Abstract
Background
Abnormalities in the circadian clockwork often characterize patients with major depressive and bipolar disorders. Circadian clock genes are targets of interest in these patients. CRY2 is a circadian gene that participates in regulation of the evening oscillator. This is of interest in mood disorders where a lack of switch from evening to morning oscillators has been postulated.
Principal Findings
We observed a marked diurnal variation in human CRY2 mRNA levels from peripheral blood mononuclear cells and a significant up-regulation (P = 0.020) following one-night total sleep deprivation, a known antidepressant. In depressed bipolar patients, levels of CRY2 mRNA were decreased (P = 0.029) and a complete lack of increase was observed following sleep deprivation. To investigate a possible genetic contribution, we undertook SNP genotyping of the CRY2 gene in two independent population-based samples from Sweden (118 cases and 1011 controls) and Finland (86 cases and 1096 controls). The CRY2 gene was significantly associated with winter depression in both samples (haplotype analysis in Swedish and Finnish samples: OR = 1.8, P = 0.0059 and OR = 1.8, P = 0.00044, respectively).
Conclusions
We propose that a CRY2 locus is associated with vulnerability for depression, and that mechanisms of action involve dysregulation of CRY2 expression.
Introduction
Rhythms that approximate the 24-hour day-night cycle or light-dark transitions are called circadian. Abnormalities of the circadian pacemaker system is often seen in mood disorders (i.e. major depressive and bipolar disorders) as evidenced with sleep stage abnormalities and the clinical efficacy of sleep deprivation [1] as well as with the therapeutic mechanisms of lithium [2] that is prescribed primarily for bipolar disorder. Approximately one tenth of all mood disorders follow a seasonal pattern and area hence categorized as seasonal affective disorder (SAD) [3]. Season-bound mood episodes may occur in both depressive and bipolar disorders and emerge in any season, but the most common type is winter depression, a condition in which major depressive episodes routinely occur in the wintertime and remit the following summer [4]. In one study as much as 93% of the winter depression cases had a diagnosis of bipolar disorder [5] although other studies tend to show a greater proportion of unipolar recurrent major depression among winter depression cases [4]. Depressive episodes in winter depression are highly recurrent and appear to be clearly endogenous as there is no psychosocial factor that would account for their onset. For bipolar type 1 disorder, the heritability estimate is very high [6]. Therefore, winter depression, whether part of major depressive disorder or bipolar disorder, as well as bipolar type 1 disorder, with or without a seasonal pattern, provide excellent models for studying the molecular mechanisms of mood disorder [7], [8].
Advances and delays in phase (the location within a cycle at a particular time) and reduced amplitudes (intensities of the oscillations) have been reported in adult patients [9]–[11], while children with winter depression have circadian rhythms that are accurate in phase but low in amplitude [12]. This agrees with the hypothesis that not only phase shifts but also amplitude attenuations [13] contribute to the pathogenesis, and suggests that there are circadian clockwork abnormalities having relevance to the daily reset and synchronization. This is further supported by findings of the seasonal changes in sensitivity to light exposure in winter depression, these patients having supersensitivity to light in terms of melatonin suppression during winter [14], abnormal melatonin levels in patients with seasonal or non-seasonal depressive disorder [15], and abnormalities in circadian alignments in patients with bipolar disorder [16]. Therefore, the internal misalignment (i.e. the sleep-wake cycle is no longer in phase with the circadian rhythms) may account for the pathogenesis of mood disorders in general. Light exposures and sleep manipulations in patients with mood disorder are tools for exploration and elucidation of the mechanisms driving the circadian and seasonal clockworks.
CRY proteins [17] differ from many transcription factors that take part in the circadian clockwork, since they have no PAS domains. This suggests that they are unique in the core of the circadian clockwork where they act [18]. Both CRY1 and CRY2 operate in the retina and non-visual light detection pathways in a manner that is important for the internal alignment [19]–[24]. Of these two, CRY2 is highly expressed in the brain in particular and has a dose-dependent inhibitory effect on the activated ARNTL, whereas both CRY2 and CRY1 repress all four combinations of the ARNTL (ARNTL2) – CLOCK (NPAS2) protein heterodimers that act as transcriptional activators in the core of the circadian clockwork [25]. These basic findings give a rationale for the CRY2 gene as a target of high interest and relevance in our study.
Here, we assessed CRY2 gene expression in eight healthy volunteers for 48 hours and in 13 patients with bipolar disorder before and after a one-night sleep deprivation, and we report that CRY2 mRNA levels are reduced and unresponsive to sleep deprivation in depressed patients with bipolar disorder. To determine whether CRY2 genetic variation is associated with depression, we analyzed circadian clock gene variants in two separate population-based samples, a Swedish sample and a Finnish sample, and report that the CRY2 gene is associated with winter depression.
Results
CRY2 Expression Analysis
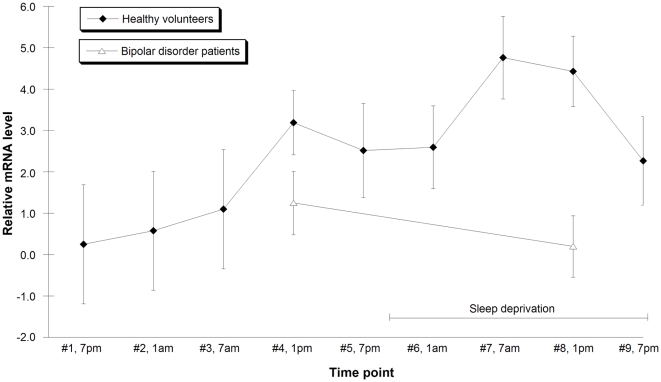
CRY2 mRNA levels displayed marked diurnal variation in healthy controls (n = 8). In addition, total sleep deprivation induced a 2.0-fold increase in CRY2 mRNA levels (P = 0.020) in the controls (Figure 1). The control gene GAPDH mRNA levels did not show diurnal variation and were not changed by the effect of sleep deprivation (P = 0.66).
Figure 1. CRY2 gene expression in healthy volunteers and bipolar disorder patients.
CRY2 mRNA expression was not changed by sleep deprivation in bipolar disorder patients (triangle) and was significantly decreased compared to healthy volunteers (square) during sleep deprivation. Blood draw 1 (x-axis) was at 7 p.m., then there was a blood draw every 6 hours. Sleep deprivation was at time points #6, #7, #8 and #9. Bars indicate mean and error bars indicate SEM.
In samples from patients in a depressive state of bipolar disorder (n = 13), sleep deprivation did not induce any increase in CRY2 mRNA levels. CRY2 mRNA expression was significantly decreased in the samples from patients as compared with the samples from healthy controls during sleep deprivation (P = 0.041 for the diagnosis and sleep deprivation interaction using ANOVA; P = 0.029 for the difference between patients and controls during sleep deprivation in post hoc test). GAPDH showed similar mRNA levels in the samples from controls to those in the bipolar disorder samples (P = 0.12).
Genetic Association Analysis
Analyses were performed on two samples of clinical cases with a diagnosis of winter depression (DSM-IV) and healthy screened controls (Swedish sample: 118 cases and 1011 controls, Finnish sample: 86 cases and 1096 controls). CRY2 allele frequency was significantly associated with winter depression in the Swedish sample, as shown with three SNPs (rs10838524 risk allele A, rs10838527 risk allele G, and rs3824872 risk allele A) having OR = 1.6, OR = 2.1 and OR = 1.8 (P = 0.0017, P = 0.00074 and 0.00070, respectively). The association to rs10838524 was confirmed in the Finnish sample (OR = 1.7, P = 0.0020) (Table 1) and indicated that homozygosity of the minor allele (overall minor allele frequency of about 0.5) was increased in the cases in both population samples (Table 2). The associated SNPs (rs10838527 and rs3824872) were present in LD block 1 (rs7123390, rs10838527, rs3824872) in both samples (Figures 2A and 2B).
Table 1. SNP allele frequency association analysis.
| SNP | Location | Alleles | MAF: A/U | OR [95% CI] a | P value a | OR [95% CI] b | Empirical P b,c |
| Swedish sample | |||||||
| rs10838524 | Intron 1 | A*/G | 0.54/0.45 | 1.65 [1.21–2.25] | 0.0017 | 1.41 [1.07–1.86] | 0.013 |
| rs7123390 | Intron 7 (50 bp from exon7) | A/G* | 0.28/0.28 | 0.97 [0.69–1.37] | 0.88 | 1.00 [0.73–1.40] | 0.96 |
| rs10838527 | Exon 12 in 3′ UTR | G/A* | 0.13/0.09 | 2.11 [1.37–3.26] | 0.00074 | 1.73 [1.15–2.61] | 0.010 |
| rs3824872 | Downstream 3′ UTR | A/C* | 0.27/0.20 | 1.76 [1.27–2.43] | 0.00070 | 1.49 [1.09–2.04] | 0.014 |
| Finnish sample | |||||||
| rs10838524 | Intron 1 | G/A* | 0.60/0.47 | 1.69 [1.21–2.36] | 0.0020 | 1.72 [1.24–2.39] | 0.0021 |
| rs7123390 | Intron 7 (50 bp from exon7) | A/G* | 0.19/0.29 | 0.60 [0.40–0.91] | 0.015 | 0.59 [0.40–0.87] | 0.016 |
| rs10838527 | Exon 12 in 3′ UTR | G/A* | 0.089/0.13 | 0.68 [0.39–1.19] | 0.18 | 0.68 [0.39–1.19] | 0.18 |
| rs3824872 | Downstream 3′ UTR | A/C* | 0.20/0.26 | 0.70 [0.47–1.05] | 0.089 | 0.70 [0.47–1.05] | 0.089 |
Alleles, minor allele first.
OR, proportion of minor versus major allele among affected (A)/proportion of minor versus major allele among non-affected (U).
Logistic regression with gender as covariate. P<0.05 are shown.
No covariate.
Point-wise P-value from 10,000 permutations with no covariate (EMP1).
Ancestral allele in CEU population data (CEPH (Utah residents with ancestry from northern and western Europe)) from www.hapmap.org.
Table 2. Genotype association analysis of the SNPs with suggestive allele frequency association.
| SNP | Cases aa/ab/bb (%) | Cases n | Controls aa/ab/bb (%) | Controls n | Cochran-Armitage trend P value | Minor allele dominant P | Minor allele recessive P |
| Swedish sample | |||||||
| rs10838524 | 30/48/22 | 113 | 20/51/29 | 906 | 0.014 | 0.14 | 0.011 |
| rs10838527 | 3.5/20/76 | 113 | 0.52/16/84 | 951 | 0.0085 | 0.048 | 0.010 |
| rs3824872 | 13/28/58 | 113 | 4.2/32/64 | 927 | 0.014 | 0.27 | 0.000041 |
| Finnish sample | |||||||
| rs10838524 | 38/43/18 | 76 | 22/49/29 | 1 039 | 0.0015 | 0.047 | 0.0014 |
| rs7123390 | 3.8/32/64 | 78 | 7.9/42/52 | 1 066 | 0.014 | 0.019 | 0.27 |
Allele ‘a’ is the minor allele.
Note that the identity of the rs10838524 minor allele differs between the Swedish and Finnish samples.
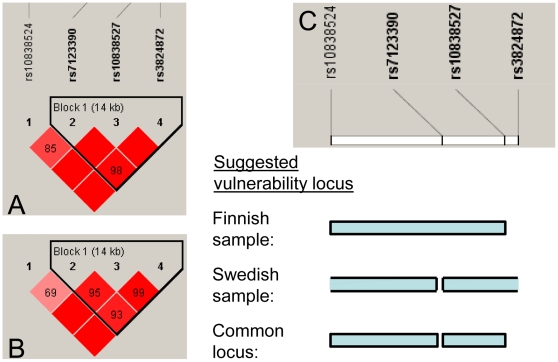
Figure 2. The suggested vulnerability locus in CRY2.
LD structure of the SNPs analyzed in A) the Swedish sample, and B) the Finnish sample. C) Scheme of the suggested location of the functional variant in the Finnish sample, in the Swedish sample, and the common locus relevant under the assumption of the same functional variant in the two samples. An open end of locus box (seen for the Swedish sample) indicates that this is not the locus border. The numbers in the squares of A and B) represent the pair-wise D' value, empty squares stand for D' = 1. Pink-red color indicates a pair-wise LOD ≥2 with redness proportional to D'. Haplotype blocks are formed if 95% of comparisons are “strong LD”, i.e. the 95% CI of D' is within [0.7–0.98].
Four haplotypes were identified and a significant difference in distribution of haplotypes between the cases and controls was found for the Swedish sample (χ2 = 8.7, P = 0.034) and the Finnish sample (χ2 = 14.6, P = 0.0022; see Supplemental data Table S1). A haplotype of the risk alleles GGA was more frequent in the Swedish cases compared with controls (OR = 1.7, P = 0.012) and even more so when adding the rs10838524 risk allele to the haplotype to be AGGA (OR = 1.8, P = 0.0059; Table 3). The block 1 (rs7123390, rs10838527, rs3824872) haplotype GAC which in the Swedish sample has the protective alleles from rs10838527 and rs3824872 was a risk haplotype in the Finnish sample (OR = 1.8, P = 0.00010), and similarly so when adding the rs10838524 allele G to the haplotype to be GGAC (OR = 1.8, P = 0.00044; Table 3). The Finnish data suggest a vulnerability locus increasing the risk for winter depression upstream rs10838527. This was further supported by the fact that changing only the rs7123390 allele in the Finnish risk haplotype (GAC) turned it into a protective haplotype (AAC) in the Finnish sample (OR = 0.61, P = 0.012), and the fact that the AGAC was non-existent of in the samples. Considering only the Swedish data the vulnerability locus could reside on either side of, but not exactly at rs7123390. If assuming the same functional variation (although probably opposite risk alleles in Swedes and Finns), the Swedish and the Finnish samples together hence suggest the vulnerability locus to reside in-between rs7123390 and rs10838527 or upstream rs7123390 (Figure 2C).
Table 3. Haplotype association analysis.
| SNPs | Haplotype | Frequency cases | Frequency controls | OR [95% CI] | P value |
| Swedish sample | |||||
| rs7123390-rs10838527-rs3824872 | GGA | 0.14 | 0.090 | 1.68 [1.13–2.53] | 0.012 |
| rs7123390-rs10838527-rs3824872 | GAC | 0.44 | 0.52 | 0.74 [0.56–0.97] | 0.032 |
| rs10838524-rs7123390-rs10838527-rs3824872 | AGGA | 0.14 | 0.082 | 1.82 [1.17–2.63] | 0.0059 |
| rs10838524-rs7123390-rs10838527-rs3824872 | GGAC | 0.44 | 0.51 | 0.75 [0.57–0.98] | 0.048 |
| Finnish sample | |||||
| rs7123390-rs10838527-rs3824872 | AAC | 0.21 | 0.29 | 0.61 [0.42–0.90] | 0.012 |
| rs7123390-rs10838527-rs3824872 | GAC | 0.59 | 0.45 | 1.84 [1.35–2.55] | 0.00010 |
| rs10838524-rs7123390-rs10838527-rs3824872 | AAAC | 0.20 | 0.27 | 0.67 [0.45–0.98] | 0.046 |
| rs10838524-rs7123390-rs10838527-rs3824872 | GGAC | 0.59 | 0.45 | 1.76 [1.31–2.47] | 0.00044 |
Odds ratio (OR): the ratio specific haplotype versus all other haplotypes among the cases, relative to the ratio specific haplotype versus all other haplotypes among the controls.
Discussion
Our key findings herein are that CRY2 gene variation and expression levels are associated with depression. CRY2 mRNA levels are lowered in blood mononuclear cells from depressed patients with bipolar disorder after total sleep deprivation in comparison to healthy controls, and CRY2 gene variation was associated with winter depression in both Swedish and Finnish patients. Differences in CRY2 risk haplotypes were observed between the Swedes and Finns. Overall there are genetic differences between these populations [26]. The fact that Swedes and Finns have different histories in terms of going through genetic bottle-necks that could have affected selection in operation at the CRY2 locus with regard to depression vulnerability, would possibly explain the differences in CRY2 risk haplotypes seen between the two samples, and explain a putative allelic heterogeneity (different vulnerability alleles in Swedes and Finns) of the functional CRY2 polymorphism. Assuming the same functional variant in the Swedish and the Finnish samples, the two samples would assist in narrowing the interval of that functional variant (Fig. 2C). The risk haplotypes spanned from CRY2 intron 1 to 3′UTR, and pointed at a location of a potential function variation somewhere upstream exon 12 in 3′UTR (rs10838527) from the Finnish data, with a distance from the intron 1 marker rs10838524 to rs10838527 corresponding to ∼33 kb (NCBI build 130). The vulnerability locus from the Swedish data overlapped the Finnish locus, but for at the intron 7 marker rs7123390, and extended downstream rs10838527.
Human CRY2 mRNA levels have been demonstrated to undergo circadian oscillation in fibroblasts [27] and in hematopoietic stem cells [28]. Our results herein add on these findings and demonstrate the effect of sleep deprivation on the circadian oscillations of CRY2 mRNA in human peripheral blood mononuclear cells. Our data indicate that depression in bipolar disorder is related to lowered levels of CRY2 mRNA. Sleep deprivation led to CRY2 mRNA increase in controls whereas depressed bipolar individuals were non-responsive.
CRY2 has been purified from human cells and its properties have been described [29], but the understanding of CRY2 regulation and CRY2 functions still need elucidation. Earlier studies have demonstrated that CRY2 transcription is driven by the canonical circadian circuit but through a unique mechanism of action not yet characterized [30]–[32]. CRY2 is unique among the canonical genes of the circadian clock, since experimental findings of its effects on the circadian clockwork violate the predictions from a theory on the roles of PER1, PER2, CRY1 and CRY2 [33]–[36]. Deletion of Cry2 gene lengthened the circadian period by approximately 48 min (from 23.7 to 24.5 hours) [37] and in fact strengthened circadian amplitudes and restored the lost rhythms in Per2 mutant mice [38], [39]. These findings fit in the view that activation of Per1 and Per2 genes occurs in the morning, whereas activation of Cry1 and Cry2 genes occurs in the evening [40]. Abnormalities in CRY2 regulation would therefore leave the morning oscillator intact, which agrees with findings in patients with winter depression [41] and those with bipolar disorder [42]. However, disruption of CRY2 regulation may compromise the evening oscillator and the subsequent proper oscillator reactions to light-dark transitions [43]. Cry2 null mutants are less decelerated, or are more accelerated, by light exposure than Cry1 null mutants [36]. The former phenotype does match with that seen in winter depression [44] and, to some extent, in bipolar type 1 disorder [45].
The action of melatonin on Cry1/Cry2 expression is hypothesized to form the basis of refractory reactions to stimulation with light [40], and the subsequent subsensitive or supersensitive responses to light exposure. Phase control through the melatonin-guided interval of Per1/Per2 to Cry1/Cry2 expression peaks may have relevance to mood disorders, as the internal alignment and the melatonin signal (amplitude and phase) are abnormal in patients with bipolar disorder and those with winter depression in particular [15], [16], [46]. Our findings now suggest that a dawn component [47], [48] and a dusk component (herein), i.e. PER2 and CRY2 variants respectively, are affected in winter depression. Such influence may well contribute to phase angle differences [49] and entrainment errors [11] that have been found in patients with winter depression.
To sum up, the phenotype of the Cry2 knockout mice together with mechanistic data are in line with the finding in this report suggesting that CRY2 has a role in winter depression.
Our current findings add to earlier gene expression studies in blood that have reported a number of potential peripheral biomarkers for bipolar disorder [50], [51], and earlier gene expression findings in brain indicating that clock genes are implicated in a model of bipolar disorder, the first report being [52], followed by [53] for CRY2. Circadian genes with a polymorphism previously reported to be associated to human bipolar disorder include CLOCK [54], and NR1D1, ARNTL and PER3 [55]–[57]. Moreover, ARNTL, RORA, RORB and RXRG were associated with bipolar disorder in a meta-analysis integrating data from genome-wide association studies and human and animal model expression studies [51]. PER2, NPAS2 and ARNTL genetic variation have been found associated with winter depression [47], [48], [58], and recent experimental data demonstrated these three proteins to regulate transcription of MAOA and to have subsequent influence on depressive behavior [59].
Other circadian genes were tested for association to winter depression in parallel with CRY2. The CRY2 finding reached significance even after correction for multiple testing considering other circadian gene SNPs tested for association to winter depression in parallel with CRY2. The corrected threshold for significance corresponded to a nominal P-value of 0.001 and was calculated applying a Bonferroni correction considering the partial LD between markers [60], [61]. As a novel finding we report here that CRY2 expression is reduced and non-responsive to sleep deprivation in depressed patients with bipolar disorder, suggesting that CRY2 plays a role in bipolar disorder.
There are some limitations in our work. A limitation in the CRY2 expression analysis was the few time points for the cases, since they do not allow analysis of the CRY2 rhythm and possible phase shift among the cases. However, irrespectively of whether there was a phase shift or not among the cases, CRY2 mRNA levels were significantly lower in cases compared to controls after sleep deprivation, which suggests that CRY2 levels is a trait marker that is non-responsive to the antidepressant sleep deprivation. Though, we cannot exclude that the CRY2 mRNA levels in the patients were influenced by medication. All the patients were on an SSRI together with a mood stabilizer. Lithium is known to affect the circadian clock through inhibition of GSK3 [2], and there are reports of SSRI influencing the circadian rhythms [62]. Since all controls and 10 of the patients were studied during the summer, the season of the examinations does not likely explain the CRY2 mRNA level difference between patients and controls. A limitation in the genetic analysis was the number of patients. However, the cases and the controls in both the Swedish and the Finnish materials represented ethnically homogeneous populations, and the controls were extensively scored to exclude any mental illness, reducing bias due to the ethnic variation and dilution of genetic effects due to disorder heterogeneity among the controls.
In conclusion, we show data suggesting that variation in CRY2 links to depression. CRY2 mRNA expression levels were lowered in PBMCs from depressed patients with bipolar disorder. In two ethnically homogeneous population-based samples, CRY2 SNPs were associated with winter depression. Although this study contains a replication of the genetic findings, it remains limited due to the lack of replication of the expression findings, thus warranting further studies.
Methods
CRY2 Expression Study
Informed consent was obtained from each participant using an approved University of California Institutional Review Board (IRB) protocol.
Thirteen patients (10 men, 3 women, out of which 11 with European, one with Asian and one with African decent) with bipolar type 1 disorder according to DSM-IV criteria, aged 40.2 years on average (SD = 13.4, range = 18–57), were enrolled in a sleep deprivation study at University of California Irvine (UCI) and San Diego (UCSD). During the study, all the patients were depressed, with the score on the 21-item Hamilton Depression Rating Scale [63] being 17.3 (SD = 6.4) on average. Eight healthy volunteers (4 women, 4 men, all with European decent), aged 23.6 years on average (SD = 5.9, range = 19–34 years), served as controls. Both the patients and the controls were hospitalized at the sleep research center at University of California at Irvine Medical Center (UCIMC) for 48 hours and deprived of sleep for 21 hours after an overnight stay (Figure 2). The season for the study of the patients was summer (n = 10) and winter (n = 3), and all controls were studied in the summer. All the patients were on an SSRI and either lithium (n = 8), valproate (n = 1) or lamotrigine (n = 4). None of the controls was on medication.
For the controls, venous blood samples were drawn at 9 different times, beginning at 7 p.m. and every 6 hours thereafter. For the patients, the blood was drawn at 1 p.m. on the days before and after sleep deprivation, corresponding to the time points #4 and #8 for the control samples. The patient blood was drawn in standard Vacutainer tubes without additive (10 ml, Becton Dickinson, Franklin Lakes, NJ, USA). From each control the blood was collected into both a standard acid citrate dextrose (ACD) tube (Becton Dickinson, Franklin Lakes, NJ, USA) as well as a Tempus Blood RNA Tube (Applied Biosystems, Foster City, CA, USA) in the same blood draw, yielding the total of 25–35 ml of whole blood. To check for potential difference in RNA degradation between control and bipolar disorder samples, the level of the house-keeping transcript GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was analyzed with qPCR (data in the results section). To further rule out possible bias in comparison between the patients and controls due to type of collection tube, we compared CRY2 and GAPDH mRNA levels using qPCR between the Tempus tube and the ACD tube from the controls collected at the same blood draw. The CRY2 and GAPDH mRNA levels were not different between tube types (data not shown).
Peripheral Blood Mononuclear Cells (PBMC) Isolation and RNA Extraction
All samples from both the patients and the controls were prepared similarly. Within a few minutes in room temperature after blood draw, the whole blood samples were layered onto Ficoll (Amersham Biosciences, Piscataway, NJ, USA), and PBMC were separated by density gradient centrifugation at 2500 rpm at room temperature for 20 min. The resulting ‘buffy’ coat at the interface was added to 5 ml phosphate buffered saline (PBS) at pH of 7.4 (Invitrogen, Carlsbad, CA, USA) and cell counts were taken. Cells were centrifuged at 1000 rpm for 10 min at room temperature. The resulting pellets were resuspended in 1 ml Trizol and stored at minus 80°C. Total RNA was extracted using the standard Trizol isolation protocol (Invitrogen, Carlsbad, CA, USA). The RNA was resuspended in 50 µL DEPC water, cleaned by passing over silica-based mini-spin columns (Qiagen RNeasy PlusMini Kit, Valencia, CA, USA) and analyzed for quality and quantity on a 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA) and concentration was adjusted to 1 µg/µl.
Real Time Quantitative PCR (qPCR)
The total RNA (1 µg RNA) from both the patients and the controls were identically synthesized into complementary DNA (cDNA) using Oligo d(T)16 primer and TaqMan Reverse Transcription Reagents and (Applied Biosystems, Foster City, CA, USA) at 25°C for 10 min, 48°C for 30 min and 95°C for 5 min. The quantitative PCR (qPCR) was performed on an ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) with triplicates for each cDNA, using Robot Biomek3000 (Beckman Coulter, Fullerton, CA, USA), with all samples run in the same plates with same normalization procedures. The reaction was performed in 12.5 µl consisting of 6.25 µl 2xSYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA); 2.5 pmol of each primer; 2 µl 1∶10 dilution of cDNA template (corresponding approximately to 4 ng RNA). The thermal cycling profiles were 50°C for 2 min (incubation), 95°C for 10 min (activation), 45 cycles at 95°C for 15 sec (denaturation) and 60°C for 1 min (annealing/extension), then by dissociation step as 95°C for 15 sec, 60°C for 15 sec, and 95°C for 15 sec. The CRY2 primer sequences spanned exon 6–7 junction within target sequence (Affymetrix probeset 3329058, transcript ID 3329029) and were: forward 5′-CCTACCTGCGCTTTGGTTGT-3′, reverse 5′-TGCTGTTCCGCTTCACCTTT-3′. The primers were pre-tested using brain cDNA, genomic DNA, no temple control (NTC) and RT minus on the ABI 7900HT. The results ensured good amplification of cDNA, and that neither residual genomic DNA nor primer-dimer contributed to measurements.
Genetic Association Study
The local ethic committees (National Public Health Institute, Karolinska Institutet, and University of Umeå) approved the study protocol, and all the participants signed an informed consent after the protocol had been fully explained.
Patients were recruited from outpatient services at which senior specialists in psychiatry, familiar with our research program, were working. All patients met the DSM-IV diagnostic criteria for major depressive disorder with the seasonal (winter) pattern [64]. The consensus diagnosis of two independent psychiatrists was required for inclusion for each patient. Patients were from Sweden (118 patients: 13.6% men, 86.4% women) and Finland (86 patients: 29.1% men, 70.9% women), both being of local origin.
Controls matched for ethnicity and nationality (1011 Swedes: 39.2% men, 60.8% women; and 1096 Finns: 29.0% men, 71.0% women) had no current or past psychotic or mood disorder as assessed with interviews or self-rated questionnaires and were representative of the Swedish and Finnish adult populations. These population-based studies have been described more in detail earlier [65]–[67]. The Swedish cases and controls lived in Västerbotten and Stockholm areas. The Swedish population has no strong internal genetic borders [24]. The Finnish cases lived in the capital area (Helsinki and its surroundings), with inhabitants from all the regions in Finland, and the controls were collected from the nation-wide health examination study.
Single-Nucleotide Polymorphism (SNP) Selection and Genotyping
Haplotype-tagging SNPs covering the variation in CRY2 were selected using the HapMap database [68], applying the cut-off values of 0.8 for r2 and of 0.1 for the minor allele frequency (MAF). The genotyping was performed using the SEQUENOM iPLEX Application with the MassARRAY System (Sequenom, Inc., San Diego, CA, USA). To control for quality, 2.5% of the samples were genotyped in duplicates. The genotypes of these replicas were all in agreement.
The four CRY2 SNPs genotyped fulfilled the criteria for SNP association test including a success rate of at least 90% of the reactions, and the Hardy-Weinberg equilibrium (HWE) with P>0.05 among the controls. The DNA samples from 10 Swedish cases and 53 controls, and 5 Finnish cases and 24 controls were excluded from analysis, since genotyping of them failed in more than 20% of a separate set of 111 SNP assays.
Statistical Analyses
For the RNA analysis, the cycle threshold (Ct) was determined approximately in the middle of exponential phase of the amplification, and generally default condition on the software SDS 2.3 was used. The average Ct value was accepted if the CV was lower than 2% and SD was lower than 0.39. The Ct values for CRY2 and the house-keeping gene GAPDH were normalized using the housekeeping genes C1ORF82 and TFG. The selection of C1ORF82 and TFG and the normalization method was based on the GENORM [69]. Within each of the case and the control groups, correlation for CRY2 and GAPDH expression levels to age was tested and showed no significant correlation. Among controls, the CRY2 and GAPDH expression levels were compared between pre-sleep deprivation time points (pooled time points from #1 to #4) and sleep deprived time points (pooled time points from #6 to #9), where the individual was used as a factor in a mixed model ANOVA.
CRY2 and GAPDH expression levels comparison between bipolar patients (time point 1 p.m.) and controls (pooled time points from #1 to #4, and from #6 to #9) was done using ANOVA with the covariates gender, age and sleep deprivation (yes/no). Significant effect in ANOVA (p<0.05) was followed-up with post hoc test.
The SNPs were analyzed for allele frequency differences between the cases and controls in each population sample using logistic regression. Gender was used as a covariate, since women were overrepresented among Swedish cases compared with controls. To obtain empirical significance values, permutation tests with up to 10,000 permutations were performed. There was a similarity between the gender corrected effect size and the effect size when no gender was corrected for, especially in the Finnish sample (Table 1). Hence, the gender was not generally controlled for in the subsequent analyses. However, suggestive genotypic association findings in the Swedish sample were verified using logistic regression with the gender as a covariate, and displayed a very minor gender effect. The power was ≥0.8 to detect allele frequency differences at odds ratio (OR) of ≥1.8 in the Swedish sample and at OR of ≥2.0 in the Finnish sample when the MAF was of 0.1 to 0.3 and the non-corrected P was of <0.05 (http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html). Those SNPs whose allele frequency difference was indicative (P<0.05) were analyzed for genotype association. Calculations were performed using the PLINK program, version 1.04 [70].
The linkage disequilibrium (LD) measure D' was calculated between the SNPs using the Haploview program, version 3.2 [71], which applies an approach similar to the partition-ligation-expectation-maximization algorithm [72]. Haplotype blocks were constructed using the LD block parameters [73], and the D' confidence interval algorithm in the Haploview program. Tests for haplotype frequency difference between the cases and controls were calculated for the haplotype blocks harboring the SNPs having P of <0.05 for allele association using the Haploview program. The SNP rs10838524 in CRY2 was included in haplotype analysis even though it was not included in any block but was nearby and associated as a single SNP with winter depression.
Supporting Information
All the detected haplotypes of the CRY2 block.
(0.04 MB DOC)
Footnotes
Competing Interests: Dr. Vawter is a co-founder of a diagnostic testing company AbaStar MDx, Dr. Kelsoe is consultant for AstraZeneca, on speakers bureau for Merck, and founder, board member and hold equity in diagnostic testing company Psynomics, all in compliance with UCSD guidelines. Dr. Bunney is a member of the scientific advisory boards of NeoSync, Inc., CNS Response, Inc. and Thuris Corporation and consultant for NeuDezine. None of these commercial affiliations played any role in this study.
Funding: This study was supported in part by grants from Academy of Finland (#201097 and #210262, www.aka.fi) and The Finnish Medical Foundation to Dr. Partonen, from Academy of Finland (#203425, www.suomenlaaketieteensaatio.fi) and Helsinki University Central Hospital (TYH538) to Dr. Paunio, from the Swedish Research Council (2005-6245, 2006-4670, www.vr.se), the Stockholm County Council (ALF, www.forskningsstod.sll.se) and Karolinska Institutet Foundations (www.ki.se) to Drs. Lavebratt and Schalling, by Public Health Service research grant M01 RR00827 from the National Center for Research Resource (www.ncrr.nih.gov) to UCI, Biomarker research grant MH 024370 to Dr. Vawter from the National Institute of Mental Health (www.nimh.nih.gov), and from the William Lion Penzner Foundation to Drs. Vawter and Bunney. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Germain A, Kupfer DJ. Circadian rhythm disturbances in depression. Hum Psychopharmacol. 2008;23:571–585. doi: 10.1002/hup.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- 3.Faedda GL, Tondo L, Teicher MH, Baldessarini RJ, Gelbard HA, et al. Seasonal mood disorders: patterns of seasonal recurrence in mania and depression. Arch Gen Psychiatry. 1993;50:17–23. doi: 10.1001/archpsyc.1993.01820130019004. [DOI] [PubMed] [Google Scholar]
- 4.Partonen T, Lönnqvist J. Seasonal affective disorder. Lancet. 1998;352:1369–1374. doi: 10.1016/S0140-6736(98)01015-0. [DOI] [PubMed] [Google Scholar]
- 5.Rosenthal NE, Sack DA, Gillin JC, Lewy AJ, Goodwin FK, et al. Seasonal affective disorder: a description of the syndrome and preliminary findings with light therapy. Arch Gen Psychiatry. 1984;41:72–80. doi: 10.1001/archpsyc.1984.01790120076010. [DOI] [PubMed] [Google Scholar]
- 6.Kieseppä T, Partonen T, Haukka J, Kaprio J, Lönnqvist J. High concordance of bipolar I disorder in a nationwide sample of twins. Am J Psychiatry. 2004;161:1814–1821. doi: 10.1176/ajp.161.10.1814. [DOI] [PubMed] [Google Scholar]
- 7.Bunney WE, Bunney BG. Molecular clock genes in man and lower animals: possible implications for circadian abnormalities in depression. Neuropsychopharmacology. 2000;22:335–345. doi: 10.1016/S0893-133X(99)00145-1. [DOI] [PubMed] [Google Scholar]
- 8.McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114:222–232. doi: 10.1016/j.pharmthera.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewy AJ, Sack RL, Miller LS, Hoban TM. Antidepressant and circadian phase-shifting effects of light. Science. 1987;235:352–354. doi: 10.1126/science.3798117. [DOI] [PubMed] [Google Scholar]
- 10.Souêtre E, Salvati E, Belugou JL, Pringuey D, Candito M, et al. Circadian rhythms in depression and recovery: evidence for blunted amplitude as the main chronobiological abnormality. Psychiatry Res. 1989;28:263–278. doi: 10.1016/0165-1781(89)90207-2. [DOI] [PubMed] [Google Scholar]
- 11.Teicher MH, Glod CA, Magnus E, Harper D, Benson G, et al. Circadian rest-activity disturbances in seasonal affective disorder. Arch Gen Psychiatry. 1997;54:124–130. doi: 10.1001/archpsyc.1997.01830140034007. [DOI] [PubMed] [Google Scholar]
- 12.Glod CA, Teicher MH, Polcari A, McGreenery CE, Ito Y. Circadian rest-activity disturbances in children with seasonal affective disorder. J Am Acad Child Adolesc Psychiatry. 1997;36:188–195. doi: 10.1097/00004583-199702000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Czeisler CA, Kronauer RE, Mooney JJ, Anderson JL, Allan JS. Biologic rhythm disorders, depression, and phototherapy: a new hypothesis. Psychiatr Clin North Am. 1987;10:687–709. [PubMed] [Google Scholar]
- 14.Thompson C, Stinson D, Smith A. Seasonal affective disorder and season-dependent abnormalities of melatonin suppression by light. Lancet. 1990;336:703–706. doi: 10.1016/0140-6736(90)92202-s. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasan V, Pandi-Perumal SR, Trakht I, Spence DW, Hardeland R. Pathophysiology of depression: role of sleep and the melatonergic system. Psychiatry Res. 2009;165:201–214. doi: 10.1016/j.psychres.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Lamont EW, Legault-Coutu D, Cermakian N, Boivin DB. The role of circadian clock genes in mental disorders. Dialogues Clin Neurosci. 2007;9:333–342. doi: 10.31887/DCNS.2007.9.3/elamont. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, et al. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 18.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 19.Miyamoto Y, Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci U S A. 1998;95:6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang N, Schleicher E, Kacprzak S, Bouly JP, Picot M, et al. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol. 2008;6:e160. doi: 10.1371/journal.pbio.0060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cameron MA, Barnard AR, Hut RA, Bonnefont X, van der Horst GT, et al. Electroretinography of wild-type and Cry mutant mice reveals circadian tuning of photopic and mesopic retinal responses. J Biol Rhythms. 2008;23:489–501. doi: 10.1177/0748730408325874. [DOI] [PubMed] [Google Scholar]
- 22.Buijs RM, Wortel J, Van Heerikhuize JJ, Feenstra MG, Ter Horst GJ, et al. Anatomical and functional demonstration of a multisynaptic suprachiasmatic nucleus adrenal (cortex) pathway. Eur J Neurosci. 1999;11:1535–1544. doi: 10.1046/j.1460-9568.1999.00575.x. [DOI] [PubMed] [Google Scholar]
- 23.de la Iglesia HO, Schwartz WJ. A subpopulation of efferent neurons in the mouse suprachiasmatic nucleus is also light responsive. Neuroreport. 2002;13:857–860. doi: 10.1097/00001756-200205070-00024. [DOI] [PubMed] [Google Scholar]
- 24.Luo AH, Aston-Jones G. Circuit projection from suprachiasmatic nucleus to ventral tegmental area: a novel circadian output pathway. Eur J Neurosci. 2009;29:748–760. doi: 10.1111/j.1460-9568.2008.06606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dardente H, Fortier EE, Martineau V, Cermakian N. Cryptochromes impair phosphorylation of transcriptional activators in the clock: a general mechanism for circadian repression. Biochem J. 2007;402:525–536. doi: 10.1042/BJ20060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lappalainen T, Hannelius U, Salmela E, von Döbeln U, Lindgren CM, et al. Population structure in contemporary Sweden – a Y-chromosomal and mitochondrial DNA analysis. Ann Hum Genet. 2009;73:61–73. doi: 10.1111/j.1469-1809.2008.00487.x. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki K, Nagase T, Mesaki M, Narukawa J, Ohara O, et al. Phosphorylation of clock protein PER1 regulates its circadian degradation in normal human fibroblasts. Biochem J. 2004;380:95–103. doi: 10.1042/BJ20031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsinkalovsky O, Smaaland R, Rosenlund B, Sothern RB, Hirt A, et al. Circadian variations in clock gene expression of human bone marrow CD34+ cells. J Biol Rhythms. 2007;22:140–150. doi: 10.1177/0748730406299078. [DOI] [PubMed] [Google Scholar]
- 29.Ozgur S, Sancar A. Purification and properties of human blue-light photoreceptor cryptochrome 2. Biochemistry. 2003;42:2926–2932. doi: 10.1021/bi026963n. [DOI] [PubMed] [Google Scholar]
- 30.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10:1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 31.Mirsky HP, Liu AC, Welsh DK, Kay SA, Doyle FJ., 3rd A model of the cell-autonomous mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106:11107–11112. doi: 10.1073/pnas.0904837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 34.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 35.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spoelstra K, Daan S. Effects of constant light on circadian rhythmicity in mice lacking functional cry genes: dissimilar from per mutants. J Comp Physiol A. 2008;194:235–242. doi: 10.1007/s00359-007-0301-3. [DOI] [PubMed] [Google Scholar]
- 37.Thompson CL, Sancar A. Photolyase/cryptochrome blue-light photoreceptors use photon energy to repair DNA and reset the circadian clock. Oncogene. 2002;21:9043–9056. doi: 10.1038/sj.onc.1205958. [DOI] [PubMed] [Google Scholar]
- 38.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oster H, Yasui A, van der Horst GT, Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes Dev. 2002;16:2633–2638. doi: 10.1101/gad.233702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lincoln GA, Andersson H, Hazlerigg D. Clock genes and the long-term regulation of prolactin secretion: evidence for a photoperiod/circannual timer in the pars tuberalis. J Neuroendocrinol. 2003;15:390–397. doi: 10.1046/j.1365-2826.2003.00990.x. [DOI] [PubMed] [Google Scholar]
- 41.Koorengevel KM, Beersma DG, den Boer JA, van den Hoofdakker RH. A forced desynchrony study of circadian pacemaker characteristics in seasonal affective disorder. J Biol Rhythms. 2002;17:463–475. doi: 10.1177/074873002237140. [DOI] [PubMed] [Google Scholar]
- 42.Elsass P, Mellerup ET, Rafaelsen OJ, Theilgaard A. Lithium effects on time estimation and mood in manic-melancholic patients: a study of diurnal variations. Acta Psychiatr Scand. 1979;60:263–271. doi: 10.1111/j.1600-0447.1979.tb00274.x. [DOI] [PubMed] [Google Scholar]
- 43.Daan S, Albrecht U, van der Horst GT, Illnerová H, Roenneberg T, et al. Assembling a clock for all seasons: are there M and E oscillators in the genes? J Biol Rhythms. 2001;16:105–116. doi: 10.1177/074873001129001809. [DOI] [PubMed] [Google Scholar]
- 44.Thompson C, Childs PA, Martin NJ, Rodin I, Smythe PJ. Effects of morning phototherapy on circadian markers in seasonal affective disorder. Br J Psychiatry. 1997;170:431–435. doi: 10.1192/bjp.170.5.431. [DOI] [PubMed] [Google Scholar]
- 45.Nurnberger JI, Jr, Adkins S, Lahiri DK, Mayeda A, Hu K, et al. Melatonin suppression by light in euthymic bipolar and unipolar patients. Arch Gen Psychiatry. 2000;57:572–579. doi: 10.1001/archpsyc.57.6.572. [DOI] [PubMed] [Google Scholar]
- 46.Wehr TA, Duncan WC, Jr, Sher L, Aeschbach D, Schwartz PJ, et al. A circadian signal of change of season in patients with seasonal affective disorder. Arch Gen Psychiatry. 2001;58:1108–1114. doi: 10.1001/archpsyc.58.12.1108. [DOI] [PubMed] [Google Scholar]
- 47.Partonen T, Treutlein J, Alpman A, Frank J, Johansson C, et al. Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann Med. 2007;39:229–238. doi: 10.1080/07853890701278795. [DOI] [PubMed] [Google Scholar]
- 48.Lavebratt C, Sjöholm LK, Partonen T, Schalling M, Forsell Y. PER2 variantion is associated with depression vulnerability. Am J Med Genet B Neuropsychiatr Genet Aug. 2009;19 doi: 10.1002/ajmg.b.31021. [DOI] [PubMed] [Google Scholar]
- 49.Lewy AJ, Lefler BJ, Emens JS, Bauer VK. The circadian basis of winter depression. Proc Natl Acad Sci U S A. 2006;103:7414–7419. doi: 10.1073/pnas.0602425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanazawa T, Chana G, Glatt SJ, Mizuno H, Masliah E, et al. The utility of SELENBP1 gene expression as a biomarker for major psychotic disorders: replication in schizophrenia and extension to bipolar disorder with psychosis. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:686–689. doi: 10.1002/ajmg.b.30664. [DOI] [PubMed] [Google Scholar]
- 51.Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- 52.Niculescu AB, 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, et al. Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics. 2000;4:83–91. doi: 10.1152/physiolgenomics.2000.4.1.83. [DOI] [PubMed] [Google Scholar]
- 53.Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, et al. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9:1007–1029. doi: 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- 54.Shi J, Wittke-Thompson JK, Badner JA, Hattori E, Potash JB, et al. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am J Med Genet Part B Neuropsychiatr Genet. 2008;147B:1047–1055. doi: 10.1002/ajmg.b.30714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mansour HA, Wood J, Logue T, Chowdari KV, Dayal M, et al. Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. 2006;5:150–157. doi: 10.1111/j.1601-183X.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- 56.Nievergelt CM, Kripke DF, Barrett TB, Burg E, Remick RA, et al. Suggestive evidence for association of the circadian genes PERIOD3 and ARNTL with bipolar disorder. Am J Med Genet. B Neuropsychiatr Genet. 2006;141:234–241. doi: 10.1002/ajmg.b.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kripke DF, Nievergelt CM, Joo EJ, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythm. 2009;7:2. doi: 10.1186/1740-3391-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, et al. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology. 2003;28:734–739. doi: 10.1038/sj.npp.1300121. [DOI] [PubMed] [Google Scholar]
- 59.Hampp G, Ripperger JA, Houben T, Schmutz I, Blex C, et al. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol. 2008;18:678–683. doi: 10.1016/j.cub.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 60.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 62.Sprouse J, Braselton J, Reynolds L. Fluoxetine modulates the circadian biological clock via phase advances of suprachiasmatic nucleus neuronal firing. Biol Psychiatry. 2006;60:896–899. doi: 10.1016/j.biopsych.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 63.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.American Psychiatric Association. Washington, DC: American Psychiatric Press; 1994. Diagnostic and Statistical Manual of Mental Disorders, 4th ed.886 [Google Scholar]
- 65.Lundberg I, Damström Thakker K, Hällström T, Forsell Y. Determinants of non-participation, and the effects of non-participation on potential cause-effect relationships, in the PART study on mental disorders. Soc Psychiatry Psychiatr Epidemiol. 2005;40:475–483. doi: 10.1007/s00127-005-0911-4. [DOI] [PubMed] [Google Scholar]
- 66.Pirkola SP, Isometsä E, Suvisaari J, Aro H, Joukamaa M, et al. DSM-IV mood-, anxiety- and alcohol use disorders and their comorbidity in the Finnish general population–results from the Health 2000 Study. Soc Psychiatry Psychiatr Epidemiol. 2005;40:1–10. doi: 10.1007/s00127-005-0848-7. [DOI] [PubMed] [Google Scholar]
- 67.Perälä J, Suvisaari J, Saarni SI, Kuoppasalmi K, Isometsä E, et al. Lifetime prevalence of psychotic and bipolar I disorders in a general population. Arch Gen Psychiatry. 2007;64:19–28. doi: 10.1001/archpsyc.64.1.19. [DOI] [PubMed] [Google Scholar]
- 68.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.1–0034.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 72.Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All the detected haplotypes of the CRY2 block.
(0.04 MB DOC)