, Van Der Mei HC. Physico-Chemical Interactions In Initial Microbial Adhesion And Relevance For Biofilm Formation. Adv Dent Res. 1997;11:24–32. doi: 10.1177/08959374970110011301. [DOI] [PubMed] [Google Scholar]
- 6.Zaura-Arite E, Van Marle J, Ten Cate JM. Conofocal Microscopy Study Of Undisturbed And Chlorhexidine-Treated Dental Biofilm. J Dent Res. 2001;80:1436–1440. doi: 10.1177/00220345010800051001. [DOI] [PubMed] [Google Scholar]
- 7.Listgarten MA, Mayo HE, Tremblay R. Development Of Dental Plaque On Epoxy Resin Crowns In Man. A Light And Electron Microscopic Study. J Periodontol. 1975;46:10–26. doi: 10.1902/jop.1975.46.1.10. [DOI] [PubMed] [Google Scholar]
- 8.Listgarten MA. Structure Of The Microbial Flora Associated With Periodontal Health And Disease In Man. A Light And Electron Microscopic Study. J Periodontol. 1976;47:1–18. doi: 10.1902/jop.1976.47.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Palmer RJ, Jr, Gordon SM, Cisar JO, Kolenbrander PE. Coaggregation-Mediated Interactions Of Streptococci And Actinomyces Detected In Initial Human Dental Plaque. J Bacteriol. 2003;185:3400–3409. doi: 10.1128/JB.185.11.3400-3409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Ahmad A, Wunder A, Auschill TM, Follo M, Braun G, et al. The In Vivo Dynamics Of Streptococcus Spp., Actinomyces Naeslundii, Fusobacterium Nucleatum And Veillonella Spp. In Dental Plaque Biofilm As Analysed By Five-Colour Multiplex Fluorescence In Situ Hybridization. J Med Microbiol. 2007;56:681–687. doi: 10.1099/jmm.0.47094-0. [DOI] [PubMed] [Google Scholar]
- 11.Wecke J, Kersten T, Madela K, Moter A, Göbel UB, et al. A Novel Technique For Monitoring The Development Of Bacterial Biofilms In Human Periodontal Pockets. FEMS Microbiol Lett. 2000;191:95–101. doi: 10.1111/j.1574-6968.2000.tb09324.x. [DOI] [PubMed] [Google Scholar]
- 12.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. SILVA: A Comprehensive Online Resource For Quality Checked And Aligned Ribosomal RNA Sequence Data Compatible With ARB 1. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz P, Hooper SD, Kyrpides NC. Focus: Synergistetes. Environ Microbiol. 2009;11:1327–1329. doi: 10.1111/j.1462-2920.2009.01949.x. [DOI] [PubMed] [Google Scholar]
- 14.Vartoukian SR, Palmer RM, Wade WG. Diversity And Morphology Of Members Of The Phylum “Synergistetes” In Periodontal Health And Disease. Appl Environ Microbiol. 2009;75:3777–3786. doi: 10.1128/AEM.02763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolenbrander PE, London J. Adhere Today, Here Tomorrow: Oral Bacterial Adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao Y, Berg EA, Costello CE, Troxler RF, Oppenheim FG. Identification Of Protein Components In Human Acquired Enamel Pellicle And Whole Saliva Using Novel Proteomics Approaches. J Biol Chem. 2003;278:5300–5308. doi: 10.1074/jbc.M206333200. [DOI] [PubMed] [Google Scholar]
- 17.Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, et al. Molecular Characterization Of Subject-Specific Oral Microflora During Initial Colonization Of Enamel. Appl Environ Microbiol. 2006;72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Helmerhorst EJ, Leone CW, Troxler RF, Yaskell T, et al. Identification Of Early Microbial Colonizers In Human Dental Biofilm. J Appl Microbiol. 2004;97:1311–1318. doi: 10.1111/j.1365-2672.2004.02420.x. [DOI] [PubMed] [Google Scholar]
- 19.Hannig C, Hannig M, Rehmer O, Braun G, Hellwig E, et al. Fluorescence Microscopic Visualization And Quantification Of Initial Bacterial Colonization On Enamel In Situ. Arch Oral Biol. 2007;52:1048–1056. doi: 10.1016/j.archoralbio.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 20.Dige I, Raarup MK, Nyengaard JR, Kilian M, Nyvad B. Actinomyces Naeslundii In Initial Dental Biofilm Formation. Microbiology. 2009;155:2116–2126. doi: 10.1099/mic.0.027706-0. [DOI] [PubMed] [Google Scholar]
- 21.Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, et al. Bacterial Interactions And Successions During Plaque Development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 22.Lai CH, Listgarten MA, Rosan B. Immunoelectron Microscopic Identification And Localization Of Streptococcus Sanguis With Peroxidase-Labeled Antibody: Localization Of Streptococcus Sanguis In Intact Dental Plaque. Infect Immun. 1975;11:200–210. doi: 10.1128/iai.11.1.200-210.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moter A, Göbel UB. Fluorescence In Situ Hybridization (FISH) For Direct Visualization Of Microorganisms. J Microbiol Methods. 2000;41:85–112. doi: 10.1016/s0167-7012(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 24.Auschill TM, Arweiler NB, Netuschil L, Brecx M, Reich E, et al. Spatial Distribution Of Vital And Dead Microorganisms In Dental Biofilms. Arch Oral Biol. 2001;46:471–476. doi: 10.1016/s0003-9969(00)00136-9. [DOI] [PubMed] [Google Scholar]
- 25.Netuschil L, Reich E, Unteregger G, Sculean A, Brecx M. A Pilot Study Of Confocal Laser Scanning Microscopy For The Assessment Of Undisturbed Dental Plaque Vitality And Topography. Arch Oral Biol. 1998;43:277–285. doi: 10.1016/s0003-9969(97)00121-0. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi N, Yamada T. Glucose And Lactate Metabolism By Actinomyces Naeslundii. Crit Rev Oral Biol Med. 1999;10:487–503. doi: 10.1177/10454411990100040501. [DOI] [PubMed] [Google Scholar]
- 27.Guggenheim M, Shapiro S, Gmür R, Guggenheim B. Spatial Arrangements And Associative Behavior Of Species In An In Vitro Oral Biofilm Model. Appl Environ Microbiol. 2001;67:1343–1350. doi: 10.1128/AEM.67.3.1343-1350.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyss C. Dependence Of Proliferation Of Bacteroides Forsythus On Exogenous N-Acetylmuramic Acid. Infect Immun. 1989;57:1757–1759. doi: 10.1128/iai.57.6.1757-1759.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma A, Inagaki S, Sigurdson W, Kuramitsu HK. Synergy Between Tannerella Forsythia And Fusobacterium Nucleatum In Biofilm Formation. Oral Microbiol Immunol. 2005;20:39–42. doi: 10.1111/j.1399-302X.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 30.Socransky SS, Haffajee AD. Periodontal Microbial Ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- 31.Noiri Y, Ozaki K, Nakae H, Matsuo T, Ebisu S. An Immunohistochemical Study On The Localization Of Porphyromonas Gingivalis, Campylobacter Rectus And Actinomyces Viscosus In Human Periodontal Pockets. J Periodontal Res. 1997;32:598–607. doi: 10.1111/j.1600-0765.1997.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 32.Noiri Y, Li L, Ebisu S. The Localization Of Periodontal-Disease-Associated Bacteria In Human Periodontal Pockets. J Dent Res. 2001;80:1930–1934. doi: 10.1177/00220345010800101301. [DOI] [PubMed] [Google Scholar]
- 33.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, et al. Combination Of 16S Rrna-Targeted Oligonucleotide Probes With Flow Cytometry For Analyzing Mixed Microbial Populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kempf VA, Trebesius K, Autenrieth IB. Fluorescent In Situ Hybridization Allows Rapid Identification Of Microorganisms In Blood Cultures. J Clin Microbiol. 2000;38:830–838. doi: 10.1128/jcm.38.2.830-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, et al. Variations Of Bacterial Populations In Human Feces Measured By Fluorescent In Situ Hybridization With Group-Specific 16S Rrna-Targeted Oligonucleotide Probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wildeboer-Veloo AC, Harmsen HJ, Welling GW, Degener JE. Development Of 16S Rrna-Based Probes For The Identification Of Gram-Positive Anaerobic Cocci Isolated From Human Clinical Specimens. Clin Microbiol Infect. 2007;13:985–992. doi: 10.1111/j.1469-0691.2007.01803.x. [DOI] [PubMed] [Google Scholar]
- 37.Gmür R, Wyss C, Xue Y, Thurnheer T, Guggenheim B. Gingival Crevice Microbiota From Chinese Patients With Gingivitis Or Necrotizing Ulcerative Gingivitis. Eur J Oral Sci. 2004;112:33–41. doi: 10.1111/j.0909-8836.2004.00103.x. [DOI] [PubMed] [Google Scholar]
- 38.Daly K, Shirazi-Beechey SP. Design And Evaluation Of Group-Specific Oligonucleotide Probes For Quantitative Analysis Of Intestinal Ecosystems: Their Application To Assessment Of Equine Colonic Microflora 8. FEMS Microbiol Ecol. 2003;44:243–252. doi: 10.1016/S0168-6496(03)00032-1. [DOI] [PubMed] [Google Scholar]
- 39.Moter A, Hoenig C, Choi BK, Riep B, Göbel UB. Molecular Epidemiology Of Oral Treponemes Associated With Periodontal Disease. J Clin Microbiol. 1998;36:1399–1403. doi: 10.1128/jcm.36.5.1399-1403.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gmür R, Lüthi-Schaller H. A Combined Immunofluorescence And Fluorescent In Situ Hybridization Assay For Single Cell Analyses Of Dental Plaque Microorganisms. J Microbiol Methods. 2007;69:402–405. doi: 10.1016/j.mimet.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 41.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer KH. Application Of A Suite Of 16S Rrna-Specific Oligonucleotide Probes Designed To Investigate Bacteria Of The Phylum Cytophaga-Flavobacter-Bacteroides In The Natural Environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 42.Sunde PT, Olsen I, Göbel UB, Theegarten D, Winter S, et al. Fluorescence In Situ Hybridization (FISH) For Direct Visualization Of Bacteria In Periapical Lesions Of Asymptomatic Root-Filled Teeth. Microbiology. 2003;149:1095–1102. doi: 10.1099/mic.0.26077-0. [DOI] [PubMed] [Google Scholar]
- 43.Gmür R, Thurnheer T. Direct Quantitative Differentiation Between Prevotella Intermedia And Prevotella Nigrescens In Clinical Specimens. Microbiology. 2002;148:1379–1387. doi: 10.1099/00221287-148-5-1379. [DOI] [PubMed] [Google Scholar]
- 44.Züger J, Lüthi-Schaller H, Gmür R. Uncultivated Tannerella BU045 And BU063 Are Slim Segmented Filamentous Rods Of High Prevalence But Low Abundance In Inflammatory Disease-Associated Dental Plaques. Microbiology. 2007;153:3809–3816. doi: 10.1099/mic.0.2007/010926-0. [DOI] [PubMed] [Google Scholar]
- 45.Kampinga GA, Bollen AE, Harmsen HJ, De Vries-Hospers HG. [Meningitis After A Superficial Dog Bite]. Ned Tijdschr Geneeskd. 2002;146:73–76. [PubMed] [Google Scholar]
- 46.Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. ARB: A Software Environment For Sequence Data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]