Abstract
Neural cell fate programs must generate an enormous number of neurons with distinct adult functions. The decision to choose one neuronal subtype from two alternatives--a binary fate decision--is one way to diversify neuronal subtypes during nervous system development. Recent progress has been made in describing the genetic programs that define late-stage neuronal identity. Here, we review mechanisms that control how such fate decisions generate two different post-mitotic, terminally differentiated neuronal subtypes. We survey examples from C. elegans and Drosophila that demonstrate different modes of binary neuronal fate specification that depend on cell division, lineage, stochastic gene expression, or extracellular signals. Comparison of these strategies reveals that, although organisms use diverse approaches to generate neural diversity, some common themes do exist.
Introduction
The nervous system is composed of an enormous number of neuronal subtypes. Neural diversity is reflected at the molecular and cellular levels by differences in gene expression profiles, axodendritic morphology, neurotransmitter receptors, or sensory receptor expression. Together these properties determine the final functional identity of an individual neuron.
What are the cell fate programs that coordinate neuronal specification and differentiation during development? Early fate decisions in mammalian neural tissues such as motor neuron specification in the spinal cord rely on the interpretation of molecular gradients that control the spatial expression of transcription factors in neural progenitors[1,2]. These subtypes can be further subdivided by progressive intrinsic genetic programs as a result of the initial transcription factor activities[3]. In other systems, such as the mammalian retina, neural precursors can undergo progressive specialization by changes in fate competence over time to produce diverse neuron types such as rod, cones, interneurons and retinal ganglion cells[4,5]. In the mouse olfactory system, an olfactory sensory neuron can choose from over 1,300 different olfactory receptor genes to produce the many olfactory sensory neuron subtypes that respond to distinct odors[6]. Studies of the above systems have yielded important conceptual paradigms for neural fate specification. However, complex neural fate programs can be considered collections of individual fate decisions integrated over time, so understanding simple cell fate decisions--one neuron and one decision at a time--should greatly advance our knowledge of how neuron diversity is achieved.
The simplest fate decision is a binary decision, when a cell chooses between one of only two possible fates. Binary fate decisions feature prominently in neural development, largely because the inherently binary process of cell division is used to couple neural proliferation and differentiation. For example, a mitotic neural progenitor can generate one daughter cell that differentiates into a post-mitotic neuron, while the other daughter cell continues to divide to produce more neural cell types. The literature on binary fate decisions that maintain a renewable source of dividing stem cells, or neuroblasts, has been reviewed recently[7,8], however, and will not be discussed further here. In this review, we focus instead on genetic mechanisms of a different type of binary fate decision: How does a precursor neuron choose which of two post-mitotic, terminally differentiated subtypes to become?
Binary fate decisions for differentiation of a neuron often contain several requirements. First, the precursor cell usually passes through an equipotent competence window, even if for a short time. Thus the induction step for one fate is often coupled to an exclusion step of both the precursor stage and the alternate fate. Second, the differentiated neuron often must continuously repress the alternate fate after the fate is specified, and maintenance mechanisms are therefore necessary. We will discuss one maintenance mechanism—the bistable loop—that appears particularly appropriate for binary fate decisions. Finally, a binary fate choice that leads to a terminal, post-mitotic fate is often the final step for expressing genes that determine a neurons physiological function.
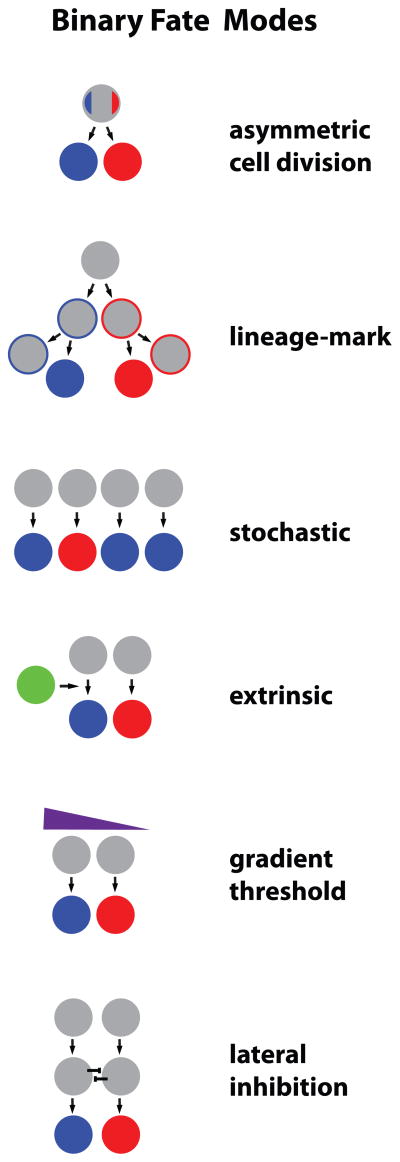
We use examples of fate decisions in C. elegans and Drosophila to illustrate the molecular mechanisms of four different modes of binary fate choices: a decision made from asymmetric division of a mother cell into two distinct post-mitotic cells, an intrinsic lineage-dependent binary decision, a stochastic decision, and a binary fate choice instructed by an extrinsic signal onto an otherwise equipotent precursor (Figure 1). Several common mechanistic concepts emerge, such as bistable feedback loops and the use of general “pre-specification” instructions that can be combined with specific neuronal class information to make a subtype decision.
Figure 1. General Modes of Binary Cell Fate Decisions.
Gray circles represent undifferentiated cells prior to fate choice; Blue and Red circles are differentiated cells that have acquired fate subtypes.
Asymmetric cell division instructs terminal fate
C. elegans has long been a workhorse for investigations into neural cell fate decisions. As the complete lineage is available for each of its 959 cells, the history of each cell with a defined identity can be traced back to the one-cell zygote, thus making it especially appropriate for studies of asymmetric binary fate decisions[9,10]. Second, the nervous system has 302 neurons whose individual subtypes can be labeled with GFP reporters and visualized in living adults. This makes the worm amenable to large-scale forward genetic screens designed to rapidly identify changes in neuronal cell fate and discover genes that are required for the terminal differentiation program of a particular neuron. A number of general and specific neural fate determinant genes have been identified through such screens, including “terminal selector genes”[11]. These are transcription factors that directly activate gene batteries that terminally differentiate and endow that neuron with subtype-specific features.
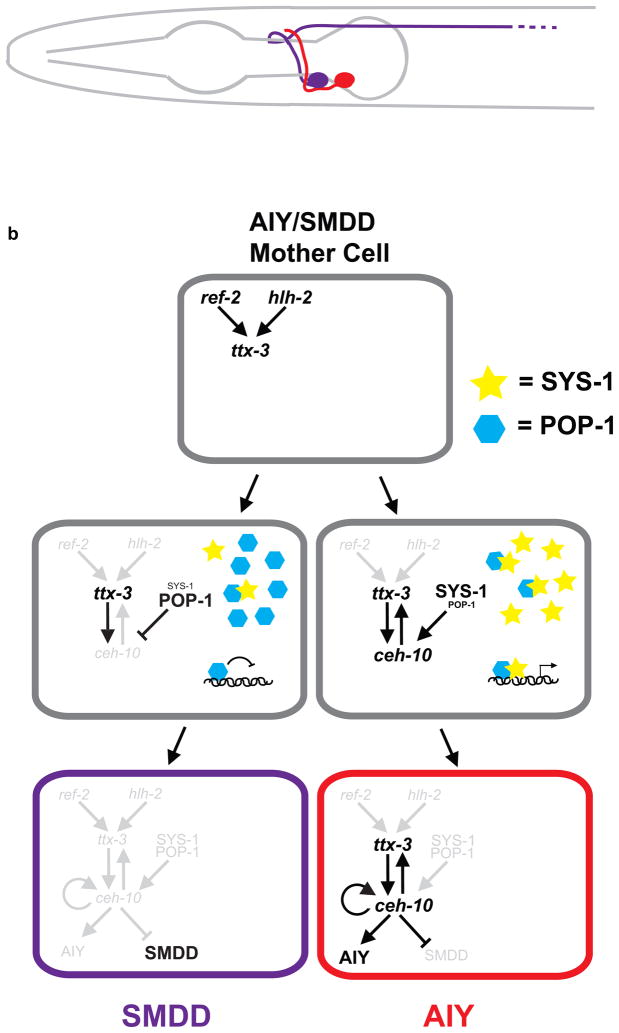
One binary fate decision in the C. elegans nervous system is the specification of two post-mitotic neurons, AIY and SMDD, upon division of the AIY/SMDD mother precursor cell. AIY is a cholinergic interneuron that integrates taste, smell, and temperature inputs[12]. It is molecularly defined by at least 40 terminal differentiation genes, all regulated by the paired-type homeoprotein CEH-10 and the LIM-homeoprotein TTX-3[13]. These transcription factors bind as a complex to the “AIY” cis-regulatory motif to directly regulate many genes that are expressed specifically in AIY. Mutants for ttx-3 or ceh-10 lose AIY neuron fate but maintain the general neuronal properties of the cell; hence the TTX-3/CEH-10 complex acts as the terminal selector for AIY fate[14]. SMDD is a motor neuron/interneuron that innervates muscle in the head, but its genetic specification is not well understood.
How is the binary fate choice between AIY and SMDD fates decided? A recent study [15] demonstrated that AIY fate is induced by direct coupling of asymmetric cell division with the cell terminal differentiation program. Initially, certain genes are expressed in the mother cell and, immediately after cell division, in both daughter cells. For example, the zinc finger transcription factor ref-2 is transiently expressed in the AIY/SMDD mother cell and acts with the bHLH transcription factor hlh-2 to directly activate ttx-3 expression. ref-2 expression is lost after the AIY/SMDD mother cell cleavage while ttx-3, which is initially expressed in both AIY and SMDD daughter cells, becomes restricted to the AIY neuron. In contrast, ceh-10, which requires ttx-3 for its expression, is expressed only in AIY. The expression of ref-2 and hlh-2 only in the mother cell and ttx-3 in both daughter cells suggests these factors are not the singular determinants of ceh-10 expression and AIY fate[15] (Figure 2).
Figure 2. AIY and SMDD terminal differentiation is induced by an asymmetric cell division.
a) Schematic of lateral view of C. elegans head showing positions of left adult AIY (Red) and SMDD (Purple) neurons with projections.
b) Mechanism of asymmetric cell division for AIY and SMDD fate. Wnt signaling induces a higher SYS-1/POP-1 ratio in the posterior daughter cell (presumptive AIY), which causes POP-1, in conjunction with TTX-3, to activate expression of CEH-10 in posterior cell only. See text for details.
Instead, the molecular difference between AIY and SMDD daughter cells that leads to ceh-10 expression and AIY fate is caused by the Wnt asymmetry pathway during the cell division[15]. This pathway, or Wnt/MAPK pathway, regulates asymmetric cell divisions during C. elegans development by responding to polarized Wnt inputs that result in asymmetric distribution of transcriptional factors in the daughter cells[16,17]. Wnt asymmetry pathway target genes are activated in only one daughter cell, as determined by the ratio of two transcription factors, POP-1 and SYS-1, whose nuclear export and import is regulated by upstream Wnt pathway components[18–20]. A current model posits that when POP-1 (TCF) protein levels are low, most POP-1 binds its co-activator, SYS-1(β-catenin), leading to activation of target genes. When POP-1 levels are high, excess POP-1 not bound to SYS-1 acts as a repressor. POP-1/SYS-1 ratios are different in nuclei of the AIY and SMDD daughter cells during cell division, with low POP-1/high-SYS-1 in AIY and high POP-1/low SYS-1 in SMDD, but this asymmetry disappears shortly after the cells have separated[15] (Figure 2). Mutants that lead to high levels of POP-1 in both nuclei remove the asymmetry--they have no ceh-10 expression and no AIY neurons and instead have two SMDD-like cells. Intriguingly, the Wnt asymmetry input and ttx-3 expression are integrated at the ceh-10 promoter, where both POP-1 and TTX-3 are bound. This directly connects the asymmetry information to terminal selector gene expression that instructs AIY differentiation. Thus, the Wnt asymmetry pathway provides the generic instructions to the dividing mother cell to make a binary fate choice, while ref-2, hlh-2, and ttx-3 act as specificity factors to coordinate these instructions with their neural subtype information. This coordination allows efficient induction of the differentiated fate immediately upon entering the post-mitotic state, precludes the need for later fate signals on each cell, and minimizes the chance of error in fate specification. Interestingly, the Wnt asymmetry pathway is required in the separate instances of AIN and ASER neuron fate determination as well[15]. It will be of great interest to figure out whether a similar Wnt asymmetry/terminal selector gene integration occurs in these neurons.
What is the direct source of the Wnt signal for SMDD/AIY fate determination? The Wnt asymmetry pathway is used frequently to produce asymmetric cell divisions for fate specification in the early embryo, where an Anterior-Posterior (A-P) gradient of Wnt defines a clear A-P axis for mitotic blastomeres. Lit-1, a kinase that regulates nuclear export of POP-1, is required for up to six successive binary fate decisions in the early embryo, and is also required for the SMDD/AIY decision, which is also along an A-P axis[15,21]. Looking further upstream, activated Frizzled (a Wnt receptor) is sufficient to induce pathway activation. However, reciprocal loss-of-function experiments removing Frizzled or Wnt ligands have not yet been performed, and it is thus unclear from where the Wnt source emanates for this decision.
One other unanswered question remains: how is SMDD specified? TTX-3 is only expressed transiently in the SMDD daughter after the mother cell division, and POP-1 likely represses most target genes rather than activates them. Is there a terminal selector gene(s) that defines SMDD fate, analogous to TTX-3 and CEH-10? Or is the SMDD fate determined by a ‘default state’ neuronal lineage enacted before the AIY/SMDD mother cell was born? Identification of SMDD-specific adult gene expression and analysis of GFP-reporters for these genes in various mutants will help answer this question.
“Pre-specifying” a late fate through an early event
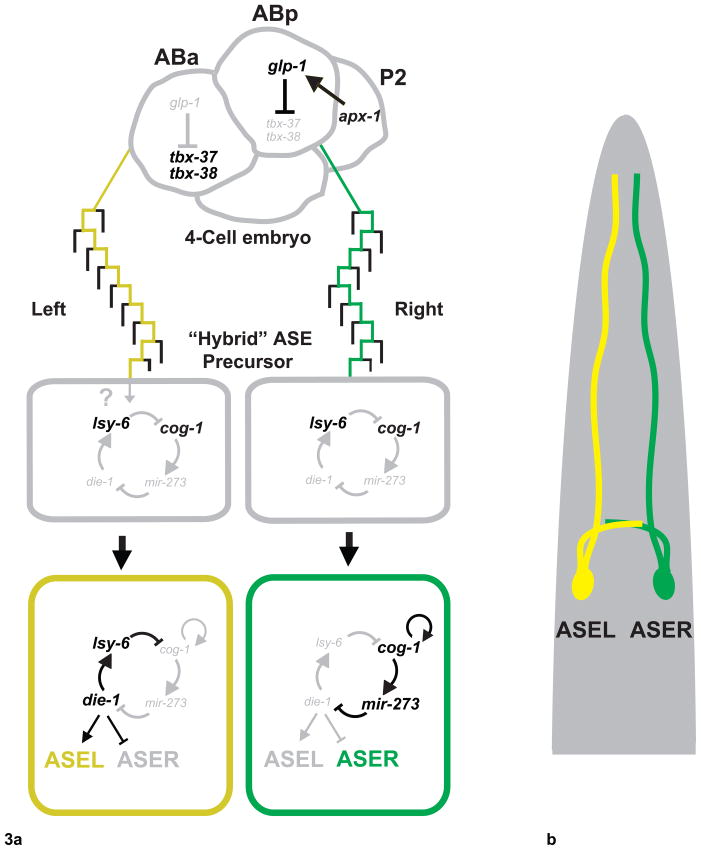
Another C. elegans binary fate decision exists to differentiate the morphologically bilaterally symmetric ASE gustatory pair of neurons that are involved in chemosensation. ASEL (left) and ASER (right) are found on opposite sides of the worm with similar axon/dendritic morphology and symmetric synaptic connectivity. Interestingly, the two neurons contain different chemosensory receptors and neuropeptides and have unique roles in chemosensation[22,23]. The ASEL/R laterality is regulated by a complex gene network, wherein two transcription factors and two microRNAs form a bistable feedback loop whose output specifies ASEL or ASER fate[24,25]. In ASEL, the microRNA lsy-6 suppresses translation of the transcription factor cog-1, which allows die-1 to promote expression of ASEL-specific terminal differentiation genes. In ASER, cog-1 activates mir-273, which then downregulates die-1 translation (Figure 3). The expression of the key regulatory factors becomes restricted to ASE subtypes only in post-mitotic neurons (ASER: cog-1, mir-273; ASEL: lsy-6, die-1). Mutations in these genes result in transformation of ASEL or ASER to the opposite fate. The two post-mitotic ASE neurons initially express genes common to both subtypes in what is proposed to be an equipotent “hybrid” state, which later switches to terminal ASEL/R states[25] (Figure 3). However, ASER and ASEL are not sister cells from the same mother cell, but result from a general left-right lineage patterning event that diverged nine cell divisions before. A series of recent genetic and laser ablation experiments determined that post-mitotic ASEL/R handedness arises from an asymmetry present in the 4-cell embryo stage when the lineages of the right and left ASE segregate[26]. This asymmetry is “remembered” in each ASE precursor through 9 cell divisions and is only interpreted when post-mitotic ASE neurons traverse the hybrid state and terminally differentiate into ASEL and ASER (Figure 3).
Figure 3. Bilaterally asymmetric ASEL/ASER post-mitotic differentiation induced by an early embryonic event.
a) In 4-cell embryo, Delta/Notch signaling (apx-1 to glp-1) induces fate difference between ABa and ABp blastomeres. ASEL always descends from ABa blastomere lineage, while ASER always descends from ABp blastomere. Forked pathway indicates cell divisions along anterior/posterior axis. At a hybrid state, ASE-R/L both express specific subtype genes. A bistable feedback loop then mediates terminal differentiation of ASE-R or L fates.
b) Schematic of C. elegans head showing position of adult ASEL (yellow) and ASER (Green) neurons with projections.
To study the source of the ASEL/R binary fate decision, the lineage of ASE neurons was altered in the early embryo and the effects on post-mitotic ASEL or ASER subtype fate was analyzed[26]. The C. elegans embryo develops in a sequence of blastomere divisions, beginning with an asymmetric division of the zygote that produces two cells which divide to become four cells. At the 4-cell stage embryo, the anterior and posterior AB cells, ABa and ABp, have distinct identities due to a Delta signal to ABp from the neighboring P2 cell. ABa is the ancestor of ASEL, while ABp will eventually give rise to ASER. Remarkably, this lineage relationship is impervious to change. Early blastomeres were genetically manipulated at the 4-cell stage such that, later in embryogenesis, the ancestors of ASEL, were generated ectopically on the right side of the worm (the opposite side). Yet these cells still specified the “left” ASEL subtype rather than switching to ASER. The same was true when ectopic ASER-ancestors were positioned on the worm’s left side. Note that in some of the experiments, most other cells on the left and right sides remained wild-type, as only specific lineages leading to ASE neurons were altered. Regardless of how the downstream lineage was altered, ABa cells always gave rise to ASEL neurons and ABp cells always gave rise to ASER[26]. Therefore, ASEL and ASER post-mitotic fates depend on the identity of the 4-cell stage blastomeres (ABa and ABp) from which they descended.
These observations raise the question of how identity is propagated from the 4-cell stage through post-mitotic terminal differentiation programs in the ASE lineage. The 4-cell stage Notch/Delta signal (GLP-1/APX-1 in worms) mediates repression of two T-box genes (tbx-37, tbx-38) in the ABp cell only, which will give rise to ASER[27] (Figure 3). The T-box genes are expressed only transiently in the ABa blastomere, so the events that ultimately lead to the differentiation of ASE into Left and Right subtypes are unknown. The T-box genes might cause an epigenetic mark on chromatin that is remembered later, or induce a sequence of genetic events that traverse 9 cell divisions before instructing the ASE neuron to finally differentiate from its hybrid state to the terminal state. Alternatively, the “hybrid” state—defined by bilateral ASE expression of subtype specific markers that are later confined to either subtype—might in fact already express yet-to-be-discovered subtype specific transcription factors, microRNAs, or other fate determinants, and thus would not be a true equipotent hybrid state with regards to ASEL versus ASER fates. The Wnt asymmetry pathway does not appear to be involved as POP-1 mutants have no effect on ASEL vs. ASER subtype fate[26]. Future studies on the events between the 4-cell stage asymmetry and induction of the lsy-6/cog-1/die-1/mi-273 bistable feedback loop will further elucidate the full mechanism of the ASEL/R binary fate choice.
Stochastic Choices
While C. elegans is well-suited for studies of neural fate decisions due to its invariant lineage, the Drosophila eye has the opposite attraction for the developmental biologist: the post-mitotic photoreceptor neurons are recruited to clusters directly from a field of undifferentiated, multi-potent precursor cells and become specified by lineage-independent signals within each cluster. The result of a fully specified photoreceptor cluster is an ommatidium, or unit eye, which contains 8 photoreceptors and 11–13 accessory cells. An adult retina comprises 800 ommatidia.
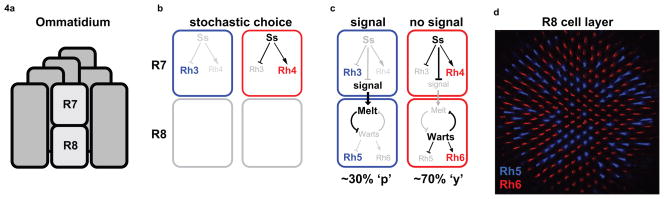
Although most binary cell fate decisions are specified by a deterministic mechanism with predictable outcomes, some stem from random fate decisions that are stabilized and transformed into robust developmental outcomes. The stochastic distribution of photoreceptor subtypes in Drosophila color vision is a prime example of a stochastic binary neuronal fate choice with consequences for the behavior of the animal. Two of the eight photoreceptor neurons within each ommatidium, R7 and R8, lie in the same optical path, one beneath the other, and express different color-sensing photopigment proteins, or Rhodopsins (Rh). R7 or R8 can each express one of two Rhs, but the two cells together coordinate their opsin expression so that R7/R8 pairs exist as two main subtypes: pale (p), wherein R7 expresses UV-sensitive Rh3 and R8 expresses Blue-Rh5 (pR7/pR8), and yellow (y), wherein R7 expresses another UV-sensitive Rh4 and R8 expresses green-Rh6 (yR7/yR8)[28,29]. These subtypes are distributed randomly across the retina in a roughly 30:70 pale:yellow ratio (Figure 4). The analysis of the molecular mechanisms of the binary fate decisions that produce these photoreceptor subtypes has led to several exciting discoveries as to how a post-mitotic, differentiated neuron determines its final functional identity.
Figure 4. Photoreceptor neuron subtype specification for Drosophila Color Vision.
a) A single ommatidium contains 8 photoreceptors. In the middle, R7 and R8 are involved in color vision.
b) R7 subtypes are specified by stochastic expression of the transcription factor spineless (ss).
c) R8 subtypes are specified by an instructive signal from R7. A bistable transcriptional feedback loop in R8 mediates the Rhodopsin output. ‘p’ is ‘pale’ subtype, ‘y’ is ‘yellow’ subtype.
d) Fly retina showing a random mosaic of photoreceptor subtypes with antibody stains of R8 Rhodopsins: Rh6 (Red) in ‘y’ subtype, Rh5 (Blue) in ‘p’ subtype.
R7 cells express Rh3 or Rh4 in a binary but stochastic fate decision that is mediated by the PAS- bHLH transcription factor, Spineless (Ss), which induces expression of Rh4[30] (Figure 4b). 70 percent of R7s express Ss and Rh4; however, which R7s contain Ss is entirely random and varies among individual flies as well as between opposite retinas of a single fly. Ss also regulates the coordinate subtypes in the neighboring R8 cell by repressing an instructive signal from R7 to R8 to induce Rh5 in R8. In ss mutants, all R7 cells are of the pR7 subtype, lose expression of Rh4 and instead express Rh3 as a default state[30]. As a consequence, R8s express Rh5 (Figure 4c). Therefore, stochastic induction of Ss not only controls a cell-autonomous fate decision, but also coordinates the binary fate choice in a neighboring post-mitotic neuron.
The mechanism for how Ss becomes stochastically expressed remains a mystery. Transcription of the gene could be a direct source of stochasticity through noisy or variable expression, or it could be downstream of other stochastically expressed factors. It is important to stress that Ss is not induced from a lineage decision, but results from an autonomous decision after R7 is specified. It remains possible that some stochastically induced asymmetry is ‘marked’ during the R7 differentiation process and still exists in the post-mitotic cell, thus influencing Ss expression. The maintenance of the R7 subtype fate decision is also not well understood, and could be linked to stabilizing the induction of a stochastic fate mechanism, perhaps by Ss positively regulating itself once its expression reaches a certain threshold.
Perhaps the Ss stochastic mechanism relates to the fact that, while Ss expression in an individual R7 photoreceptor is random, its expression across an entire retinal population of R7s occurs consistently in about 70% of the cells. The bias towards yR7/yR8 subtypes being present in around ~2/3 of ommatidia does not deviate strongly even in distantly related dipteran species [31], suggesting that there is some evolutionary pressure for the Rhodopsin ratio manifested within the stochastic Ss expression mechanism. Further insights into Ss expression awaits enhancer analysis and identification of upstream regulators.
Fate imposed: interpreting a signal
Extrinsic fate decisions, which are influenced by cell non-autonomous signals, can also instruct an equipotent precursor to become one neuron subtype or the other. This type of mechanism occurs in the R8 photoreceptor neuron, which receives communication from the R7 photoreceptor so that the pR7/pR8 and yR7/yR8 rhodopsin pairing is properly established. When R8s receive no signal from R7 (as in sevenless mutants that have no R7 cells, for example), almost all R8s become yR8s and express Rh6[28]. If all R7 send the signal (such as in ss mutants), then most R8s express Rh5 and become pR8s (Figure 4c). This R7 to R8 signal is interpreted and transduced into a robust output by two genes in the R8 photoreceptor, the kinase Warts and PH-domain containing Melted[32] (Figure 4c). Interestingly, in other contexts warts acts as a tumor suppressor[33,34], and melted as a positive regulator of growth involved in the Insulin/TOR pathway[35]. warts is expressed in yR8, which are post-mitotic cells, and is necessary and sufficient to induce yR8 and Rh6 fates. Conversely, melted is expressed in pR8 and is necessary and sufficient to induce pR8 and Rh5 fates. Moreover, warts and melted repress each other’s transcription in a double negative, or bistable, transcriptional feedback loop whose outcome is expression of either Rh5 or Rh6[32].
Several facets of the mechanism for R8 subtype specification are unresolved. Because the R8 fate is determined from an extrinsic signal, and that signal is determined in the stochastically defined R7 subtype, R8 must be initially competent to become either subtype. The molecular nature of the signal remains elusive, but its identification might reveal how the loop is triggered to push the R8 subtype loop towards yR8 or pR8. Once the loop is biased, the cross-repression in the bistable loop might act as a mechanism to maintain R8 subtype fate once it is fully specified, and thus the induction and maintenance mechanism of R8 subtype fate can be coupled. In mitotic tissues, Warts acts in a tumor suppressor signaling network[36], so it is important to know if other genes in the Warts signaling network are part of the post-mitotic fate decision and the bistable loop. Additionally, neither warts nor melted are transcription factors, so the ways by which they regulate each other’s transcription will be crucial to understand the full regulatory network with the R8 subtype fate decision. As we have seen with the bistable loop in ASEL/R neurons, the genetic networks responsible for a simple binary fate decision can involve many genes and extensive regulatory interactions.
Conclusions
The four examples of binary neural fate decisions discussed in this review exemplify the diversity of strategies cells use to select one fate over another. Nevertheless, a few mechanistic themes are shared:
Reciprocal repression between opposing fate determinants is an effective means to simultaneously promote one fate and exclude the other. The bistable feedback loops used in the ASEL/R and R8 photoreceptor subtype decisions are examples where key regulators of alternate fates repress each other to inhibit the other fate while reinforcing their own. This subsequent reinforcement of expression of the “winning” factor, either through the presence of a permissive activator or through self-activation, makes bistable feedback loops effective at transducing a weak or transient signal into a robust specification output[37], as might occur in the R8 photoreceptor subtype decision. In addition, transcriptional bistable loops can serve both the induction and maintenance phases of cell fate. For example, the Warts signaling pathway that induces the yR8 neuron subtypes and expression of Rh6 is also required in adults to maintain repression of the Rhodopsin of the alternative fate, Rh5 (DJ & CD, unpublished). Thus, a differentiated neuron can maintain its functional subtype for the life of the animal without the need to employ an entirely new maintenance program.
A comparison between the asymmetric division that produces AIY and SMDD and the lineage-based strategy that differentiates ASE neurons emphasizes two central themes. First, both decisions integrate fate instructions specific to a neuron class with fate information coming from a non-specific, asymmetric cell division. Once the ASE cells become post-mitotic, their fate is restricted by general ASE factors that limit their fate potential to two options, ASEL or ASER. When the “asymmetry mark” is interpreted at the time of ASE terminal differentiation, other genetic programs have endowed both ASEs with pan-neuronal properties, sensory neuronal properties, and perhaps even some neuronal subclass properties. Similarly, during AIY/SMDD specification, the asymmetry information provided by Wnt signaling intersects with AIY/SMDD neuron class-specific input (in the form of TTX-3 transcription factor) to activate the critical regulatory gene for AIY fate, ceh-10. This strategy can allow for greater combinations of fates, since it employs general aspects of development (cell division, lineage, etc.) to transmit information for a specialized purpose.
Second, having a relatively small number of neurons and invariant lineage affords C. elegans the ability to use series of binary cell divisions or embryonic lineage marks as primary fate mechanisms. Worm development must be streamlined, such that fate decisions directed by strict lineages and cell divisions can reduce variability among animals, as well as minimize mistakes within an animal. The number of mitotic divisions before a worm neuron reaches post-mitotic maturity is also relatively small. In organisms with far more tissue, like vertebrates, cells proliferate longer and the resulting cell fields require positional information, gradients, and inductive signals to coordinate specific cell fates. Vertebrates also use the strategy of controlling cell fates by cell division or lineage, just less predominantly than worms. As animal brains increase in complexity, their neural diversity expands as well. It is interesting to compare the worm, which has 302 neurons and ~900 transcription factors, to even the fruit fly, which also has about 900 transcription factors, but over 100,000 neurons, and at least over a thousand neural subtypes. Thus, the tendency of an organism to prefer some modes of neural specification over others, given its needs, is not surprising.
Stochastic mechanisms are found in several well-studied non-neuronal fate decisions and have been extensively studied in Notch/Delta signaling between cells, and more recently with competence of Bacillus subtilis to incorporate foreign DNA[38]. The selection of olfactory receptors in mouse olfactory receptor neurons is also a stochastic process, though in a large multi-outcome choice, where a stochastic choice seems appropriate. Why use a stochastic mechanism for a binary fate decision, such as in R7 subtype determination in the Drosophila eye, rather than one of the other modes? One reason may be that R7 is a post-mitotic cell specified independent of lineage, and is surrounded by other cells whose fates are also lineage-independent. This environment makes any earlier, determinate extra-cellular signal (like a gradient or ommatidia-to-ommatidia induction) reaching a random distribution of precisely 70% of R7 photoreceptors highly unlikely. It seems that stochastic expression of the fate switch, Ss, is one of the few mechanisms that meets the above constraints.
The study of more individual cases might provide insight into more complex systems like the mammalian spinal cord, where a hierarchy of signals followed by cross-repressive interactions between pairs of transcription factors give rise to multiple motor neuron progenitor identities. These studies might also identify more general mechanisms of cell fate decisions. Furthermore, owing to recent advances in induced pluripotent stem cell (iPS) and embryonic stem cell technology, the ability to engineer specific neuronal populations for basic research, tissue regeneration, or treatment of neurodegenerative diseases will also require knowledge of specific neural fate decisions[39,40].
Acknowledgments
We apologize to all whose work we inadvertently omitted. We would like to thank Robert Johnston and Daniel Vasiliauskas for helpful discussions and comments on the manuscript. David Jukam is supported by a New York University Dean’s Dissertation Award. Claude Desplan is supported by the National Institutes of Health (NIH) grant R01 EY13012.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 2.Price SR, Briscoe J. The generation and diversification of spinal motor neurons: signals and responses. Mech Dev. 2004;121:1103–1115. doi: 10.1016/j.mod.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 3.Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 4.Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 6.Fuss SH, Ray A. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol Cell Neurosci. 2009;41:101–112. doi: 10.1016/j.mcn.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 9.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 10.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 11.Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A. 2008;105:20067–20071. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 13.Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- 15**.Bertrand V, Hobert O. Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans. Dev Cell. 2009;16:563–575. doi: 10.1016/j.devcel.2009.02.011. This study demonstrates, in remarkable mechanistic detail, how asymmetric cell division and lineage information are integrated to instruct AIY vs. SMDD neuron differentiation in C. elegans. The authors use genetics and promoter analysis to show how Wnt asymmetry pathway and AIY/SMDD mother cell transcription factors directly activate ceh-10 expression in AIY to instruct the AIY differentiation program. They provide evidence that this precise integration strategy might occur in other C. elegans binary neural fate decisions as well. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips BT, Kimble J. A new look at TCF and beta-catenin through the lens of a divergent C. elegans Wnt pathway. Dev Cell. 2009;17:27–34. doi: 10.1016/j.devcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizumoto K, Sawa H. Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol. 2007;17:465–473. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Huang S, Shetty P, Robertson SM, Lin R. Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and its coactivator beta-catenin SYS-1. Development. 2007;134:2685–2695. doi: 10.1242/dev.008268. [DOI] [PubMed] [Google Scholar]
- 19.Kidd AR, 3rd, Miskowski JA, Siegfried KR, Sawa H, Kimble J. A beta-catenin identified by functional rather than sequence criteria and its role in Wnt/MAPK signaling. Cell. 2005;121:761–772. doi: 10.1016/j.cell.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 20.Phillips BT, Kidd AR, 3rd, King R, Hardin J, Kimble J. Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:3231–3236. doi: 10.1073/pnas.0611507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaletta T, Schnabel H, Schnabel R. Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature. 1997;390:294–298. doi: 10.1038/36869. [DOI] [PubMed] [Google Scholar]
- 22.Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 23.Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci U S A. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarin S, O’Meara MM, Flowers EB, Antonio C, Poole RJ, Didiano D, Johnston RJ, Jr, Chang S, Narula S, Hobert O. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics. 2007;176:2109–2130. doi: 10.1534/genetics.107.075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci U S A. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poole RJ, Hobert O. Early embryonic programming of neuronal left/right asymmetry in C. elegans. Curr Biol. 2006;16:2279–2292. doi: 10.1016/j.cub.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 27.Good K, Ciosk R, Nance J, Neves A, Hill RJ, Priess JR. The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development. 2004;131:1967–1978. doi: 10.1242/dev.01088. [DOI] [PubMed] [Google Scholar]
- 28.Salcedo E, Huber A, Henrich S, Chadwell LV, Chou WH, Paulsen R, Britt SG. Blue- and green-absorbing visual pigments of Drosophila: ectopic expression and physiological characterization of the R8 photoreceptor cell-specific Rh5 and Rh6 rhodopsins. J Neurosci. 1999;19:10716–10726. doi: 10.1523/JNEUROSCI.19-24-10716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- 30.Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franceschini N, Kirschfeld K, Minke B. Fluorescence of photoreceptor cells observed in vivo. Science. 1981;213:1264–1267. doi: 10.1126/science.7268434. [DOI] [PubMed] [Google Scholar]
- 32**.Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. Mikeladze-Dvali et al show how a post-mitotic neuron can interpret a non-autonomous signal to become one of two subtypes. They reveal that a bistable transcriptional feedback loop between two genes controls the decision to express either Rhodopsin5 or Rhodopsin6 in the R8 photoreceptor. They suggest that bistable feedback loops might serve as an efficient way to transform a weak or non-autonomous signal into a robust fate outcome. [DOI] [PubMed] [Google Scholar]
- 33.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 34.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 35.Teleman AA, Chen YW, Cohen SM. Drosophila Melted modulates FOXO and TOR activity. Dev Cell. 2005;9:271–281. doi: 10.1016/j.devcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- 37.Ferrell JE., Jr Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 38.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 40.Karumbayaram S, Novitch BG, Patterson M, Umbach JA, Richter L, Lindgren A, Conway AE, Clark AT, Goldman SA, Plath K, et al. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells. 2009;27:806–811. doi: 10.1002/stem.31. [DOI] [PMC free article] [PubMed] [Google Scholar]