Abstract
Phagocytes are the most important early components of the immune response, programmed to recognize, engulf and destroy immune complexes (formed when antibodies recognize their specific antigens), foreign particles, bacteria, mycobacteria, apoptotic cells, etc. Neutrophils, monocytes, macrophages and dendritic cells all participate in this process. Flow cytometry permits observation of phagocytes which have responded and, with the appropriate fluorescent probes, of the environment in the phagosome which has enclosed the foreign matter. This article gives the background and the protocols for performing such studies.
Introduction
Phagocytosis, a vital component of the primary immune response, is a complex process by which a cell, having recognized a target via specific plasma membrane receptors which initiated appropriate signals, uses the outer plasma membrane to enclose one or more target-receptor complexes in a pocket called a phagosome which seals itself off from the surrounding medium. The target can be a foreign particle, a bacterium or other organism, an apoptotic cell or a cell fragment. The target may or may not be coated by opsonins in the blood stream which specific receptors on the cell then recognize. The process of phagocytosis is beginning to be understood for the main phagocytes, i.e. neutrophils, monocytes, macrophages and dendridic cells, with neutrophils being the first line of defense. Efficient phagocytosis is essential for the subsequent destruction of the foreign entity since the fusion of the phagovacuole with lysosomal organelles within the cell (for neutrophils, aka polymorphonuclear leukocytes or PMN, these were called azurophilic and secondary granules in the older literature) releases bactericidal/microbicidal peptides and proteins into the newly formed phagosome and initiates the formation of reactive oxygen species (e.g. superoxide, peroxide, O and OH free radicals, aka ROS) whose role in the destruction of the target entity is critical. [For some very recent reviews see Steinberg B.E. and Grinstein, S. 2009, Krajewski, A. et al. 2009, Fuhrmann, S. et al 2008 , Nathan, C. 2006, Underhill, D.M. and Ozinsky, A. 2002, ]. This chapter will focus on neutrophils and the entities they may encounter in the blood where they defend the host from microorganisms. The protocols described here are readily adaptable to other phagocytes and stimuli.
Phagocytosis was first described by Mechnikov 125 years ago and was, for a long time, the main way in which functionality of phagocytic cells was evaluated. In more recent times it has become clear that, even in normal phagocytic cells where phagocytosis has proceeded, the enclosed organism is not always killed. Thus certain mycobacteria and some other organisms may not only evade destruction but even multiply within the PMN, monocytes or macrophages which have engulfed them [Huynh, K.K. and Grinstein, S. 2007, Bellaire, B.H et al. 2005, Ramachandra, L. et al. 2005, Levitz, S.M. et al. 1999]. In some instances (e.g. Mycobacterium avium, Mycobacterium tuberculosis) the phagosome does not undergo the acidification which normally accompanies the phagosome-lysosome fusion and creates the correct conditions for killing [Huynh, K.K. and Grinstein, S. 2007, Bellaire, B.H et al. 2005, Ramachandra, L. et al. 2005, Voyich, J.M.et al. 2005], in others (e.g. Cryptococcus neoformans [Levitz, S.M.et al. 1999] the compartment hyperacidifies and the organism survives. Alternatively the phagosome-lysosome fusion may be impaired or the phagosome itself may never close. Any of these perturbations can lead to survival of the phagocytized entity. The mechanisms by which such evasions of the normal phagocytic killing processes occur are varied and, as yet, are still being investigated and debated. These observations have rekindled an interest in phagocytosis and its measurement [Krajewski et al. 2009, Miksa et al. 2009].
Although high throughput fluorescence microscopy has also been used [Steinberg and Grinstein, 2009], the ready availability of multilaser flow cytometers has made it possible to monitor the phagocytic process in a specific cell population on a cell-by-cell basis, to evaluate which cells respond, whether signaling occurs, via which receptor, whether and which bactericidal processes (ROS, lytic enzyme activity) are functioning, and in some cases to characterize the conditions within the phagosome [Miksa et al. 2009, Elbim, C. and Lizard, G. 2009, Fuhrmann et al. 2008, Heinzelmann et al. 1997]. This is especially effective in PMN and most difficult in live (unfixed) macrophages which are strongly adhesive and clumpy when activated, making studies of single cells in a flow cytometer much more difficult by frequent clogging unless they have been fixed. Therefore fluorescence microscopy, imaging cytometry or multiwell plates are sometimes used in investigations of macrophage phagocytosis, its mechanisms and its consequences. The following discussion and protocols will focus on flow cytometry of phagocytosis by polymorphonuclear leukocytes (neutrophils, PMN).
Direct flow cytometric evaluation of PMN phagocytic capability has most frequently been effected by engulfment of a fluorescently labeled target such as dextran or latex beads [Savina et al. 2006], e coli particles [Kaur I. et al. 2008, Kaur I. et al. 2005, Kaur I. et al. 2004], immune complexes [Strohmeier et al. 1995 a,b, Brunkhorst et al. 1991, 1992, Ryan et al. 1990], or organisms [Miksa et al. 2009, Ramachandra et al. 2005, Heinzelmann et al. 1997]. While a flow cytometer reports only cell-associated fluorescence so that any entity in suspension can be ignored, it cannot distinguish between phagocytized and surface-bound but not enclosed targets. It is therefore important to quench the fluorescence of any external bound particle, a feat accomplishable by adding trypan blue [Seetoo et al. 1997] though ethidium bromide [Heinzelmann et al. 1997] or an anti-fluorescein antibody have also been used.
The most common labels, added as N-hydroxysuccinimide esters (SE) which react covalently with –NH2 groups, have been fluorescein derivatives (e.g. fluorescein, dicarboxyfluorescein, Oregon Green™, dihydrodichlorofluorescein) excitable with a 488nm visible laser. All SE react almost as readily with HOH as with -NH2 and it is therefore vital to prepare the stock solutions in totally dry DMSO, at a high enough concentration so that the carrier DMSO does not damage the stimulus being labeled. The fluoresceins have pKs between 5.5 and 6.5 and so can also indicate pH, especially in the acidic direction, though their sensitivity decreases rapidly above pH 7. Very recently a new probe, pHrodo™, has been developed by Molecular Probes/Invitrogen (now called LifeTechnologies). Opposite to the fluoresceins, its fluorescence increases with decreasing pH, with good sensitivity up to pH 8, as we have shown by labeling Molecular Probes/Invitrogen’s pHrodo™Bioparticles with OregonGreen™-SE (Fig.1). While the enhanced pH sensitivity of pHrodo™ is a great advantage in evaluating the pH within a phagosome, and while we find that trypan blue quenches its fluorescence and those of the fluoresceins, the excitation wave length, 565±3 nm, for pHrodo™ requires a special and different (561nm “yellow” or a tunable dye) laser which many routine flow cytometers do not yet have.
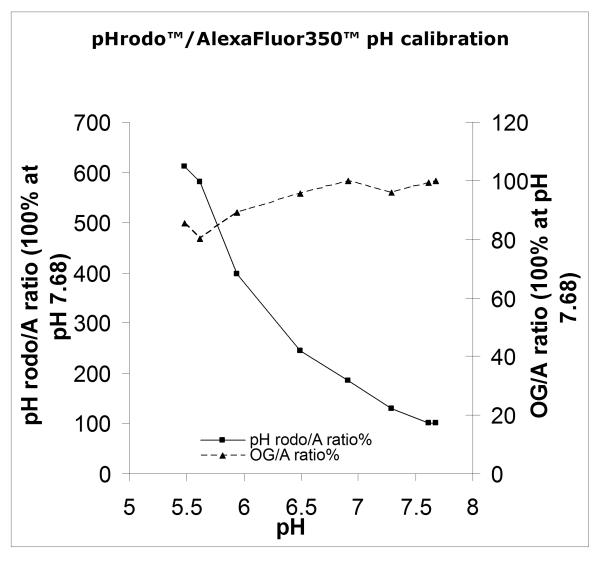
Figure 1.
pHrodo™Bioparticles (Invitrogen/Molecular Probes) were labeled according to Protocol 1 with OregonGreen™ SE and AlexaFluor350™ SE. The fluorescence of a 1μg/ml suspension in PBS at various pH was then measured in a Hitachi 4500 spectrofluorimeter (lexc 565, 488 and 357, lem 590, 530 and 450, respectively). The ratio of F590/F450 and F530/F450 was then calculated. The fluorescence of AlexaFluor350™ did not vary detectably over the pH range. Because the ratios were too disparate to permit reasonable comparison, the % of the ratio at each pH was calculated and is plotted here, the ratios at pH7.68 being arbitrarily defined as 100%.
The pH sensitivity of a probe does not matter as much if, rather than the early events (<2-3 minutes) which we study, the investigator’s interest lies in the eventual phagosomal conditions (after 1 to several hours) when focus is on identifying or sorting and studying cells that have undergone a phagosomal change (e.g. pH, ROS, lytic enzyme), and therefore have responded, vs those that have not. This approach, seeking the eventual conditions or results of processing within the phagosome, has been used by a number of laboratories seeking to identify important steps or participants in phagocytosis-to-bactericidal/mycobicidal processing [Miksa et al. 2009, Bellaire et al. 2005]. By then the pHp has reached an equilibrium dictated by the combination of oxidative products and granule contents which have been injected or generated within the phagosome; thus such studies permit distinction between different cells’ eventual functionality but do not identify whether, for example, cells which failed to change did so because they had no early signals or because they failed to generate ROS or because their granules failed to fuse with or to empty into the phagovacuole, etc.
It should be noted that, as we showed twenty years ago, one can investigate the phagosomal environment not only with respect to pH but also with respect to ROS presence, using DCF™, a reduced dihydrodichlorofluorescein, as label for the target, in our case initially high valency immune complexes [Herrmann et al. 2007, Strohmeyer et al. 1995a,b, Brunkhorst et al. 1991, 1992, Ryan et al., 1990]. Similarly, by using a self-quenching lytic substrate Bodipy™-labeled stimulus (now called DQ™ by Molecular Probes)[Herrmann et al., 2007, Seetoo et al. 1997], one can follow the release of PMN lytic enzymes (mainly elastase) into the phagosome. Again our main interest has been in early kinetics within the phagosome as PMN processing of targets proceeds, while others have been more focused on the eventual conditions or in using the latter to distinguish between functional PMN, or even to identify reacting phagocytes in whole blood samples [Kaur et al., 2008, 2005, 2004]. The following protocols describe our current procedures; they are generic since concentrations and conditions are adjusted (“tweaked”) each preparation of labeled organisms.
Generic protocols for OG™, DCF™, pHrodo™ and/or Alexa350™ labeling of phagocyte targets
Protocol 1: Labeling
Prepare a stock solution of each probe’s N-hydroxysuccinimide ester (SE) in dry dimethylsulfoxide (DMSO). Because it is important that all water be excluded keep the bottle of DMSO in a closed jar containing Drierite, and remove the desired volume of solvent quickly. The stock concentration of SE should be such that the desired final concentration of DMSO to achieve 2-5μM SE in the labeling buffer does not exceed 1%. When more than 1 label is to be covalently bound to the same organism or particle, additions should be made in the order of increasing affinity and greater fluorescence yield and stocks must be more concentrated in order not to exceed the DMSO/buffer limit of 1%.
2 mg (or protein equivalent) of the desired stimulus is dissolved in 2mL of phosphate-buffered saline (PBS) pH 9. If DCF™ SE is being used the buffer should be de-aerated under vacuum or have N2 bubbled through it for 30 minutes.
As described in 1. above, the N-hydroxysuccinimidyl esters (SE) of the desired fluorescent probes are then added sequentially, allowing 15 min. of incubation with rocking, at room temperature, between additions. The tube should be covered in foil to prevent photobleaching.
For example, if triple labeling of an organism is desired, to 2mL of the protein suspension in PBS pH 9 add 2μL of dichlorodihydrofluorescein N-hydroxysuccinimide ester (DCF™SE, Invitrogen/Molecular Probes #D2935) stock solution (250mM in dry DMSO) or 2μL of OregonGreen™SE (OG™SE, Invitrogen/Molecular Probes #O6147) stock solution (50mM in dry DMSO). {NB Since both probes are fluorescein derivatives with nearly identical excitation and emission spectra they cannot be used simultaneously}. Incubate 15 minutes at room temp., rocking, covered with foil.
Then add 20μL of pHrodo™SE stock solution (50mM in dry DMSO, Invitrogen/Molecular Probes #P36600).). Incubate 15 minutes at room temp., rocking, covered with foil.
Lastly, add 3.34μL Alexa Fluor™ 350 carboxylic acid succinimidyl ester (Alexa Fluor350™SE, Invitrogen/Molecular Probes A10168) stock solution (150mM in dry DMSO, ) incubate overnight at 4°C, shaking or rocking, covered with foil.
Add 4 mL of PBS, no glucose, pH 8 and vortex well.
Centrifuge at 3000rpm (approx 1400xg) in a desktop centrifuge for 10 minutes, 4°C. Not all of the organisms may pellet and therefore some will be discarded with the supernatant. The final concentration of stimulus stock will need to be adjusted after several washes (see 16. below).
Discard supernatant.
Resuspend pellet in 4mL PBS, no glucose, pH 8.
Recentrifuge a 2nd time at 3000rpm, 10 minutes, 4°C
Discard supernatant.
Resuspend pellet in 4mL PBS, no glucose, pH 8.
Recentrifuge a 3rd time at 3000rpm, 10 minutes, 4°C.
Discard supernatant.
Resuspend pellet in PBS, no glucose, pH 7.4. Adjust the final concentration of labeled stimulus so that the volume needed to stimulate the PMN, which are usually at 1-2 million/ml, is less than 5% of the total volume (e.g. less than 50μl/ml of PMN suspension).
Store at 4°C until ready to use. Freezing is not recommended.
To use, vortex well.
Protocol 2: Addition of organisms/targets to cells
In an actual experiment (Fig. 2) the chosen amount of labeled targets is added to the selected volume of cells in a stirred thermostated suspension. For example:
1 million cells/ml of buffer are equilibrated at 37oC with stirring and a baseline flow cytometric value is obtained.
The desired volume (not more than 50μl is recommended) of target suspension is added to the cells at time = 0.
After a chosen time or, in our experiments, continually, the fluorescence of all the probes is recorded at the relevant wave lengths. The narrower the band pass on the filters used and the longer the Stokes shift between excitation and emission wave lengths, the better since the contribution of the scatter peak will be reduced or eliminated.
If only the fully phagocytized and enclosed particles are of interest, trypan blue can be used to quench the fluorescence of bound but not enclosed targets over a broad range of probes{3) below}.
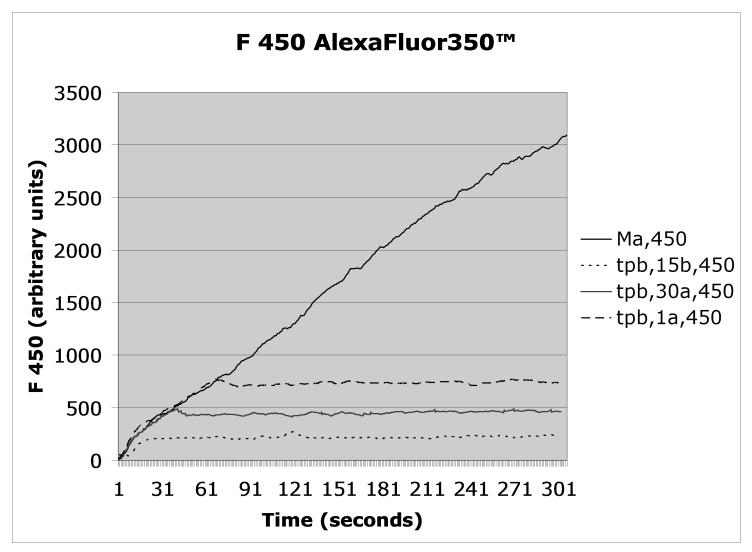
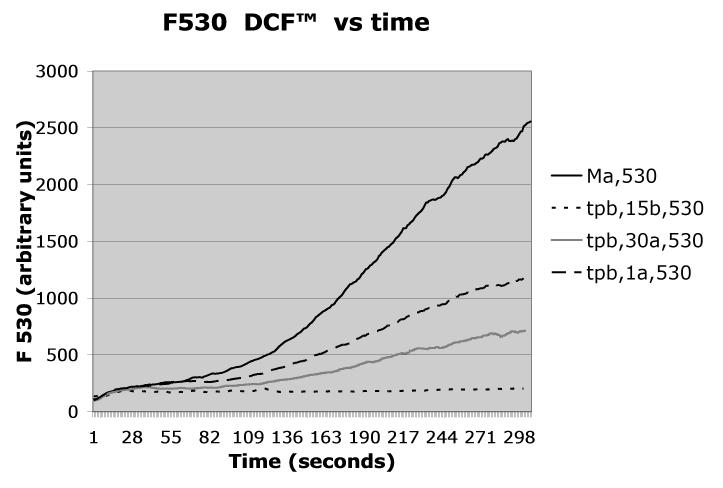
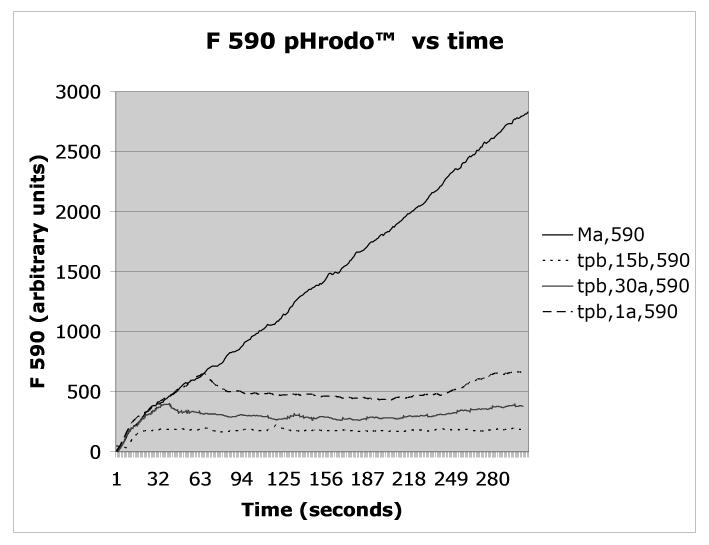
Figure 2.
Mycobacterium avium was triple labeled with AlexaFluor350™/pHrodo™/DCF™ according to Protocol 1, opsonized with autologous serum, and added to PMN at t=0 according to Protocol 2. Fluorescence emission was recorded continuously at 450nm (Alexafluor 350), 530 nm (DCF), or 590nm (pHrodo), a shown in the respective panels. As indicated in the figures’ legends, trypan blue (tpb) was added according to Protocol 3 to distinguish between enclosed (non-quenched) and bound but not enclosed (exposed to trypan blue, therefore quenched) organisms either 15 seconds before (15b) or at the indicated times after the Mycobacterium avium suspension (one second after = 1a, 30 seconds after = 30a). The control, i.e., unquenched, fluorescence is indicated as Ma.
Protocol 3: Distinction between externally cell-bound and phagosome-enclosed organisms/targets
To quench the fluorescence of bound but not enclosed particles or organisms, add 25 μl of 0.4% trypan blue per ml of PMN (1 million cells/ml) at the chosen time and record the residual fluorescence.
A control experiment without addition of trypan blue should be run.
A second control in which the trypan blue is added before the targets is also necessary so that intrinsic cell fluorescence can be determined and corrected for, i.e. when the fluorescence of the probes bound to the organisms are totally quenches.
Commentary
We have used all of these fluorescent labels, DCF™ and DQ™ having first been developed in our labs in collaboration with Molecular Probes, and present here generic optimal protocols which work well for each of the N-hydroxysuccinimide esters. We have found that they may need to be tweaked, with small changes, depending on the entity or organism to which the probes have been covalently attached and to the phagocytes being studied. Thus, while the covalent linkage requires the unprotonated NH2 group and therefore an alkaline pH, we found that some bacteria, for example, do not tolerate pH>8 and therefore we use bicarbonate buffer at pH 8, lengthening the labeling time, rather than phosphate-buffered saline (PBS, pH 9). Most immunoglobulins also do not tolerate pH 9 without changes that affect their properties but, if their specific antigen is less pH sensitive (e.g. bovine serum albumin), we can label the antigen and form the requisite immune complexes afterwards [Ryan et al. 1990, Brunkhorst et al. 1991,1992, Strohmeyer et al. 1995a,b, Herrmann et al. 2007]. For example, the OxyBursts™ are DCF™- or Rhosamine™-BSA/antiBSA complexes [Ryan et al. 1990]). When the esters of these reduced probes, indicators of ROS, are used, it is vital that all buffers be freed of dissolved O2 (vacuum or N2 bubbling) and that the labeling be performed under an N2 atmosphere.
As indicated in Protocol 1, the same stimulus can be labeled with several different probes, maximizing the amount of information that can be obtained by flow cytometry when the instrument has the necessary lasers. Fig.1 is an example of such multiple labeling: Molecular Probes’ pHrodo™E coli Bioparticles™ were suspended in PBS, pH 9, and OG™-NHS was added according to the protocol below; fluorescence was then measured at various pH (N.B. in the absence of PMN). Overlap of the probes’ fluorescence maxima should be avoided. Furthermore, when phagocytes are being examined, a suitable external quenching agent to distinguish surface-bound to PMN phagosome-enclosed stimuli will still be needed for each probe. The order of addition of the succinimide esters also needs to be adapted to the individual situation since their affinity for -NH2 and their fluorescence yield varies, so that the one giving the lowest yield should be added first when the largest number of ligandable -NH2 groups is available but too much labeling risks internal quenching. Thus, whether adding single or multiple probe esters, we routinely perform a dose-response test to find the optimal amount which will give us detectable indication in the linear region of fluorescence vs dose.
As indicated in Protocol 2, the labeled organisms/targets are added to the cell suspension at the desired time and observation on the flow cytometer is made either continuously, as we do, or can be made after a desired time interval.. Typical data from one of our experiments are given in Figure 2 for opsonized DCF™/pHrodo™/AlexaFluor350™ (i.e. triple) labeled Mycobacterium avium, opsonized with homologous serum and added to human neutrophils.
The effect of quenching the fluorescences with trypan blue, as described in Protocol 3 above, is also shown in Figure 2 when trypan blue (tpb) is added 15 sec. before, or 30 or 60 sec. after the organism.
The protocols above have been developed in our laboratory as best for our systems; they may need some adjustments of pH, quantity of reagent and/or incubation time when applied to a different organism/target acting on different cells.
Acknowledgements
I would like to thank the National Institutes of Health for their support of my research via HL076463 and DK31056. I am especially grateful to John Bernardo and Heidi Long for our many years of productive and enjoyable collaboration and the many students, fellows and technicians whose endeavors in my laboratory have inspired these studies and helped to develop these protocols.
References
- Bellaire BH, Roop RM, II, Cardelli JA. Opsonized virulent Brucella abortus replicates within nonacidic, endoplasmic reticulum-negative, LAMP-1-positive phagosomes in human monocytes. Infection and Immunity. 2005;73:3702–3713. doi: 10.1128/IAI.73.6.3702-3713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkhorst BA, Lazzari KG, Strohmeier G, Weil G, Simons ER. Calcium changes in immune complex-stimulated human neutrophils. Simultaneous measurement of receptor occupancy and activation reveals full population stimulus binding but subpopulation activation. J. Biol. Chem. 1991;266:13035–13043. [PubMed] [Google Scholar]
- Brunkhorst BA, Strohmeier G, Lazzari K, Weil G, Melnick D, Fleit HB, Simons ER. Differential roles of Fc gamma RII and Fc gamma RIII in immune complex stimulation of human neutrophils. J. Biol. Chem. 1992;267:20659–20666. [PubMed] [Google Scholar]
- Elbim C, Lizard G. Flow cytometric investigation of neutrophil oxidative burst and apoptosis physiological and pathological situation. Cytometry Part A. 2009;75A:475–481. doi: 10.1002/cyto.a.20726. [DOI] [PubMed] [Google Scholar]
- Fuhrmann S, Streitz M, Kern F. How Flow Cytometry is changing the study of TB immunology and clinical diagnosis. Cytometry, Part A. 2008;73A:1100–1106. doi: 10.1002/cyto.a.20614. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Bernardo J, Long HJ, Seetoo K, McMenamin ME, Batista EL, Jr., Van Dyke TE, Simons ER. Sequential chemotactic and phagocytic activation of human polymorphonuclear neutrophils. Infection and Immunity. 2007;75:398–3998. doi: 10.1128/IAI.00388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzelmann M, Herzig DO, Swain B, Mercer-Jopnes MA, Bergamini TM, Polk HC., Jr. Phagocytosis and oxidative burst response of planktonic Staphylococcus epidermidis RP62A and its non-slime-prouding variant in human neutrophils. Clin. and Diagnostic Lab. Immunol. 1997;4:705–710. doi: 10.1128/cdli.4.6.705-710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KK, Grinstein S. Regulation of vacuolar pH and ts modulation by some microbial species. Microbiology and Molecular Biology Reviews. 2007;71:452–462. doi: 10.1128/MMBR.00003-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur I, Simons ER, Kapadia AS, Ott CM, Pierson DL. Effect of spaceflight on ability of monocytes to respond to endotoxins of gram-negative bacteria. Clinical & Vaccine Immunology. 2008;15:1523–8. doi: 10.1128/CVI.00065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur I, Simons ER, Castro VA, Ott CM, Pierson DL. Changes in monocyte functions of astronauts. Brain, Behavior, & Immunity. 2005;19:547–54. doi: 10.1016/j.bbi.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Kaur I, Simons ER, Castro VA, Ott CM, Pierson DL. Changes in neutrophil functions in astronauts. Brain, Behavior, & Immunity. 2004;18:443–50. doi: 10.1016/j.bbi.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Krajewski A, Garg M, De M, Chandawarkar RY. Phagocytosis: Reemerging roles for a primitive function. Plastic and Reconstructive Surgery. 2009;123:834–847. doi: 10.1097/PRS.0b013e318199f01d. [DOI] [PubMed] [Google Scholar]
- Levitz SM, Nong S-H, Seetoo KF, Harrison TS, Speizer RA, Simons ER. Cryptococcus neoformans resides and thrives in an acidic phagolysosome of human macrophages. Infection and Immunity. 1999;67:885–890. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksa M, Komura H, Wu R, Shah KG, Wang P. A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J. Immunol. Methods. 2009;342:71–77. doi: 10.1016/j.jim.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. Neutrophils and Immunity: Challenges and opportunities. Nature Reviews Immunology. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- Ramachandra L, Smialek JL, Shank SS, Convery M, Boom WH, Harding CV. Phagosomal processing of Mycobacterium tuberculosis antigen 85B is modulated independently of mycobacterial viability and phagosome maturation. Infection and Immunity. 2005;73:1097–1105. doi: 10.1128/IAI.73.2.1097-1105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan TC, Weil GJ, Newburger PE, Haugland R, Simons ER. Measurement of superoxide release in the phagovacuoles of immune complex-stimulated human neutrophils. J. Immunol. Methods. 1990;130:223–233. doi: 10.1016/0022-1759(90)90052-w. [DOI] [PubMed] [Google Scholar]
- Savina A, Jancic C, Hughues S, Guermonprez P, Vargas P, Moura IC, Lennon-Dumenil A-M, Seabra MC, Raposo G, Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Seetoo KF, Schonhorn JE, Gewirtz AT, Zhou MJ, McMenamin ME, Delva L, Simons ER. A cytosolic calcium transient is not necessary for degranulation or oxidative burst in immune complex-stimulated neutrophils. J. Leukoc. Biol. 1997;62:329–340. doi: 10.1002/jlb.62.3.329. [DOI] [PubMed] [Google Scholar]
- Steinberg BE, Grinstein S. Analysis of macrophage phagocytosis: Quantitative assays of phagosome formation and maturation using high throughput fluorescence microscopy. Methods in Molecular Biology. 2009;531:45–56. doi: 10.1007/978-1-59745-396-7_4. [DOI] [PubMed] [Google Scholar]
- Strohmeier GR, Brunkhorst BA, Seetoo KF, Bernardo J, Weil GJ, Simons ER. Neutrophil functional responses depend on immune complex valency. J. Leukoc. Biol. 1995;58:403–414. doi: 10.1002/jlb.58.4.403. [10] [DOI] [PubMed] [Google Scholar]
- Strohmeier GR, Brunkhorst BA, Seetoo KF, Meshulam T, Bernardo J, Simons ER. Role of the Fc gamma R subclasses Fc gamma RII and Fc gamma RIII in the activation of human neutrophils by low and high valency immune complexes. J. Leukoc. Biol. 1995;58:415–422. doi: 10.1002/jlb.58.4.415. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A. Phagocytosis of microbes: Complexity in action. Ann.Rev.Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Sais-Salin B, Porcella SF, Long D, Dorward DW, Gerdner DJ, Kreiswirth BN, Musser JM, DeLeo FR. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]