Overview
Werner syndrome protein (WRN) is one of a family of five human RecQ helicases implicated in the maintenance of genome stability. The conserved RecQ family also includes RecQ1, Bloom syndrome protein (BLM), RecQ4, and RecQ5 in humans (see additional reviews in this issue), as well as Sgs1 in Saccharomyces cerevisiae, Rqh1 in Schizosaccharomyces pombe, and homologs in Caenorhabditis elegans, Xenopus laevis, and Drosophila melanogaster [1]. Defects in three of the RecQ helicases, RecQ4, BLM, and WRN, cause human pathologies linked with cancer predisposition and premature aging [1–5]. Mutations in the WRN gene are the causative factor of Werner syndrome (WS). WRN is one of the best characterized of the RecQ helicases and is known to have roles in DNA replication and repair, transcription, and telomere maintenance [1–6]. Studies both in vitro and in vivo indicate that the roles of WRN in a variety of DNA processes are mediated by post-translational modifications, as well as several important protein-protein interactions [1, 2, 7]. Many of these functions of WRN in genome maintenance, as well as the clinical characteristics of WS, have been recently reviewed [8–12]. In this work, we will summarize some of the early studies on the cellular roles of WRN and highlight the recent findings that shed some light on the link between the protein and its cellular functions with the disease pathology.
Molecular Genetics of Werner Syndrome (WS)
WS is a rare autosomal recessive progeroid disorder characterized by the development of cataracts, changing of skin conditions, bird-like facies, atypical short stature, and premature graying or thinning of the hair [9]. Patients also often develop hypogonadism, osteoporosis, diabetes mellitus, artherosclerosis [13], and cancers, particularly sarcomas [14]. Onset of symptoms usually occurs in the third decade of life, and health subsequently declines with median age at death between 47–54 years [13]. Because WS presents with early-onset of conditions commonly seen in the aged, it is a good model system for the study of mechanisms of normal aging [15, 16].
WRN Biochemistry
The causative factor of the majority of WS is mutation in the WRN gene, which codes for a member of the highly conserved RecQ family of helicases. While several different mutations within the gene are seen in WS, most result in production of truncated WRN protein [13]. Human WRN possesses 3′–5′ATP-dependent helicase, 3′–5′ exonuclease, and single-stranded DNA annealing activities [17–23]. Early gel filtration chromatography studies indicated that purified full length WRN exists as a trimer [24], and a fragment containing only the exonuclease domain equilibrates between a trimeric and hexameric form [25]. On the other hand, current electron microscopy data indicates that WRN is likely found as a dimer in solution, yet behaves as a tetramer in complex with DNA [26]. Still, while unwinding DNA, WRN acts as a monomer [27]. Together these results suggest that WRN’s oligomeric state may be dependent on whether or not it is catalytically active and how it is interacting with DNA.
Orthologs of human WRN have been identified recently and characterized in other eukaryotes, including Caenorhabditis elegans (CeWRN-1) [28, 29], Drosophila melanogaster (DmWRN) [30–32], and Xenopus laevis (focus forming activity 1 (FFA-1)) [33]. Xenopus FFA-1 is active as a helicase, while the exonuclease has not yet been investigated [33]. Similarly, the CeWRN-1 is active as a 3′–5′ATP-dependent helicase, though it lacks the 3′–5′ exonuclease [28]. In contrast, in Drosophila DmWRN is active as a 3′–5′ exonuclease [30] but the WRN helicase homolog has yet to be identified [31, 32]. Nevertheless, cellular studies indicate that depletion or mutation of WRN increases sensitivity to DNA damage and results in increased genome instability [29–32]. While the enzymatic activities of the WRN orthologs in Xenopus, C. elegans, and Drosophila may not be entirely conserved with the human WRN, each is clearly important for genome maintenance.
WRN is active in resolving a variety of DNA substrates, including forks, flaps, displacement loops (D-loops), bubbles, Holliday junctions, and G-quadruplexes (G4), all of which represent intermediates in DNA replication and repair processes [34, 35]. The roles of WRN in these pathways will be described later in detail. Importantly, the enzymatic activity of WRN on DNA substrates can be modulated by post-translational modifications [7]. The helicase and exonuclease activities of WRN are inhibited by oxidation [36]. WRN catalytic activity is also inhibited by both DNA-PK serine/threonine phosphorylation [37, 38] and c-Abl tyrosine phosphorylation [39]. Conversely, p300 acetylation stimulates WRN helicase and exonuclease [40]. Additionally, WRN is subject to sumoylation, although the affect of this on its activities has yet to be determined [41]. Presumably, post-translational modification of WRN regulates its roles in multiple DNA transactions.
Epigenetic Regulation and Cancer
Recent studies have also shown an epigenetic component of WRN regulation [42, 43]. Epigenetic regulation, specifically hypermethylation of tumor suppressor genes, is tightly linked with cancer [44]. In primary tumors, hypermethylation of the WRN promoter is common and correlates with lower levels of WRN protein expression. This hypermethylation inactivates the gene and results in increased chromosomal instability [42]. Inactivation of WRN in cancer cells increases susceptibility to the cytotoxic effects of topoisomerase inhibitors commonly used as chemotherapeutic agents [42, 45]. Moreover, knockdown of WRN protein expression induces cell death and growth arrest in several human cancer cell lines, consistent with a tumor-suppressor like role of WRN. Conversely, following WRN depletion, cells that persist in growth have detectable levels of WRN protein indicating that WRN may also promote tumor proliferation [43]. It is likely that the roles of WRN in preserving genome stability and facilitating DNA repair must be balanced to allow for normal cell growth while protecting against cancer formation [43]. Together, these results suggest that WRN is a potential target for anti-tumor therapies [42, 43, 45].
In addition to the general association of WRN protein expression with cancer, recent analyses of WRN single nucleotide polymorphisms (SNPs) show a connection with cancer susceptibility [46, 47]. An increased risk of breast cancer is seen among Chinese patients with the WRN Leu1074Phe SNP [46] and among German patients with the WRN Cys1367Arg SNP [47]. Yet, Cys1367Arg and Leu1074Phe SNPs do not notably affect the helicase or exonuclease activity of WRN [48, 49]. Yet, it is possible that these polymorphisms interrupt the C-terminal WRN protein interaction with mediators of damage signaling and DNA repair, including p53 and BRCA1 [50, 51], thus contributing to cancer susceptibility [47]. On the other hand, WRN Cys1367Arg SNP in a Taiwanese population is not associated with prevalence of breast cancer [52]. Also, analysis of Brazilian glioma patients did not detect any links between WRN Cys1367Arg SNP and development of gliomas [53]. Taken together these results suggest that correlation of WRN SNPs with cancer susceptibility is dependent on racial background and also on type of cancer.
Nevertheless, WRN SNPs are correlated with other pathologies characteristic of the WS phenotype. The WRN Cys1367Arg SNP may confer protection for diabetes mellitus [54], sarcomas [55], and non-Hodgkin lymphoma [56]. Moreover, both WRN Cys1367Arg and Leu1074Phe are associated with protection against cardiovascular disease [57, 58]. Healthy individuals with WRN 1367Cys and 1074Phe display increased levels of plasminogen activator inhibitor type 1 (PAI-1), an inhibitor of fibrinolysis [59]. Additionally, WRN regulates expression of PAI-1 [59]. Consistent with this, PAI-1 levels are elevated in WS cells [60], which may correlate with occurrence of artherosclerosis in WS. More work is needed in this area to detect associations between WRN SNPs and aspects of the aging phenotype.
Atypical Werner Syndrome
While Werner syndrome is most often associated with mutations of the WRN gene, a select number of cases have been identified in which patients are normal for WRN but instead have mutations in the lamin A/C gene (LMNA). This condition is referred to as atypical WS [61]. Patients with atypical WS display some of the common WS clinical features, including bird-like facies, osteoporosis, diabetes mellitus, and artherosclerosis [61–64]. Initial analysis of the genetic causes of atypical WS, revealed a set of heterozygous missense mutations within conserved residues of the LMNA gene [61, 63]. Recent studies of patients with atypical WS detect two novel mutations in LMNA: a previously unidentified missense mutation [64] and a deletion mutation [62]. While it is clear that LMNA mutations are linked with atypical WS, the mechanisms for the biological manifestation of the LMNA gene mutations has yet to be elucidated. Moreover, the molecular basis for the pathologies associated with WS also requires further study. Consequently, investigation of the functions of WRN is important for understanding both the progression of WS, as well as the progression of the normal aging processes.
Participation of WRN in DNA Replication and Cell Cycle Progression
To understand the link between the pathology of WS and the cellular roles of WRN, studies have been focused on investigating the biological consequences of WRN depletion on cell survival and cell cycle progression. Collectively, these cellular analyses reveal a role of WRN in DNA replication [6, 12]. Early examination of cells derived from WS patients showed a delay in S-phase progression compared to normal cells [65–69]. The delay has been attributed to either decreased rates of DNA extension [70] and replication fork propagation [68] or to disruptions in replication initiation or origin firing [66, 71–73]. More recent studies also demonstrate that replication fork elongation is slowed in WRN depleted fibroblasts treated with the replication inhibitors methylmethane sulfonate (MMS) and hydroxyurea (HU). Moreover, in the WRN depleted cells there is a more pronounced cell cycle delay in S-phase and G2-phase following exposure to MMS and HU [74].
Recovery from Replication Fork Stalling
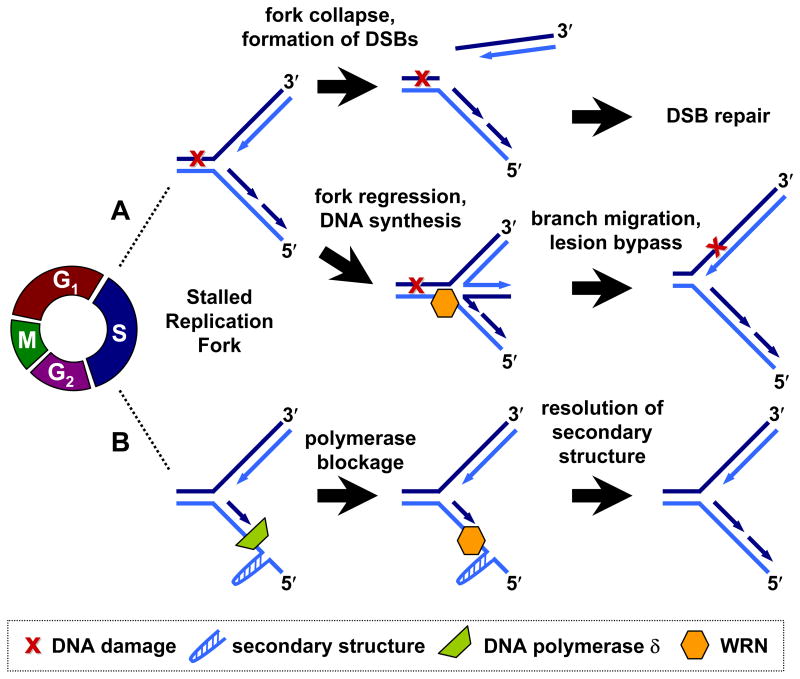
Several groups have shown that WRN cells are sensitive to treatment with replication inhibitors and DNA damaging agents that cause replication fork stalling [65, 67, 75–77]. The focus of many current investigations has largely been on the response of WRN deficient cells to these replication disruptions, and the role of WRN in recovery from replication-dependent DNA damage (Figure 1). During replication, topoisomerases relieve supercoiling in the DNA that occurs as a result of strand separation [78, 79]. Incomplete topoisomerase release from DNA, such as occurs upon treatment with topoisomerase inhibitors, leads to formation of covalent topoisomerase-DNA complexes that can pose a barrier to replication and can result in the formation of strand breaks [79, 80]. When exposed to topoisomerase I inhibitor topotecan (TPT), cells with a knockdown of WRN have a greater arrest in S-phase and inhibition of replication compared to control cells. This effect is specific for topoisomerase I inhibitors since the effects are not seen when cells depleted of WRN are treated with the topoisomerase II inhibitor etoposide (ETO). In WRN knockdown versus wild-type cells, there is an increased propensity for conversion of TPT-induced single strand breaks (SSBs) into double strand breaks (DSBs), suggesting that WRN prevents SSBs at replication forks from being converted into DSBs [81]. In addition, DSBs accumulate in WRN deficient cells in response to HU treatment, which induces replication fork stalling. These DSBs form as a result of collapsed replication forks, as indicated by proliferating cell nuclear antigen (PCNA) release from chromatin during S-phase. In the absence of WRN, stalled replication forks are processed via a compensatory pathway, which can be dependent on Mus81 endonuclease, causing DSB formation [82]. Together the results indicate that WRN functions in protecting cells from DSB formation that can occur as a result of replication fork stalling and collapse.
Figure 1. Roles of WRN in S-phase at stalled replication forks.
A) WRN may participate in repair of double-strand breaks (DSBs) following DNA damage-induced replication fork collapse. Alternatively, WRN may function to regress the fork and allow for synthesis bypass of DNA damage. B) Secondary structures which block DNA polymerases may be resolved by WRN. (See text for details.)
Moreover, WRN is involved in the response to replication fork stalling induced by agents that generate crosslinks within the DNA. Chromium is an environmental genotoxin known to affect DNA replication through formation of interstrand crosslinks that inhibit polymerase elongation of the DNA [83, 84] and also through creation of DSBs during S-phase that can lead to replication fork collapse [83, 85, 86]. Cells depleted of WRN are hypersensitive to chromium exposure, showing increased cell cycle arrest and cell death compared to wild-type cells. In WRN deficient cells exposed to chromium, WRN colocalizes with damage sites, as detected by phosphorylated histone H2AX (γ-H2AX) foci. These cells display a longer recovery time for stalled replication forks and repair of DSBs than control cells. Altogether the results indicate that WRN is involved in the recovery and/or repair of chromium-induced replication stress and DNA damage during replication [87]. One potential avenue of WRN participation in the restart of damage-induced stalled replication forks is through processing of regressed forks [88]. When damage is encountered at the fork, replication halts and the fork regresses into a chicken foot structure. In this structure, the lagging strand serves as a template for leading strand synthesis. Subsequently, WRN can mediate reverse branch migration of the chicken foot to bypass the damage, which can then be repaired by alternate pathways (Figure 1) [88].
S-Phase Checkpoints
Signaling checkpoints are activated in response to DNA damage at stalled forks to arrest replication and facilitate repair [89]. During S-phase, this signaling is mediated by the ataxia telangiectasia mutated (ATM) and ATM and Rad3 related protein (ATR) kinases [90]. While ATM is primarily involved in the response to DSBs [91, 92], ATR more frequently facilitates the response to stalled replication forks [93]. During S-phase WRN interacts with both ATM and ATR. ATM and WRN colocalize with replication foci during S-phase [94]. Yet, the association of WRN with the S-phase checkpoint is dependent on formation of DSBs. Activation of ATM and its downstream checkpoint targets is impaired in WRN deficient cells exposed to psoralen plus UVA (PUVA), which generates interstrand crosslinks and subsequent DSBs during S-phase [94]. Therefore, in response to replication-dependent interstrand crosslink-induced DSBs, WRN facilitates initiation of the intra-S-phase checkpoint through ATM activation [94].
Furthermore, WRN is phosphorylated in an ATM/ATR-dependent manner after damage associated with arrest of S-phase [95]. In addition, WRN is found in complex with members of the Rad52 epistasis group and ATR upon replication arrest caused by mitomycin C generated interstrand crosslinks [96]. Conversely, initiation of the ATR signaling cascade also occurs in the absence of WRN after c-Myc-induced DNA damage activates an intra-S-phase checkpoint [97]. Besides its roles in regulating transcription [98], c-Myc has recently been implicated in replication through its association with replication origins and through its interaction with the pre-replication complex [99]. Cells overexpressing c-Myc accumulate more DNA damage at replication sites in the absence of WRN, indicating that WRN protects against c-Myc-induced DNA damage during S-phase [97]. These results indicate that WRN plays an important role in reducing DNA damage stress sustained during S-phase. Overexpression of c-Myc leads to an accelerated S-phase, which may lead to formation of abnormal replication structures. It is conceivable that WRN may function in repair of these structures [97].
Presumably, WRN has both an indirect role in facilitating the recovery of stalled replication forks through signaling pathways and a direct role through enzymatic processes. In response to exposure to replication fork stalling by aphidicolin treatment, WRN deficient cells display an increase in fragile site instability and breaks compared to wild-type. Still, even in the absence of treatment, WRN deficient cells are more susceptible to DNA breaks suggesting that WRN is involved in maintenance of fragile sites [100]. It has been shown that fragile sites are susceptible to slowing of replication fork progression and to replication fork stalling even in the absence of treatment with replication inhibitors [101–103]. Studies show that it is the helicase activity of WRN which is responsible for the protection of genome stability at fragile sites [100]. WRN helicase may resolve DNA structures and facilitate polymerase replication at fragile sites (Figure 1) [100]. Earlier studies demonstrated that WRN does stimulate DNA polymerase δ (Pol δ) synthesis through structured regions, such as G4 containing DNA [104].
Interactions with Replication Proteins
WRN association with some of the major players in replication, specifically in lagging strand synthesis, also supports a role for it during replication; these interactions include Pol δ [105, 106], PCNA [24, 107, 108], flap endonuclease 1 (FEN1) [88, 109–113], and replication protein A (RPA) [21, 114–116]. Notably, the interaction with FEN1 implicates WRN function in replication of the lagging strand, as well as in base excision repair (discussed in a later section). However, WRN is not able to stimulate FEN1 cleavage of structured substrates [110], unlike BLM which resolves foldbacks allowing for FEN1 processing [117]. It is possible, then, that the WRN-FEN1 interaction is important either for processing other types of structured substrates during lagging strand synthesis or for alternative DNA transactions such as repair. Alternatively, WRN may resolve structured DNA which poses as a barrier to replicative polymerases, as mentioned above. In support of this notion, the Xenopus homolog of WRN FFA-1 associates both with replication foci [33, 118, 119] and also with Pol δ and RPA on chromatin upon replication blockage [120].
While it is evident from biochemical interactions and cellular studies that WRN is important for replication and S-phase progression, the effects of WRN depletion are most evident when DNA damage is associated with replication fork collapse. Consequently, although WRN has a prominent role in replication processes, it is also associated with repair of DNA damage.
Multiple Roles of WRN in DNA Repair
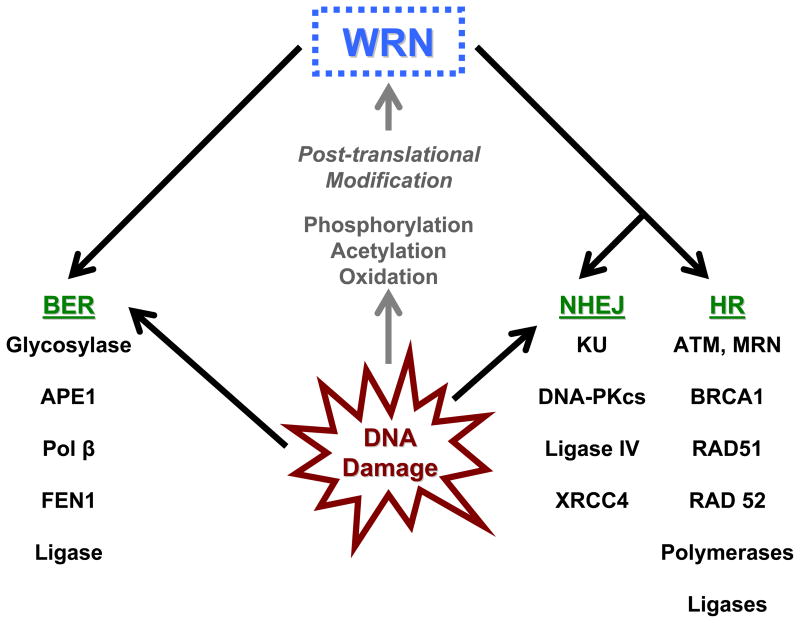
Endogenous and exogenous agents, chemicals, and ionizing radiation (IR) constantly damage cellular DNA [121], resulting in numerous DNA lesions throughout the genome. If not repaired properly, these lesions can eventually lead to point mutations, errors in DNA replication, DNA strand breaks, and induction of the stress response. Cells have different pathways to remove and/or repair different types of lesions to maintain the stability of the nuclear genome [122]. The major DNA repair pathways are base excision repair (BER), nucleotide excision repair (NER), double strand break repair (DSBR), and mismatch repair (MMR). BER repairs oxidative DNA base lesions such as 8-oxoguanine (8-oxodG), alkaline base damages, and SSBs. Bulky helix-distorting DNA lesions and crosslinks are repaired by NER. While DSBR repairs strand breaks in the DNA backbone, MMR is responsible for repairing single nucleotide mismatches and small insertion-deletion mispairs [2]. In addition to their functions in DNA replication, RecQ helicases are believed to play important roles in DNA repair processes. Among all the RecQ helicases, WRN’s involvement in various repair pathways is most prominent, as it interacts with a number of repair proteins [7]. Notably, WRN interacts with the tumor suppressor p53 [50, 123–125], which can activate repair and/or initiate apoptosis following DNA damage, and WS cells have attenuated p53-mediated apoptotic response [126]. Furthermore, WS cells are sensitive to DNA damaging agents such as camptothecin (CPT), 4-nitroquinoline 1-oxide (4-NQO), and MMS, and are slightly sensitive to IR [127–132]. They also accumulate more oxidative lesions after hydrogen peroxide treatment than normal fibroblasts [133]. The specific roles of WRN in different DNA repair pathways are discussed below (Figure 2).
Figure 2. Roles of WRN during DNA repair.
Schematic diagram depicting WRN interactions with various repair proteins in multiple pathways following DNA damage. DNA damage leads to post-translational modifications of WRN which may regulate its roles in DNA repair. (See text for details.)
Base Excision Repair (BER)
Oxidative DNA base modifications are caused by endogenous reactive oxygen species (ROS), normal by-products of mitochondrial oxidative phosphorylation and other metabolic processes, and are removed by BER [134, 135]. The involvement of WRN in BER is evident from the fact that WS cells are sensitive to hydrogen peroxide [133]. WRN siRNA treated human primary fibroblasts also accumulate increased damage after oxidative stress [136]. These in vivo studies are supported by in vitro examination of WRN knockdown cells, showing reduced BER activity [132]. WRN has also been implicated in BER of methylation-induced DNA damages and SSBs. A number of methylating agents, such as MMS [129, 132], methyl-lexitropsine, and telozolomide [137], sensitize WRN knockdown cells. WS cells are also sensitive to SSB producing agents like CPT and 4-NQO [127, 130, 131]. In brief, these results imply that WS cells are susceptible to agents that produce DNA damage, and lack of BER in these cells may be responsible for aspects of the aging phenotype.
The BER pathway proceeds either via short-patch (SP-BER), which involves single-nucleotide replacement, or via long-patch (LP-BER), which involves multiple nucleotide strand displacement. WRN physically and functionally interacts with proteins involved in both the BER pathways. DNA Polymerase β (Pol β) is mainly involved in SP-BER, though it can be involved in LP-BER as well [138, 139]. The strand displacement activity of Pol β is stimulated by the WRN helicase activity [140]. WRN helicase activity also cooperates with its exonuclease activity to assist Pol β activity at 3′-mismatches [132]. In vitro evidence indicates that WRN exonuclease can act as an autonomous proofreading enzyme for Pol β during LP-BER [141]. Furthermore, FEN1, which removes the protruding ssDNA flap generated by DNA strand displacement during LP-BER, is stimulated by WRN [110]. Apurinic/apyrimidinic endonuclease 1 (APE1), the endonuclease that incises abasic sites during BER, inhibits WRN; this interaction could possibly prevent promiscuous unwinding of DNA repair intermediates [142]. Although WRN does not appear to interact with human OGG1, the major glycosylase for 8-oxodG in human cells, it does interact in vivo and in vitro with NEIL1 [143]. NEIL1 is a human glycosylase for formamidopyrimidine (fapy) lesions [143, 143, 144]. Consistent with this interaction, fapy lesions accumulate in cells deficient in WRN [143]. WRN also functionally interacts with polyADP ribose polymerase (PARP-1) which ribosylates a large number of cellular proteins during DNA repair. The ribosylate activity is diminished in WRN-deficient cells, suggesting that PARP-1 is activated or stimulated by WRN [145].
Recent studies by Blander et al. suggested acetylation of WRN in vivo [146]. This process can either enhance the interaction of WRN with proteins that drives it to the nucleoplasm or diminishes its interaction with proteins holding it to the nucleolus. Furthermore, examination of the effect of acetylation on WRN catalytic activity and BER shows that acetylated WRN enhances the strand displacement activity of Pol β more efficiently than the unacetylated WRN [40]. The enhancement of Pol β strand displacement DNA synthesis after MMS treatment is also higher in WRN positive cells versus WRN knockdown cells. Collectively, these results suggest that acetylation is associated with activity of Pol β and that this process requires the presence of WRN [40]. Additionally, Li et al. recently showed that WRN-mediated cellular response to DNA damage is regulated by SIRT1 deacetylation [147]. Translocation of WRN from the nucleolus to the nucleoplasm is also affected by down regulation of SIRT1 [147]. The requirement of SIRT1-mediated deacetylation to stabilize WRN protein is supported by work from the Setou group [148]. They found that SIRT1 inhibitors diminish WRN levels. Likewise, WRN level is decreased in Sirt1-deficient mice [148]. Another recent study by Michishita et al. shows that at telomeric chromatin, SIRT6 deacetylates histone H3 lysine 9, which is required for the stable association of WRN [149]. This result further suggests that acetylation/deacetylation is a key mediator of WRN’s participation in DNA damage repair processes.
Double strand break repair (DSBR)
DSBs are mainly induced by IR. They are recognized by DNA damage sensing proteins, leading to formation of DNA repair foci enriched in γ-H2AX. Rapid accumulation of WRN at laser-induced DSBs has been shown, and it remains at the DSB site for at least for 4 h [150]. WRN depleted cells are very sensitive to a well known DSB generating agent, hydroquinone. A recent study by Galvan et al. has demonstrated that WRN silenced HeLa cells accumulate more DSBs upon hydroquinone treatment than WRN positive HeLa cells. Additionally, an elevated DNA damage response was indicated by more than five-fold induction of γ-H2AX compared to the control cells [151]. This result is supported by another study where short hairpin RNA was used to silence endogenous WRN levels in the human HL60 acute promyelocytic cell line. These cells are also highly susceptible to hydroquinone-induced cytotoxicity and show an elevated DNA damage response [152]. Moreover, WS cells show phenotypes such as non-homologous chromosome exchanges and large chromosomal deletions, caused by deficiency of DSBR [5]. Collectively, these results suggest an important role of WRN in DSBR.
DSBs are repaired by either non-homologous end joining (NHEJ) or homologous recombination (HR) processes. WRN is known to physically and functionally interact with two key proteins involved in NHEJ, Ku and DNA-PKcs [37, 38, 153]. In HeLa cells, Ku was identified as the tightest binding protein to the C-terminus of WRN [153]. However, Ku also interacts with the N-terminus or exonuclease terminus [154]. This protein enhances the exonuclease activity of WRN [153] and enables it to digest some substrates which WRN alone cannot process [155]. For example, 8-oxodG and 8-oxoadenine (8-oxoA) lesions block WRN exonuclease activity, but in the presence of Ku, WRN can digest strands containing these lesions [154, 156]. Recently, Bukowy et al. reported the same results with strands containing 5-hydroxy uracil (5-OHU) and 5-hydroxy cytidine (5-OHdC) lesions [157]. Physical interaction between DNA-PKcs and WRN has been reported by several groups. However, it is not clear if the interaction is direct or mediated by Ku [37, 38]. The DNA-PK complex has been shown to regulate WRN phosphorylation in vivo and in vitro [37, 38, 158]. Notably, the serine/threonine phosphorylation of WRN by the DNA-PK complex inhibits the WRN helicase and exonuclease activities. WRN also interacts with another essential factor of NHEJ, the XRCC4/Ligase IV (X4L4) complex [159]. X4L4 binds with WRN, stimulating its exonuclease activity. The interaction between X4L4 and WRN may enable WRN exonuclease to serve as a DNA end-processing factor during NHEJ [159]. Systems deficient in NHEJ core components show hypersensitivity to IR treatment. Saintigny et al. observed that SV-40 transformed WS fibroblasts display a mild, but significant sensitivity to IR, compared with the appropriate control fibroblasts [160]. A similar result was obtained by Yannone et al., comparing IR sensitivity of hTERT WS fibroblasts with WS fibroblasts expressing exogenous WRN [38]. Additionally, Sallmyr et al. found an alternative NHEJ pathway for DSB repair in chronic myeloid leukemia cells, involving DNA ligase IIIα and WRN [161]. Taken together, these results suggest that WRN is probably not an essential component of NHEJ, but is more likely acting as an accessory protein during this process and is involved in alternative NHEJ pathways.
Besides its involvement in NHEJ pathway of DSBR, WRN also physically and functionally interacts with several important proteins in the HR pathway. After replication arrest induced by HU, WRN co-localizes with Rad51, which is a key player in the strand invasion event during HR [162]. Although no direct interaction between WRN and Rad51 has been reported, WS cells show elevated levels of Rad51 foci. WRN increases the efficiency of Rad52-mediated strand annealing [163], whereas, Rad52 both inhibits and enhances WRN helicase activity in a DNA structure-dependent manner. These results suggest that Rad52 and WRN may cooperatively facilitate rescue of stalled or blocked DNA replication forks. WRN colocalizes with Rad54, another key protein in this pathway, in response to replicative stress [96]. WRN associates with the Mre11-Rad50-NBS1 complex via NBS1, a essential component of HR [164] Through this interaction, DNA unwinding by WRN is stimulated suggesting that WRN may be involved in resolving recombination intermediates. WRN also interacts physically with BRCA1, the possible HR factor, and BRCA1 stimulates WRN helicase activity [51]. Furthermore, WS cells are deficient in removal of DNA interstrand crosslinks, a process that requires recombination [51, 94, 165]. Consistent with an involvement in DSBR, there is growing evidence that WRN interacts with topoisomerases. Interplay between topo I and WRN has been reported and WRN is involved in repair of topo I-inhibitor induced DNA damage [81, 166]. Interaction between WRN and topo III is also suggested by Aggarwal et al. [167]. Their recent study demonstrated that WRN can function in a genetic pathway that affects topo III related phenotypes. Another RecQ helicase, BLM, functions in HR mediated DSB repair [168]. BLM is known to physically and functionally interact with WRN, inhibiting its exonuclease activity [169]. Thus, these two RecQ helicases could work together in DSB repair. Supporting this hypothesis, synergistically increased hypersensitivities to 4-NQO, CPT, MMS, and UV, were observed in Wrn−/−/Blm−/− double mutant chicken DT40 cells compared to the single mutants [129]. However, further studies are required to explore the associations and exact roles of these two proteins in HR.
Involvement of WRN in Transcription
The first indication of the involvement of WRN in transcription comes from the fact that purified recombinant WRN protein has been demonstrated to unwind not only a duplex DNA but also an RNA-DNA heteroduplex [23, 170]. Association of WRN with transcription was also suggested by the result that WRN overexpression affects p-53 mediated transcription [50]. Furthermore, Shiratori and colleagues have shown that WRN leaves the nucleoli upon inhibition of ribosomal RNA (rRNA) transcription following actinomycin-D treatment [171]. They have also demonstrated that the level of rRNA transcription in WS fibroblasts is significantly lower than that in wild-type fibroblasts, and the reduced level of rRNA can be rescued by expression of wild-type WRN in WS fibroblasts [171]. The functional interaction between WRN and rRNA was further supported by the observation that methylation of rRNA genes is accelerated in WS fibroblasts [172]. Furthermore, the efficiency of RNA polymerase II (RNA pol II) transcription is reduced by about 50% in permeabilized WS cells and extracts from WS cells compared to normal cells, implicating WRN in RNA pol II-dependent transcription. Additionally, RNA pol II-dependent transcription can be enhanced by the addition of recombinant WRN protein. Deficiencies in RNA polymerase I (RNA pol I) transcript level were not observed in this study [173]. In contrast, WRN was co-immunoprecipitated with an RNA pol I subunit in another study [171]. It was suggested that the ongoing transcription of RNA pol I controls nucleolar accumulation of WRN in serum-stimulated cells. Likewise, RNA pol I inhibition or serum starvation leads to translocation of WRN from the nucleolus to the nucleus [171]. A recent study by Lutomska et al. has shown a defect in the RNA pol I transcription in response to distinct growth factors in WS cells [174]. WRN also stimulates promoter clearance of RNA pol I transcription and binds to the active fraction of rDNA in quiescent cells. Furthermore, the role of WRN in the differential expressions of individual genes has been studied by Kyng et al. using a cDNA microarray system[175]. They propose a model in which WRN protein, by virtue of its helicase and transcription-activating activities, as well as its protein interactions, is directly or indirectly involved in the transcription of genes upstream in the network of aging pathways [175]. Collectively, these results indicate involvement of WRN in transcription and suggest that decreased transcription rate might be a cause of premature aging phenotypes observed in WS patients.
Importance of WRN for Telomere Maintenance
The DNA polymerases involved in DNA replication process are restricted from continuing DNA synthesis to the end of the chromosome because of their unidirectional nature [176, 177]. While prokaryotes employ a number of diverse mechanisms to deal with this ‘end replication problem’, eukaryotes have developed a unique solution involving a special structure known as the telomere [178]. Telomeres are nucleoprotein complexes situated at the ends of the linear chromosomes that prevent chromosome termini from being recognized as broken DNA ends (i.e., DSBs). In most of the organisms studied, telomeres consist of long repetitive G-rich and C-rich DNA strands, the ribonucleoprotein telomerase, and telomere binding and associated proteins [179]. Loss of telomeric repeats or loss of protection by telomere-associated proteins triggers telomere dysfunction. Apart from being subject to nucleolytic degradation and undesirable recombination mediated by HR or NHEJ, dysfunctional telomeres are also recognized by many DNA damage response proteins, including ATM, γ-H2AX, 53BP1, MDC1, and NBS1. Association of these proteins can form telomere dysfunction-induced foci and can induce cell cycle arrest, senescence, or apoptosis [180–182]. Aging [183] and premature aging syndromes [11] are also frequently associated with telomere attrition.
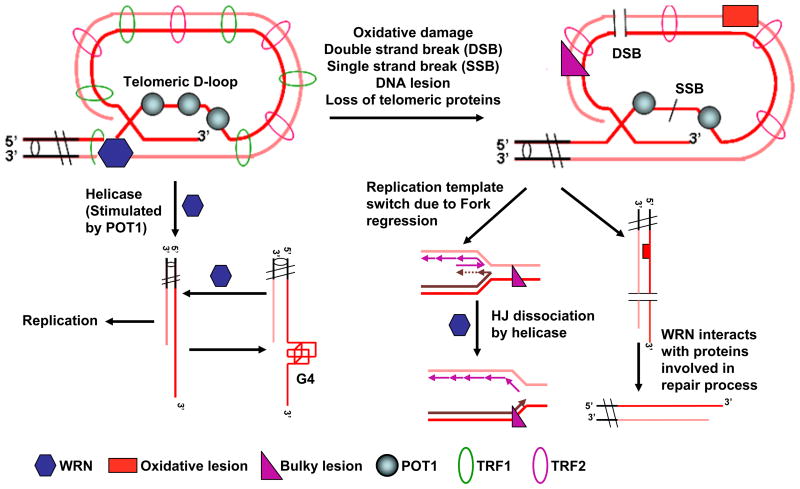
The unique sequence and structure of telomeric DNA plays an important role in maintenance of the telomere [184]. In mammals, telomeres are composed of double-stranded tandem repeat sequences followed by a single-stranded short 3′-overhang, which invades the double-stranded region to create a loop structure (Figure 3) [185]. The telomere binding and associated proteins stabilize this so-called “D-loop” (displacement loop) configuration. As the single-stranded region is G-rich, G-quadruplex (G4) structures may also arise during telomere metabolism [186]. Crystal structure analysis of G4 generated from telomeric DNA sequences [187], as well as in vivo [188] studies, provided evidence for the existence of G4 structures in the telomeric region. Recently, the formation of G4 structures was also observed within telomeric RNA (TERRA) [189].
Figure 3. Role of WRN in replication, recombination, and repair processes at telomeres.
(See text for details.)
Telomerase, which contains both telomerase RNA and a reverse transcriptase (TERT), is another crucial component at telomeres that maintains telomere length [190, 191]. Additionally, telomere-specific binding proteins and their associated proteins protect the telomeric DNA at the end of the chromosomes [192] and are collectively referred to as the ‘shelterin’ complex [180]. In mammals, this complex includes TRF1 and TRF2, proteins that bind to the double-stranded telomeric DNA, and POT1, a protein that binds to the single-stranded telomeric DNA overhang, as well as their associated proteins Rap1, TIN2, and TPP1.
Apart from the shelterin proteins, a number of DNA repair/damage checkpoint proteins associate with the telomere protein complex. These include ATM, ATR, and DNA helicases and nucleases (WRN, BLM, Apollo, EXOL1), as well as components of HR or NHEJ (KU/DNA-PKcs), DSBR (MRE11/NBS1/RAD50 complex), and NER/BER (ERCC1/XPF, PARP1/PARP2, FEN1) [2]. Many of these DNA repair/damage checkpoint proteins are actively involved in telomere maintenance, possibly by assisting in telomeric DNA repair and telomere capping. RecQ helicases, especially WRN, are known to play significant roles in proper maintenance of telomeres. WRN functionally and/or physically interacts with telomeric DNA protecting proteins POT1, TRF1 and TRF2 [193–195]. In addition, it is associated with Mre11, Rad50, and Nbs1, which are also associated with TRF2 [51]. Furthermore, WRN functionally interacts with Ku, which interacts with both TRF1 and TRF2 [156]. Moreover, interactions of WRN with telomeric D-loop and G4 structures were shown by in vitro studies [194, 196].
Besides biochemical evidence of WRN association with telomere associated proteins, there is cellular evidence of a role for WRN in telomere processing. WRN and TERT minus double mutant mice have phenotypes resembling human clinical features [197]. In vivo gene specific repair studies have shown that the extent and rate of telomeric repair is lower in WS patients [198]. This notion is further supported by accelerated telomere loss displayed in WS cells [199]. A recent study on the effect of the glucose utilization inhibitor 2-deoxy-D-glucose on gene/protein expressions of RecQ helicases and telomerase also indicates that the cooperative interaction between WRN and telomerase is advantageous for proper telomere maintenance in cells [200]. We will discuss the implications of these studies briefly.
Telomere Replication
One of the most important implications of WRN’s interaction with shelterin proteins and telomere specific DNA substrates is its involvement in telomere replication. Replication fork terminations at telomeres are particularly damaging due to the absence of any replication origins past the telomeres to account for the unreplicated portions. Early evidence of involvement of WRN in replication comes from the fact that WS patients show prolonged S-phase [67] and WRN colocalizes with RPA in cells arrested in S-phase by HU treatment [201, 202]. Although, WRN localizes predominantly to the nucleolus in telomerase-positive human cell lines, a significant fraction of WRN colocalizes with TRF1 and TRF2 at telomeric DNA in human ALT cell lines [193, 194]. This is suggestive of a role of WRN in telomeric DNA replication. However, WRN might not participate in the general telomere replication process, as it is associated with only 5% of telomeric S-phase fibroblasts [11]. Rather, it may be recruited to the replicating telomeres in response to replication stress. CO-FISH studies also suggested that WRN is involved in an alternative mechanism to resolve relatively rare, but lethal, events during telomere replication [203].
Dissociation of D-loop structures is required for replication of telomeric DNA replication. In vitro studies have shown that WRN unwinds the D-loop structures to release the invading strand, a process that is regulated by TRF1 and TRF2 [194]. WRN helicase activity at telomeric D-loop structures is enhanced by physical interaction with TRF2 [193]. Conversely, TRF1 and TRF2 limit the exonuclease activity of WRN suggesting the cooperation of these telomere binding proteins with WRN in the processing of telomeric DNA. Telomeric single strand binding protein POT1 also enhances D-loop unwinding of WRN [195] by maintaining the unwound strands in a melted state (Figure 3) [204]. WRN also has a role in restart of stalled replication forks, an important mechanism for continuing the replication process following DNA damage. WRN can efficiently unwind DNA substrates with a one nucleotide gap. More importantly, WRN enhances FEN1 activity to cleave the unwound 5′-strand in vitro. WRN and FEN-1 co-localize at PCNA foci after the induction of stalled replication forks in vivo [88]. Furthermore, formation of G4 structures is another potential block to the replication of telomeric invading strand. G4 structures are favored substrates for WRN, which can prevent replication stalling at G4 DNA [104]. It was also shown that POT1 can resolve telomeric G4 structures [205]. Thus, WRN and POT1 can cooperate with each other to resolve the G4 structures at telomere (Figure 3).
Telomeric Recombination and Repair
Alternative lengthening of telomeres (ALT) pathways involve multiple telomere binding and recombination proteins. In budding yeast, RecQ helicase Sgs1 acts in resolution of recombination intermediates, when critically short telomeres undergo recombination to restore the telomeric length. A fraction of telomeric DNA from human cell lines maintaining the telomeres by ALT co-localizes with WRN [11], and it can partially substitute the function of Sgs1 in type II ALT [206, 207]. In vivo study demonstrates that telomerase-positive WS fibroblasts develop an elevated level of telomeric t-circles in the absence of the N-terminal basic domain of TRF2. Reconstitution with WRN reduces this t-circle formation. However, mutant WRN lacking either helicase or exonuclease is not able reduce the t-circles, indicating roles for both the functions of WRN to suppress the t-circle formations in telomerase-positive cells [208]. WRN interacts physically with the Ku70/80 heterodimer and POT1, which also have crucial roles in protection of telomeres from aberrant HR [209, 210]. As WRN interacts with both these proteins, it may be also required to suppress the unusual recombination intermediates at telomeres.
In addition to recombination based repair, WRN has been implicated in oxidative DNA damage repair at telomeres (Figure 3) [11]. In vitro analysis has shown that guanine (G) has the lowest oxidation potential among the nucleobases and the GGG sequence has even lower oxidation potential [211]. Repetitive sequence of triple guanines makes telomeric DNA prone to oxidative damage, lesions such as 8-oxodG. The association between oxidative DNA damage and telomere shortening is described in several studies [212, 213]. Oxidative DNA damage is repaired by the BER process, and WRN is believed to take part in BER because it physically interacts with several of the proteins involved (see Multiple Roles of WRN in DNA Repair section). Recently, we have seen that WRN interacts with the D-loop structures containing 8-oxodG lesions, and it unwinds these more efficiently than the undamaged D-loops [214]. This result also indicates a role of WRN in resolving complex structures at damaged telomeres in order to accelerate the activities of the repair enzymes. However, it is still mostly unclear whether and how BER functions at the telomeres, and a significant amount of research is needed to assess the exact role of WRN in the repair of oxidative damage at telomeres.
Model system for human WS
Although WRN helicase has several activities in human cells, Wrn-null mice do not show any phenotypes of human WS [215]. However, studies using mice null with respect to Wrn and mTerc (encoding telomerase RNA component) revealed that the late generation mTerc−/−Wrn−/− mouse displays most of the classical clinical features resembling human WS [197, 216]. WS human fibroblasts show elevated levels of DNA-damage as detected by γH2AX and 53BP1 and a profound decline in replicative lifespan, which was also displayed by the mTerc−/−Wrn−/− mouse embryonic fibroblasts. It is important to note that these features do not represent deterioration of aging phenotypes observed in the telomerase null mouse, but reiterate the specific phenotypes of WS patients. WS patients also exhibit increased incidence of chromosomal instability and cancer. Studies using the mTerc−/−Wrn−/− double knockout mice indicate that these may be caused by aberrant recombination at telomeres [10]. Together, the results strongly support the hypothesis that Wrn-null mice do not show human WS phenotypes, as they have long telomeres, and to manifest the premature aging symptoms, critically short telomeres are required. These studies also suggest that WRN and telomerase work together to maintain the telomere integrity and WRN is strongly involved in telomere dynamics.
Comparison of WRN with other RecQ Helicases
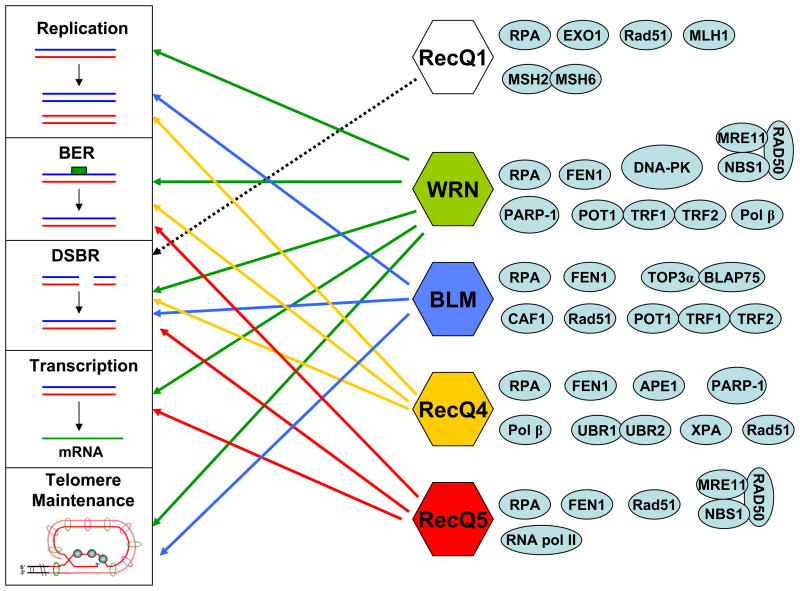
While most other eukaryotes possess one or two members of the RecQ helicase family, humans have five distinct RecQ helicases. Each of these RecQ1, WRN, BLM, RecQ4, and RecQ5 is involved in multiple pathways to maintain genome stability [1–3, 217] (see additional reviews in this issue). Accordingly, questions of redundancy can be raised. Why do humans have multiple RecQ helicases? Do they compete with each other or are their cellular roles unique? To address these questions, it is important to compare the functions of the RecQ helicases and the pathways in which they participate (Figure 4).
Figure 4. Involvement of RecQ helicases in maintenance of genomic integrity.
Association of different RecQ helicases (RecQ1, WRN, BLM, RecQ4, and RecQ5) with replication, base excision repair (BER), double strand break repair (DSBR), transcription, and telomere maintenance has been indicated by arrows. Dotted line indicates probable involvement. Some major physical and/or functional interacting proteins are also shown with each respective RecQ helicase. (See text for additional details and reviews in [1–3, 5, 217].)
The human RecQ helicases are the causative factors of a variety of human premature aging and cancer disposition pathologies. Deficiencies in WRN and BLM are linked to WS and Bloom syndrome (BS), respectively. BS is characterized by short stature, skin lesions, immunodeficiency, low fertility or infertility, diabetes, and cancer predisposition [218, 219]. Furthermore, RecQ4 deficiencies are the cause of Baller-Gerold syndrome (BGS), RAPADILINO syndrome, and Rothmund-Thomson syndrome (RTS) [220–224]. BGS patients have short stature, abnormal skull and brain growth (craniosynostosis), and absent development or underdevelopment of the radius (radial aplasia or hypoplasia) [221]. RAPADILINO is typified by radial and patellar hypoplasia, cleft palate, diarrhoea, dislocated joints, little size, limb malformation, slender nose, normal intelligence, and predisposition to lymphoma and osteosarcoma [225]. RTS presents with developmental abnormalities, growth deficiencies, proneness to cancer, predominantly osteosacrcomas, and premature aging, including development of cataracts and hair loss [226, 227]. To date, no human pathologies have been found with deficiencies in either RecQ1 or RecQ5. Still, while some of the clinical features of each of the RecQ associated diseases overlap, some are unique, suggesting that the RecQ helicases may have differential cellular roles that translate into the distinct symptoms.
Enzymatically all the RecQ helicases are similar, carrying out 3′-5′ ATP-dependent unwinding and DNA strand annealing activities [217]. However, the substrate specificity of each of the helicases is varied. WRN, BLM, RecQ1, and RecQ5 are capable of readily unwinding forked substrates with shorter regions of duplex DNA and longer duplexes in the presence of RPA [21, 114–116, 228–231]. Likewise, it has been shown recently that RecQ4 also shows unwinding on fork substrates [232, 233] which is stimulated by RPA (Rossi et al., submitted for publication). In contrast, only WRN and BLM can effectively resolve G4 DNA [34, 234, 235], while RecQ1 [236] and RecQ4 cannot (Ghosh and Bohr, unpublished data). Additionally, WRN and BLM readily unwind telomeric D-loops, while RecQ5 displays weak activity on these substrates [214]. RecQ5 displays a similarly weak activity on Holliday junctions [237], which are resolved efficiently by WRN, BLM, and RecQ1 [34, 236, 238]. However, immobile Holliday junctions are only unwound by RecQ1 not BLM [236]. It is evident from these examples that the RecQ family members are structure specific helicases with some overlapping and some unique substrate specificities.
Moreover, the effectiveness of WRN and BLM in processing complex structures like G4 suggests that they may play a more prominent role in facilitating DNA replication and repair through specialized regions of the genome, such as telomeres. Alternatively, WRN, BLM, and RecQ1 resolution of Holliday junctions could indicate a favored role in homologous recombination pathways. Still, RecQ4 and RecQ5 preferential unwinding of simpler replication fork substrates could be indicative of a preferred role in replication processes over the other RecQ helicases [1–3, 6]. Nevertheless, a comprehensive study comparing the activity of all the RecQ helicases on a panel of substrates representing intermediates of DNA replication and repair processes has not been conducted. Because helicase activity has now been detected in all of the human RecQ family members, such research is merited and may provide insight into which specific pathways each of the RecQ helicases function. It is a remaining challenge to determine whether there is a specific substrate, potentially one that contains DNA damage, on which some of the RecQ helicases can act with synergy.
Furthermore, analysis of the interacting partners of the RecQ helicases provides clues as to the redundancy and uniqueness of functions of the family members. As mentioned above, the activity of the RecQ helicases can be stimulated by RPA [21, 114–116, 228–231]. Since RPA is involved in both DNA replication and DNA repair [239], and the interaction is conserved among all the RecQ family members, it is not a distinguishing factor of RecQ helicase cellular functions. In addition, another highly conserved physical and functional protein interaction is FEN1. WRN, BLM, RecQ4, and RecQ5 stimulate FEN1 cleavage [109, 240, 241] (and Speina et al., submitted for publication), implicating these RecQ helicases in lagging strand replication and/or BER.
On the other hand, some protein associations are more restricted to specific RecQ helicases. For example, WRN and BLM both interact with the telomere proteins TRF1 and TRF2 and are stimulated by POT1 [193–195]. Although the interaction between these telomere binding proteins and either RecQ1, RecQ4, or RecQ5 has not been tested, it is conceivable that the associations are limited to WRN and BLM. Such a privileged interaction would suggest that WRN and BLM are uniquely involved in telomere maintenance. Moreover, the associations of WRN with DNA-PK, Ku70/80, and X4L4 imply a direct role of WRN in NHEJ, which has not been detected for the other RecQ helicases [37, 153, 154, 159]. Additionally, WRN and RecQ5 have been linked to transcription [173, 242–244]. WRN was shown to regulate RNA pol II transcription efficiency potentially by activation [173]. Conversely, RecQ5 inhibits transcription through a direct physical interaction with RNA pol II [242, 243]. These results indicate that even if the pathways in which the RecQ helicases function overlap, each helicase may have differential effects.
Generally, the RecQ helicases participate in many of the same pathways of DNA replication and repair. However, the specific mechanisms of function is not been fully elucidated. While it appears that the RecQ helicases are partly redundant, there are clear examples of distinct function. Additional studies of the biochemical and cellular functions of RecQ1, WRN, BLM, RecQ4, and RecQ5, may reveal more distinct roles of these proteins, distinguishing one from the other.
Perspectives
Although the connection between the WS phenotype and the function of the WRN protein has yet to be made, it is clear that the missing link is related to the function of WRN in DNA replication, DNA repair, transcription, and telomere maintenance. Past studies clearly indicate that WRN is important for protection of genome stability. Yet a complete understanding of all the mechanisms of WRN’s function in DNA transactions remains elusive. Future research on the deficiencies in WRN depleted cells and the roles of WRN in DNA metabolism will reveal more complete understanding of its cellular functions. It will be important to determine how the involvement of WRN in a variety of different processes is modulated, whether through post-translational modifications and/or through protein-protein interactions. Moreover, additional study of the function of WRN in comparison with BLM, RecQ1, RecQ4, and RecQ5 may reveal some unique roles, distinguishing it from the other RecQ helicases. Understanding the differences between the RecQ helicases will ultimately be necessary to address the question as to why humans have developed multiple seemingly redundant proteins.
Acknowledgments
We thank Dr. Dharmendra K. Singh and Mr. Joshua A. Sommers for helpful comments on the manuscript. This work was supported entirely by funds from the Intramural Research Program of the National Institutes of Health, National Institute on Aging. We regret that space limitations and the breadth of this field prevented us from citing every relevant reference.
Abbreviations
- γ-H2AX
phosphorylated histone H2AX
- 4-NQO
4-nitroquinoline 1-oxide
- 5-OHdC
5-hydroxycytidine
- 5-OHU
5-hydroxyuracil
- 8-oxodA
8-oxoadenine
- 8-oxodG
8-oxoguanine
- ALT
alternative lengthening of telomeres
- APE1
apurinic/apyrimidinic endonuclease 1
- ATM
ataxia telangiectasia mutated
- ATR
ATM and Rad3 related
- BER
base excision repair
- BLM
Bloom syndrome protein
- CeWRN-1
Caenhorhabditis elegans WRN
- CPT
camptothecin
- D-loop
displacement loop
- DmWRN
Drosophila melanogaster Werner syndrome protein
- DSB
double strand break
- DSBR
double strand break repair
- ETO
etoposide
- fapy
formamidopyrimidine
- FEN1
flap endonuclease 1
- FFA-1
focus forming activity 1
- G4
G-quadruplex
- HR
homolous recombination
- HU
hydroxyurea
- IR
ionizing radiation
- LMNA
lamin A/C
- MMR
mismatch repair
- MMS
methylmethane sulfonate
- NER
nucleotide excision repair
- NHEJ
non-homologous end joining
- PARP-1
poly (ADP-ribose) polymerase
- PCNA
proliferating cell nuclear antigen
- Pol β
DNA Polymerase β Pol δ, DNA Polymerase δ
- PUVA
psoralen plus UVA
- RNA pol I
RNA polymerase I
- RNA pol II
RNA polymerase II
- ROS
reactive oxygen species
- RPA
replication protein A
- rRNA
ribosomal RNA
- SNPs
single nucleotide polymorphisms
- SSB
single strand break
- TPT
topotecan
- WRN
Werner syndrome protein
- WS
Werner syndrome
- X4L4
XRCC4/Ligase IV
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009 doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 4.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–2322. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh DK, Ahn B, Bohr VA. Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology. 2009;10:235–252. doi: 10.1007/s10522-008-9205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 7.Kusumoto R, Muftuoglu M, Bohr VA. The role of WRN in DNA repair is affected by post-translational modifications. Mech Ageing Dev. 2007;128:50–57. doi: 10.1016/j.mad.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Ding SL, Shen CY. Model of human aging: recent findings on Werner’s and Hutchinson-Gilford progeria syndromes. Clin Interv Aging. 2008;3:431–444. doi: 10.2147/cia.s1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muftuoglu M, Oshima J, von KC, Cheng WH, Leistritz DF, Bohr VA. The clinical characteristics of Werner syndrome: molecular and biochemical diagnosis. Hum Genet. 2008;124:369–377. doi: 10.1007/s00439-008-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Multani AS, Chang S. WRN at telomeres: implications for aging and cancer. J Cell Sci. 2007;120:713–721. doi: 10.1242/jcs.03397. [DOI] [PubMed] [Google Scholar]
- 11.Opresko PL. Telomere ResQue and preservation--roles for the Werner syndrome protein and other RecQ helicases. Mech Ageing Dev. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Sidorova JM. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair (Amst) 2008;7:1776–1786. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Lee L, Hanson NB, Lenaerts C, Hoehn H, Poot M, Rubin CD, Chen DF, Yang CC, Juch H, Dorn T, Spiegel R, Oral EA, Abid M, Battisti C, Lucci-Cordisco E, Neri G, Steed EH, Kidd A, Isley W, Showalter D, Vittone JL, Konstantinow A, Ring J, Meyer P, Wenger SL, von HA, Wollina U, Schuelke M, Huizenga CR, Leistritz DF, Martin GM, Mian IS, Oshima J. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27:558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Futami K, Ishikawa Y, Goto M, Furuichi Y, Sugimoto M. Role of Werner syndrome gene product helicase in carcinogenesis and in resistance to genotoxins by cancer cells. Cancer Sci. 2008;99:843–848. doi: 10.1111/j.1349-7006.2008.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Goto M. Hierarchical deterioration of body systems in Werner’s syndrome: implications for normal ageing. Mech Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 17.Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 18.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-->5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner syndrome protein. II. Characterization of the integral 3′ --> 5′ DNA exonuclease. J Biol Chem. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- 20.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 21.Shen JC, Gray MD, Oshima J, Loeb LA. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26:2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. DNA helicase activity in Werner’s syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′-->5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y, Ratcliff GC, Wang H, vis-Searles PR, Gray MD, Erie DA, Redinbo MR. A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3′–5′ exonuclease and 5′-protruding strand endonuclease activities. Biochemistry. 2002;41:2901–2912. doi: 10.1021/bi0157161. [DOI] [PubMed] [Google Scholar]
- 26.Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. The Werner syndrome protein binds replication fork and Holliday junction DNAs as an oligomer. J Biol Chem. 2008;283:24478–24483. doi: 10.1074/jbc.M803370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choudhary S, Sommers JA, Brosh RM., Jr Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of werner syndrome protein. J Biol Chem. 2004;279:34603–34613. doi: 10.1074/jbc.M401901200. [DOI] [PubMed] [Google Scholar]
- 28.Hyun M, Bohr VA, Ahn B. Biochemical characterization of the WRN-1 RecQ helicase of Caenorhabditis elegans. Biochemistry. 2008;47:7583–7593. doi: 10.1021/bi800197m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SJ, Yook JS, Han SM, Koo HS. A Werner syndrome protein homolog affects C. elegans development, growth rate, life span and sensitivity to DNA damage by acting at a DNA damage checkpoint. Development. 2004;131:2565–2575. doi: 10.1242/dev.01136. [DOI] [PubMed] [Google Scholar]
- 30.Boubriak I, Mason PA, Clancy DJ, Dockray J, Saunders RD, Cox LS. DmWRNexo is a 3′-5′ exonuclease: phenotypic and biochemical characterization of mutants of the Drosophila orthologue of human WRN exonuclease. Biogerontology. 2009;10:267–277. doi: 10.1007/s10522-008-9181-3. [DOI] [PubMed] [Google Scholar]
- 31.Cox LS, Clancy DJ, Boubriak I, Saunders RD. Modeling Werner Syndrome in Drosophila melanogaster: hyper-recombination in flies lacking WRN-like exonuclease. Ann N Y Acad Sci. 2007;1119:274–288. doi: 10.1196/annals.1404.009. [DOI] [PubMed] [Google Scholar]
- 32.Saunders RD, Boubriak I, Clancy DJ, Cox LS. Identification and characterization of a Drosophila ortholog of WRN exonuclease that is required to maintain genome integrity. Aging Cell. 2008;7:418–425. doi: 10.1111/j.1474-9726.2008.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan H, Chen CY, Kobayashi R, Newport J. Replication focus-forming activity 1 and the Werner syndrome gene product. Nat Genet. 1998;19:375–378. doi: 10.1038/1263. [DOI] [PubMed] [Google Scholar]
- 34.Mohaghegh P, Karow JK, Brosh JR, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orren DK, Theodore S, Machwe A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- 36.Harrigan JA, Piotrowski J, Di NL, Levine RL, Bohr VA. Metal-catalyzed oxidation of the Werner syndrome protein causes loss of catalytic activities and impaired protein-protein interactions. J Biol Chem. 2007;282:36403–36411. doi: 10.1074/jbc.M706107200. [DOI] [PubMed] [Google Scholar]
- 37.Karmakar P, Piotrowski J, Brosh RM, Jr, Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J Biol Chem. 2002;277:18291–18302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- 38.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 39.Cheng WH, von KC, Opresko PL, Fields KM, Ren J, Kufe D, Bohr VA. Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution. Mol Cell Biol. 2003;23:6385–6395. doi: 10.1128/MCB.23.18.6385-6395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muftuoglu M, Kusumoto R, Speina E, Beck G, Cheng WH, Bohr VA. Acetylation regulates WRN catalytic activities and affects base excision DNA repair. PLoS One. 2008;3:e1918. doi: 10.1371/journal.pone.0001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woods YL, Xirodimas DP, Prescott AR, Sparks A, Lane DP, Saville MK. p14 Arf promotes small ubiquitin-like modifier conjugation of Werners helicase. J Biol Chem. 2004;279:50157–50166. doi: 10.1074/jbc.M405414200. [DOI] [PubMed] [Google Scholar]
- 42.Agrelo R, Cheng WH, Setien F, Ropero S, Espada J, Fraga MF, Herranz M, Paz MF, Sanchez-Cespedes M, Artiga MJ, Guerrero D, Castells A, von KC, Bohr VA, Esteller M. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci U S A. 2006;103:8822–8827. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Opresko PL, Calvo JP, von KC. Role for the Werner syndrome protein in the promotion of tumor cell growth. Mech Ageing Dev. 2007;128:423–436. doi: 10.1016/j.mad.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 45.Futami K, Takagi M, Shimamoto A, Sugimoto M, Furuichi Y. Increased chemotherapeutic activity of camptothecin in cancer cells by siRNA-induced silencing of WRN helicase. Biol Pharm Bull. 2007;30:1958–1961. doi: 10.1248/bpb.30.1958. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Xu Y, Tang J, Ma H, Qin J, Lu C, Wang X, Hu Z, Wang X, Shen H. A polymorphism in Werner syndrome gene is associated with breast cancer susceptibility in Chinese women. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0327-z. [DOI] [PubMed] [Google Scholar]
- 47.Wirtenberger M, Frank B, Hemminki K, Klaes R, Schmutzler RK, Wappenschmidt B, Meindl A, Kiechle M, Arnold N, Weber BH, Niederacher D, Bartram CR, Burwinkel B. Interaction of Werner and Bloom syndrome genes with p53 in familial breast cancer. Carcinogenesis. 2006;27:1655–1660. doi: 10.1093/carcin/bgi374. [DOI] [PubMed] [Google Scholar]
- 48.Bohr VA, Metter EJ, Harrigan JA, von KC, Liu JL, Gray MD, Majumdar A, Wilson DM, III, Seidman MM. Werner syndrome protein 1367 variants and disposition towards coronary artery disease in Caucasian patients. Mech Ageing Dev. 2004;125:491–496. doi: 10.1016/j.mad.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Kamath-Loeb AS, Welcsh P, Waite M, Adman ET, Loeb LA. The enzymatic activities of the Werner syndrome protein are disabled by the amino acid polymorphism R834C. J Biol Chem. 2004;279:55499–55505. doi: 10.1074/jbc.M407128200. [DOI] [PubMed] [Google Scholar]
- 50.Blander G, Kipnis J, Leal JF, Yu CE, Schellenberg GD, Oren M. Physical and functional interaction between p53 and the Werner’s syndrome protein. J Biol Chem. 1999;274:29463–29469. doi: 10.1074/jbc.274.41.29463. [DOI] [PubMed] [Google Scholar]
- 51.Cheng WH, Kusumoto R, Opresko PL, Sui X, Huang S, Nicolette ML, Paull TT, Campisi J, Seidman M, Bohr VA. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006;34:2751–2760. doi: 10.1093/nar/gkl362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding SL, Yu JC, Chen ST, Hsu GC, Shen CY. Genetic variation in the premature aging gene WRN: a case-control study on breast cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2007;16:263–269. doi: 10.1158/1055-9965.EPI-06-0678. [DOI] [PubMed] [Google Scholar]
- 53.Pinto GR, Yoshioka FK, Clara CA, Santos MJ, Almeida JR, Burbano RR, Rey JA, Casartelli C. WRN Cys1367Arg SNP is not associated with risk and prognosis of gliomas in Southeast Brazil. J Neurooncol. 2008;90:253–258. doi: 10.1007/s11060-008-9664-8. [DOI] [PubMed] [Google Scholar]
- 54.Hirai M, Suzuki S, Hinokio Y, Yamada T, Yoshizumi S, Suzuki C, Satoh J, Oka Y. WRN gene 1367 Arg allele protects against development of type 2 diabetes mellitus. Diabetes Res Clin Pract. 2005;69:287–292. doi: 10.1016/j.diabres.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 55.Nakayama R, Sato Y, Masutani M, Ogino H, Nakatani F, Chuman H, Beppu Y, Morioka H, Yabe H, Hirose H, Sugimura H, Sakamoto H, Ohta T, Toyama Y, Yoshida T, Kawai A. Association of a missense single nucleotide polymorphism, Cys1367Arg of the WRN gene, with the risk of bone and soft tissue sarcomas in Japan. Cancer Sci. 2008;99:333–339. doi: 10.1111/j.1349-7006.2007.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, Wang SS, Holford TR, Leaderer B, Yeager M, Welch R, Kang D, Boyle P, Zhang B, Zou K, Zhu Y, Chanock S, Rothman N. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Hum Genet. 2006;119:659–668. doi: 10.1007/s00439-006-0177-2. [DOI] [PubMed] [Google Scholar]
- 57.Castro E, Edland SD, Lee L, Ogburn CE, Deeb SS, Brown G, Panduro A, Riestra R, Tilvis R, Louhija J, Penttinen R, Erkkola R, Wang L, Martin GM, Oshima J. Polymorphisms at the Werner locus: II. 1074Leu/Phe, 1367Cys/Arg, longevity, and atherosclerosis. Am J Med Genet. 2000;95:374–380. [PubMed] [Google Scholar]
- 58.Ye L, Miki T, Nakura J, Oshima J, Kamino K, Rakugi H, Ikegami H, Higaki J, Edland SD, Martin GM, Ogihara T. Association of a polymorphic variant of the Werner helicase gene with myocardial infarction in a Japanese population. Am J Med Genet. 1997;68:494–498. doi: 10.1002/(sici)1096-8628(19970211)68:4<494::aid-ajmg30>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 59.Castro E, Oviedo-Rodriguez V, ngel-Chavez LI. WRN polymorphisms affect expression levels of plasminogen activator inhibitor type 1 in cultured fibroblasts. BMC Cardiovasc Disord. 2008;8:5. doi: 10.1186/1471-2261-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murano S, Nakazawa A, Saito I, Masuda M, Morisaki N, Akikusa B, Tsuboyama T, Saito Y. Increased blood plasminogen activator inhibitor-1 and intercellular adhesion molecule-1 as possible risk factors of atherosclerosis in Werner syndrome. Gerontology. 1997;43(Suppl 1):43–52. doi: 10.1159/000213885. [DOI] [PubMed] [Google Scholar]
- 61.Chen L, Lee L, Kudlow BA, dos Santos HG, Sletvold O, Shafeghati Y, Botha EG, Garg A, Hanson NB, Martin GM, Mian IS, Kennedy BK, Oshima J. LMNA mutations in atypical Werner’s syndrome. Lancet. 2003;362:440–445. doi: 10.1016/S0140-6736(03)14069-X. [DOI] [PubMed] [Google Scholar]
- 62.Doh YJ, Kim HK, Jung ED, Choi SH, Kim JG, Kim BW, Lee IK. Novel LMNA gene mutation in a patient with Atypical Werner’s Syndrome. Korean J Intern Med. 2009;24:68–72. doi: 10.3904/kjim.2009.24.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacob KN, Baptista F, dos Santos HG, Oshima J, Agarwal AK, Garg A. Phenotypic heterogeneity in body fat distribution in patients with atypical Werner’s syndrome due to heterozygous Arg133Leu lamin A/C mutation. J Clin Endocrinol Metab. 2005;90:6699–6706. doi: 10.1210/jc.2005-0939. [DOI] [PubMed] [Google Scholar]
- 64.Renard D, Fourcade G, Milhaud D, Bessis D, Esteves-Vieira V, Boyer A, Roll P, Bourgeois P, Levy N, De Sandre-Giovannoli A. Novel LMNA mutation in atypical Werner syndrome presenting with ischemic disease. Stroke. 2009;40:e11–e14. doi: 10.1161/STROKEAHA.108.531780. [DOI] [PubMed] [Google Scholar]
- 65.Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, Rabinovitch PS, Monnat RJ., Jr Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 66.Hanaoka F, Yamada M, Takeuchi F, Goto M, Miyamoto T, Hori T. Autoradiographic studies of DNA replication in Werner’s syndrome cells. Adv Exp Med Biol. 1985;190:439–457. doi: 10.1007/978-1-4684-7853-2_22. [DOI] [PubMed] [Google Scholar]
- 67.Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 68.Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner’s syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 69.Takeuchi F, Hanaoka F, Goto M, Yamada M, Miyamoto T. Prolongation of S phase and whole cell cycle in Werner’s syndrome fibroblasts. Exp Gerontol. 1982;17:473–480. doi: 10.1016/s0531-5565(82)80009-0. [DOI] [PubMed] [Google Scholar]
- 70.Fujiwara Y, Higashikawa T, Tatsumi M. A retarded rate of DNA replication and normal level of DNA repair in Werner’s syndrome fibroblasts in culture. J Cell Physiol. 1977;92:365–374. doi: 10.1002/jcp.1040920305. [DOI] [PubMed] [Google Scholar]
- 71.Fujiwara Y, Kano Y, Ichihashi M, Nakao Y, Matsumura T. Abnormal fibroblast aging and DNA replication in the Werner syndrome. Adv Exp Med Biol. 1985;190:459–477. doi: 10.1007/978-1-4684-7853-2_23. [DOI] [PubMed] [Google Scholar]
- 72.Hanaoka F, Takeuchi F, Matsumura T, Goto M, Miyamoto T, Yamada M. Decrease in the average size of replicons in a Werner syndrome cell line by Simian virus 40 infection. Exp Cell Res. 1983;144:463–467. doi: 10.1016/0014-4827(83)90425-1. [DOI] [PubMed] [Google Scholar]
- 73.Takeuchi F, Hanaoka F, Goto M, Akaoka I, Hori T, Yamada M, Miyamoto T. Altered frequency of initiation sites of DNA replication in Werner’s syndrome cells. Hum Genet. 1982;60:365–368. doi: 10.1007/BF00569220. [DOI] [PubMed] [Google Scholar]
- 74.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner’s syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol Biol Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodriguez-Lopez AM, Whitby MC, Borer CM, Bachler MA, Cox LS. Correction of proliferation and drug sensitivity defects in the progeroid Werner’s Syndrome by Holliday junction resolution. Rejuvenation Res. 2007;10:27–40. doi: 10.1089/rej.2006.0503. [DOI] [PubMed] [Google Scholar]
- 77.Salk D, Bryant E, Hoehn H, Johnston P, Martin GM. Growth characteristics of Werner syndrome cells in vitro. Adv Exp Med Biol. 1985;190:305–311. doi: 10.1007/978-1-4684-7853-2_14. [DOI] [PubMed] [Google Scholar]
- 78.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 79.Leppard JB, Champoux JJ. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. doi: 10.1007/s00412-005-0345-5. [DOI] [PubMed] [Google Scholar]
- 80.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 81.Christmann M, Tomicic MT, Gestrich C, Roos WP, Bohr VA, Kaina B. WRN protects against topo I but not topo II inhibitors by preventing DNA break formation. DNA Repair (Amst) 2008;7:1999–2009. doi: 10.1016/j.dnarep.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bridgewater LC, Manning FC, Patierno SR. Arrest of replication by mammalian DNA polymerases alpha and beta caused by chromium-DNA lesions. Mol Carcinog. 1998;23:201–206. [PubMed] [Google Scholar]
- 84.O’Brien T, Mandel HG, Pritchard DE, Patierno SR. Critical role of chromium (Cr)-DNA interactions in the formation of Cr-induced polymerase arresting lesions. Biochemistry. 2002;41:12529–12537. doi: 10.1021/bi020452j. [DOI] [PubMed] [Google Scholar]
- 85.Ha L, Ceryak S, Patierno SR. Generation of S phase-dependent DNA double-strand breaks by Cr(VI) exposure: involvement of ATM in Cr(VI) induction of gamma-H2AX. Carcinogenesis. 2004;25:2265–2274. doi: 10.1093/carcin/bgh242. [DOI] [PubMed] [Google Scholar]
- 86.Reynolds MF, Peterson-Roth EC, Bespalov IA, Johnston T, Gurel VM, Menard HL, Zhitkovich A. Rapid DNA double-strand breaks resulting from processing of Cr-DNA cross-links by both MutS dimers. Cancer Res. 2009;69:1071–1079. doi: 10.1158/0008-5472.CAN-08-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu FJ, Barchowsky A, Opresko PL. The Werner syndrome protein functions in repair of Cr(VI)-induced replication-associated DNA damage. Toxicol Sci. 2009;110:307–318. doi: 10.1093/toxsci/kfp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol Biol Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 90.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 91.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 92.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 93.Shechter D, Costanzo V, Gautier J. Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst) 2004;3:901–908. doi: 10.1016/j.dnarep.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 94.Cheng WH, Muftic D, Muftuoglu M, Dawut L, Morris C, Helleday T, Shiloh Y, Bohr VA. WRN is required for ATM activation and the S-phase checkpoint in response to interstrand cross-link-induced DNA double-strand breaks. Mol Biol Cell. 2008;19:3923–3933. doi: 10.1091/mbc.E07-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pichierri P, Rosselli F, Franchitto A. Werner’s syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene. 2003;22:1491–1500. doi: 10.1038/sj.onc.1206169. [DOI] [PubMed] [Google Scholar]
- 96.Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119:5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- 97.Robinson K, Asawachaicharn N, Galloway DA, Grandori C. c-Myc accelerates S-Phase and requires WRN to avoid replication stress. PLoS One. 2009;4:e5951. doi: 10.1371/journal.pone.0005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 99.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, la-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 100.Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 102.Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 103.Glover TW, Arlt MF, Casper AM, Durkin SG. Mechanisms of common fragile site instability. Hum Mol Genet 14 Spec No. 2005;2:R197–R205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- 104.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PM, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J Biol Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 105.Kamath-Loeb AS, Johansson E, Burgers PM, Loeb LA. Functional interaction between the Werner Syndrome protein and DNA polymerase delta. Proc Natl Acad Sci U S A. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szekely AM, Chen YH, Zhang C, Oshima J, Weissman SM. Werner protein recruits DNA polymerase delta to the nucleolus. Proc Natl Acad Sci U S A. 2000;97:11365–11370. doi: 10.1073/pnas.97.21.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- 108.Rodriguez-Lopez AM, Jackson DA, Nehlin JO, Iborra F, Warren AV, Cox LS. Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner’s syndrome, and an essential replication factor, PCNA. Mech Ageing Dev. 2003;124:167–174. doi: 10.1016/s0047-6374(02)00131-8. [DOI] [PubMed] [Google Scholar]
- 109.Brosh RM, Jr, von KC, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brosh RM, Jr, Driscoll HC, Dianov GL, Sommers JA. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 2002;41:12204–12216. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- 111.Sharma S, Sommers JA, Brosh RM., Jr In vivo function of the conserved non-catalytic domain of Werner syndrome helicase in DNA replication. Hum Mol Genet. 2004;13:2247–2261. doi: 10.1093/hmg/ddh234. [DOI] [PubMed] [Google Scholar]
- 112.Sharma S, Sommers JA, Gary RK, Friedrich-Heineken E, Hubscher U, Brosh RM., Jr The interaction site of Flap Endonuclease-1 with WRN helicase suggests a coordination of WRN and PCNA. Nucleic Acids Res. 2005;33:6769–6781. doi: 10.1093/nar/gki1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zheng L, Zhou M, Chai Q, Parrish J, Xue D, Patrick SM, Turchi JJ, Yannone SM, Chen D, Shen B. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6:83–89. doi: 10.1038/sj.embor.7400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J Biol Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 115.Doherty KM, Sommers JA, Gray MD, Lee JW, von KC, Thoma NH, Kureekattil RP, Kenny MK, Brosh RM., Jr Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J Biol Chem. 2005;280:29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- 116.Shen JC, Lao Y, Kamath-Loeb A, Wold MS, Loeb LA. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech Ageing Dev. 2003;124:921–930. doi: 10.1016/s0047-6374(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 117.Wang W, Bambara RA. Human Bloom protein stimulates flap endonuclease 1 activity by resolving DNA secondary structure. J Biol Chem. 2005;280:5391–5399. doi: 10.1074/jbc.M412359200. [DOI] [PubMed] [Google Scholar]
- 118.Chen CY, Graham J, Yan H. Evidence for a replication function of FFA-1, the Xenopus orthologue of Werner syndrome protein. J Cell Biol. 2001;152:985–996. doi: 10.1083/jcb.152.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yan H, Newport J. FFA-1, a protein that promotes the formation of replication centers within nuclei. Science. 1995;269:1883–1885. doi: 10.1126/science.7569932. [DOI] [PubMed] [Google Scholar]
- 120.Sasakawa N, Fukui T, Waga S. Accumulation of FFA-1, the Xenopus homolog of Werner helicase, and DNA polymerase delta on chromatin in response to replication fork arrest. J Biochem. 2006;140:95–103. doi: 10.1093/jb/mvj130. [DOI] [PubMed] [Google Scholar]
- 121.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 122.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, Lehmann AR, Lindahl T, Lowndes N, Sarasin A, Wood RD. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006;5:986–996. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 123.Brosh RM, Jr, Karmakar P, Sommers JA, Yang Q, Wang XW, Spillare EA, Harris CC, Bohr VA. p53 Modulates the exonuclease activity of Werner syndrome protein. J Biol Chem. 2001;276:35093–35102. doi: 10.1074/jbc.M103332200. [DOI] [PubMed] [Google Scholar]
- 124.Sommers JA, Sharma S, Doherty KM, Karmakar P, Yang Q, Kenny MK, Harris CC, Brosh RM., Jr p53 modulates RPA-dependent and RPA-independent WRN helicase activity. Cancer Res. 2005;65:1223–1233. doi: 10.1158/0008-5472.CAN-03-0231. [DOI] [PubMed] [Google Scholar]
- 125.Yang Q, Zhang R, Wang XW, Spillare EA, Linke SP, Subramanian D, Griffith JD, Li JL, Hickson ID, Shen JC, Loeb LA, Mazur SJ, Appella E, Brosh RM, Jr, Karmakar P, Bohr VA, Harris CC. The processing of Holliday junctions by BLM and WRN helicases is regulated by p53. J Biol Chem. 2002;277:31980–31987. doi: 10.1074/jbc.M204111200. [DOI] [PubMed] [Google Scholar]
- 126.Spillare EA, Robles AI, Wang XW, Shen JC, Yu CE, Schellenberg GD, Harris CC. p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes Dev. 1999;13:1355–1360. doi: 10.1101/gad.13.11.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gebhart E, Bauer R, Raub U, Schinzel M, Ruprecht KW, Jonas JB. Spontaneous and induced chromosomal instability in Werner syndrome. Hum Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- 128.Grigorova M, Balajee AS, Natarajan AT. Spontaneous and X-ray-induced chromosomal aberrations in Werner syndrome cells detected by FISH using chromosome-specific painting probes. Mutagenesis. 2000;15:303–310. doi: 10.1093/mutage/15.4.303. [DOI] [PubMed] [Google Scholar]
- 129.Imamura O, Fujita K, Itoh C, Takeda S, Furuichi Y, Matsumoto T. Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene. 2002;21:954–963. doi: 10.1038/sj.onc.1205143. [DOI] [PubMed] [Google Scholar]
- 130.Ogburn CE, Oshima J, Poot M, Chen R, Hunt KE, Gollahon KA, Rabinovitch PS, Martin GM. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum Genet. 1997;101:121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- 131.Poot M, Gollahon KA, Rabinovitch PS. Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum Genet. 1999;104:10–14. doi: 10.1007/s004390050903. [DOI] [PubMed] [Google Scholar]
- 132.Harrigan JA, Wilson DM, III, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.von Kobbe C, May A, Grandori C, Bohr VA. Werner syndrome cells escape hydrogen peroxide-induced cell proliferation arrest. FASEB J. 2004;18:1970–1972. doi: 10.1096/fj.04-1895fje. [DOI] [PubMed] [Google Scholar]
- 134.Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic Biol Med. 2002;32:804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 135.Fortini P, Pascucci B, Parlanti E, D’Errico M, Simonelli V, Dogliotti E. 8-Oxoguanine DNA damage: at the crossroad of alternative repair pathways. Mutat Res. 2003;531:127–139. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 136.Szekely AM, Bleichert F, Numann A, Van KS, Manasanch E, Ben NA, Canaan A, Weissman SM. Werner protein protects nonproliferating cells from oxidative DNA damage. Mol Cell Biol. 2005;25:10492–10506. doi: 10.1128/MCB.25.23.10492-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blank A, Bobola MS, Gold B, Varadarajan S, Kolstoe D, Meade EH, Rabinovitch PS, Loeb LA, Silber JR. The Werner syndrome protein confers resistance to the DNA lesions N3-methyladenine and O6-methylguanine: implications for WRN function. DNA Repair (Amst) 2004;3:629–638. doi: 10.1016/j.dnarep.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 138.Dianov GL, Prasad R, Wilson SH, Bohr VA. Role of DNA polymerase beta in the excision step of long patch mammalian base excision repair. J Biol Chem. 1999;274:13741–13743. doi: 10.1074/jbc.274.20.13741. [DOI] [PubMed] [Google Scholar]
- 139.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 140.Harrigan JA, Opresko PL, von KC, Kedar PS, Prasad R, Wilson SH, Bohr VA. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J Biol Chem. 2003;278:22686–22695. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- 141.Harrigan JA, Fan J, Momand J, Perrino FW, Bohr VA, Wilson DM., III WRN exonuclease activity is blocked by DNA termini harboring 3′ obstructive groups. Mech Ageing Dev. 2007;128:259–266. doi: 10.1016/j.mad.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ahn B, Harrigan JA, Indig FE, Wilson DM, III, Bohr VA. Regulation of WRN helicase activity in human base excision repair. J Biol Chem. 2004;279:53465–53474. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- 143.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza-Pinto N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr VA. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase Neil1. J Biol Chem. 2007;282:26591–26602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- 144.Imoto S, Patro JN, Jiang YL, Oka N, Greenberg MM. Synthesis, DNA polymerase incorporation, and enzymatic phosphate hydrolysis of formamidopyrimidine nucleoside triphosphates. J Am Chem Soc. 2006;128:14606–14611. doi: 10.1021/ja065525r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blander G, Zalle N, Daniely Y, Taplick J, Gray MD, Oren M. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J Biol Chem. 2002;277:50934–50940. doi: 10.1074/jbc.M210479200. [DOI] [PubMed] [Google Scholar]
- 147.Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, Ge Q, Gu W, Orren D, Luo J. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 148.Kahyo T, Mostoslavsky R, Goto M, Setou M. Sirtuin-mediated deacetylation pathway stabilizes Werner syndrome protein. FEBS Lett. 2008;582:2479–2483. doi: 10.1016/j.febslet.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 149.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 151.Galvan N, Lim S, Zmugg S, Smith MT, Zhang L. Depletion of WRN enhances DNA damage in HeLa cells exposed to the benzene metabolite, hydroquinone. Mutat Res. 2008;649:54–61. doi: 10.1016/j.mrgentox.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ren X, Lim S, Smith MT, Zhang L. Werner syndrome protein, WRN, protects cells from DNA damage induced by the benzene metabolite hydroquinone. Toxicol Sci. 2009;107:367–375. doi: 10.1093/toxsci/kfn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 154.Karmakar P, Snowden CM, Ramsden DA, Bohr VA. Ku heterodimer binds to both ends of the Werner protein and functional interaction occurs at the Werner N-terminus. Nucleic Acids Res. 2002;30:3583–3591. doi: 10.1093/nar/gkf482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li B, Comai L. Requirements for the nucleolytic processing of DNA ends by the Werner syndrome protein-Ku70/80 complex. J Biol Chem. 2001;276:9896–9902. doi: 10.1074/jbc.M008575200. [DOI] [PubMed] [Google Scholar]
- 156.Orren DK, Machwe A, Karmakar P, Piotrowski J, Cooper MP, Bohr VA. A functional interaction of Ku with Werner exonuclease facilitates digestion of damaged DNA. Nucleic Acids Res. 2001;29:1926–1934. doi: 10.1093/nar/29.9.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bukowy Z, Harrigan JA, Ramsden DA, Tudek B, Bohr VA, Stevnsner T. WRN Exonuclease activity is blocked by specific oxidatively induced base lesions positioned in either DNA strand. Nucleic Acids Res. 2008;36:4975–4987. doi: 10.1093/nar/gkn468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li B, Comai L. Displacement of DNA-PKcs from DNA ends by the Werner syndrome protein. Nucleic Acids Res. 2002;30:3653–3661. doi: 10.1093/nar/gkf488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kusumoto R, Dawut L, Marchetti C, Wan LJ, Vindigni A, Ramsden D, Bohr VA. Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry. 2008;47:7548–7556. doi: 10.1021/bi702325t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sakamoto S, Nishikawa K, Heo SJ, Goto M, Furuichi Y, Shimamoto A. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells. 2001;6:421–430. doi: 10.1046/j.1365-2443.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- 163.Baynton K, Otterlei M, Bjoras M, von KC, Bohr VA, Seeberg E. WRN interacts physically and functionally with the recombination mediator protein RAD52. J Biol Chem. 2003;278:36476–36486. doi: 10.1074/jbc.M303885200. [DOI] [PubMed] [Google Scholar]
- 164.Cheng WH, von KC, Opresko PL, Arthur LM, Komatsu K, Seidman MM, Carney JP, Bohr VA. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J Biol Chem. 2004;279:21169–21176. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- 165.Poot M, Yom JS, Whang SH, Kato JT, Gollahon KA, Rabinovitch PS. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- 166.Laine JP, Opresko PL, Indig FE, Harrigan JA, von KC, Bohr VA. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res. 2003;63:7136–7146. [PubMed] [Google Scholar]
- 167.Aggarwal M, Brosh RM., Jr Premature aging syndrome gene WRN genetically interacts with a topoisomerase. Cell Cycle. 2009;8:2143. doi: 10.4161/cc.8.14.9118. [DOI] [PubMed] [Google Scholar]
- 168.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 169.von Kobbe C, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh RM, Jr, Hickson ID, Bohr VA. Colocalization, Physical, and Functional Interaction between Werner and Bloom Syndrome Proteins. J Biol Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 170.Gray MD, Wang L, Youssoufian H, Martin GM, Oshima J. Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Experimental Cell Research. 1998;242:487–494. doi: 10.1006/excr.1998.4124. [DOI] [PubMed] [Google Scholar]
- 171.Shiratori M, Suzuki T, Itoh C, Goto M, Furuichi Y, Matsumoto T. WRN helicase accelerates the transcription of ribosomal RNA as a component of an RNA polymerase I-associated complex. Oncogene. 2002;21:2447–2454. doi: 10.1038/sj.onc.1205334. [DOI] [PubMed] [Google Scholar]
- 172.Machwe A, Orren DK, Bohr VA. Accelerated methylation of ribosomal RNA genes during the cellular senescence of Werner syndrome fibroblasts. Faseb Journal. 2000;14:1715–1724. doi: 10.1096/fj.99-0926com. [DOI] [PubMed] [Google Scholar]
- 173.Balajee AS, Machwe A, May A, Gray MD, Oshima J, Martin GM, Nehlin JO, Brosh R, Orren DK, Bohr VA. The Werner syndrome protein is involved in RNA polymerase II transcription. Mol Biol Cell. 1999;10:2655–2668. doi: 10.1091/mbc.10.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lutomska A, Lebedev A, Scharffetter-Kochanek K, Iben S. The transcriptional response to distinct growth factors is impaired in Werner syndrome cells. Experimental Gerontology. 2008;43:820–826. doi: 10.1016/j.exger.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 175.Kyng KJ, May A, Kolvraa S, Bohr VA. Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12259–12264. doi: 10.1073/pnas.2130723100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.OLOVNIKO AM. Principle of Marginotomy in Template Synthesis of Polynucleotides. Doklady Akademii Nauk Sssr. 1971;201:1496. [PubMed] [Google Scholar]
- 177.Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere End-Replication Problem and Cell Aging. Journal of Molecular Biology. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 178.Blackburn EH, Szostak JW. The Molecular-Structure of Centromeres and Telomeres. Annual Review of Biochemistry. 1984;53:163–194. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- 179.Greenberg RA. Telomeres, crisis and cancer. Curr Mol Med. 2005;5:213–218. doi: 10.2174/1566524053586590. [DOI] [PubMed] [Google Scholar]
- 180.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes & Development. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 181.di Fagagna FD, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 182.di Fagagna FD, Teo SH, Jackson SP. Functional links between telomeres and proteins of the DNA-damage response. Genes & Development. 2004;18:1781–1799. doi: 10.1101/gad.1214504. [DOI] [PubMed] [Google Scholar]
- 183.Harley CB, Futcher AB, Greider CW. Telomeres Shorten During Aging of Human Fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 184.Crabbe L, Karlseder J. In the end, it’s all structure. Curr Mol Med. 2005;5:135–143. doi: 10.2174/1566524053586527. [DOI] [PubMed] [Google Scholar]
- 185.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 186.GELLERT M, LIPSETT MN, DAVIES DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 188.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 189.Randall A, Griffith JD. Structure of long telomeric RNA transcripts: the G-rich RNA forms a compact repeating structure containing G-quartets. J Biol Chem. 2009;284:13980–13986. doi: 10.1074/jbc.M900631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wai LK. Telomeres, telomerase, and tumorigenesis--a review. MedGenMed. 2004;6:19. [PMC free article] [PubMed] [Google Scholar]
- 191.Greider CW, Blackburn EH. Telomeres, telomerase and cancer. Scientific American. 1996;274:92–97. doi: 10.1038/scientificamerican0296-92. [DOI] [PubMed] [Google Scholar]
- 192.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 193.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. Journal of Biological Chemistry. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 194.Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Molecular Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 195.Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. Journal of Biological Chemistry. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- 196.Li JL, Harrison RJ, Reszka AP, Brosh RM, Jr, Bohr VA, Neidle S, Hickson ID. Inhibition of the Bloom’s and Werner’s syndrome helicases by G-quadruplex interacting ligands. Biochemistry. 2001;40:15194–15202. doi: 10.1021/bi011067h. [DOI] [PubMed] [Google Scholar]
- 197.Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nature Genetics. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 198.Kruk PA, Rampino NJ, Bohr VA. Dna-Damage and Repair in Telomeres - Relation to Aging. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:258–262. doi: 10.1073/pnas.92.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Crabbe L, Jauch A, Naeger CM, Holtgreve-Grez H, Karlseder J. Telomere dysfunction as a cause of genomic instability in Werner syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2205–2210. doi: 10.1073/pnas.0609410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhou B, Ikejima T, Watanabe T, Iwakoshi K, Idei Y, Tanuma S, Uchiumi F. The effect of 2-deoxy-D-glucose on Werner syndrome RecQ helicase gene. FEBS Lett. 2009;583:1331–1336. doi: 10.1016/j.febslet.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 201.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Opresko PL, Cheng WH, von Kobbe C, Harrigan JA, Bohr VA. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003;24:791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- 203.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 204.Sowd G, Lei M, Opresko PL. Mechanism and substrate specificity of telomeric protein POT1 stimulation of the Werner syndrome helicase. Nucleic Acids Res. 2008;36:4242–4256. doi: 10.1093/nar/gkn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci U S A. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Cohen H, Sinclair DA. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3174–3179. doi: 10.1073/pnas.061579598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mandell JG, Goodrich KJ, Bahler J, Cech TR. Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase. J Biol Chem. 2005;280:5249–5257. doi: 10.1074/jbc.M412756200. [DOI] [PubMed] [Google Scholar]
- 208.Li B, Jog SP, Reddy S, Comai L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol Cell Biol. 2008;28:1892–1904. doi: 10.1128/MCB.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng YB, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 210.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nature Cell Biology. 2006;8:885–U162. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 211.Kawanishi S, Oikawa S, Murata M, Tsukitome H, Saito I. Site-specific oxidation at GG and GGG sequences in double-stranded DNA by benzoyl peroxide as a tumor promoter. Biochemistry. 1999;38:16733–16739. doi: 10.1021/bi990890z. [DOI] [PubMed] [Google Scholar]
- 212.Newman JPA, Banerjee B, Fang WR, Poonepalli A, Balakrishnan L, Low GKM, Bhattacharjee RN, Akira S, Jayapal M, Melendez AJ, Baskar R, Lee HW, Hande MP. Short dysfunctional telomeres impair the repair of arsenite-induced oxidative damage in mouse cells. Journal of Cellular Physiology. 2008;214:796–809. doi: 10.1002/jcp.21276. [DOI] [PubMed] [Google Scholar]
- 213.Satoh M, Ishikawa Y, Takahashi Y, Itoh T, Minami Y, Nakamura M. Association between oxidative DNA damage and telomere shortening in circulating endothelial progenitor cells obtained from metabolic syndrome patients with coronary artery disease. Atherosclerosis. 2008;198:347–353. doi: 10.1016/j.atherosclerosis.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 214.Ghosh A, Rossi ML, Croteau DL, Bohr VA. Telomeric D-loops containing 8-oxo-2-deoxyguanosine are preferred substrates for Werner and Bloom syndrome helicases and are bound by POT1. J Biol Chem. 2009 doi: 10.1074/jbc.M109.027532. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lombard DB, Beard C, Johnson B, Marciniak RA, Dausman J, Bronson R, Buhlmann JE, Lipman R, Curry R, Sharpe A, Jaenisch R, Guarente L. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol Cell Biol. 2000;20:3286–3291. doi: 10.1128/mcb.20.9.3286-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, Guarente L, Johnson FB. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol Cell Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.German J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 1993;72:393–406. [PubMed] [Google Scholar]
- 219.German J. Bloom’s syndrome. Dermatol Clin. 1995;13:7–18. [PubMed] [Google Scholar]
- 220.Dietschy T, Shevelev I, Stagljar I. The molecular role of the Rothmund-Thomson-, RAPADILINO- and Baller-Gerold-gene product, RECQL4: recent progress. Cell Mol Life Sci. 2007;64:796–802. doi: 10.1007/s00018-007-6468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kellermayer R. The versatile RECQL4. Genet Med. 2006;8:213–216. doi: 10.1097/01.gim.0000214457.58378.1a. [DOI] [PubMed] [Google Scholar]
- 222.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 223.Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 224.Van ML, Siitonen HA, Jalkh N, Chouery E, De RM, Delague V, Muenke M, Jabs EW, Cai J, Wang LL, Plon SE, Fourneau C, Kestila M, Gillerot Y, Megarbane A, Verloes A. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kaariainen H, Ryoppy S, Norio R. RAPADILINO syndrome with radial and patellar aplasia/hypoplasia as main manifestations. Am J Med Genet. 1989;33:346–351. doi: 10.1002/ajmg.1320330312. [DOI] [PubMed] [Google Scholar]
- 226.Stinco G, Governatori G, Mattighello P, Patrone P. Multiple cutaneous neoplasms in a patient with Rothmund-Thomson syndrome: case report and published work review. J Dermatol. 2008;35:154–161. doi: 10.1111/j.1346-8138.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 227.Vennos EM, Collins M, James WD. Rothmund-Thomson syndrome: review of the world literature. J Am Acad Dermatol. 1992;27:750–762. doi: 10.1016/0190-9622(92)70249-f. [DOI] [PubMed] [Google Scholar]
- 228.Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 229.Cui S, Klima R, Ochem A, Arosio D, Falaschi A, Vindigni A. Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J Biol Chem. 2003;278:1424–1432. doi: 10.1074/jbc.M209407200. [DOI] [PubMed] [Google Scholar]
- 230.Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Suzuki T, Kohno T, Ishimi Y. DNA helicase activity in purified human RECQL4 protein. J Biochem. 2009;146:327–335. doi: 10.1093/jb/mvp074. [DOI] [PubMed] [Google Scholar]
- 233.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein. RECQ4, EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 235.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 236.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 237.Ozsoy AZ, Ragonese HM, Matson SW. Analysis of helicase activity and substrate specificity of Drosophila RECQ5. Nucleic Acids Res. 2003;31:1554–1564. doi: 10.1093/nar/gkg243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 239.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 240.Schurman SH, Hedayati M, Wang Z, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, III, Croteau DL, Bohr VA. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum Mol Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM., Jr Stimulation of flap endonuclease-1 by the Bloom’s syndrome protein. J Biol Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 242.Aygun O, Svejstrup J, Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc Natl Acad Sci U S A. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Aygun O, Xu X, Liu Y, Takahashi H, Kong SE, Conaway RC, Conaway JW, Svejstrup JQ. Direct inhibition of RNA polymerase II transcription by RECQL5. J Biol Chem. 2009 doi: 10.1074/jbc.M109.015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Izumikawa K, Yanagida M, Hayano T, Tachikawa H, Komatsu W, Shimamoto A, Futami K, Furuichi Y, Shinkawa T, Yamauchi Y, Isobe T, Takahashi N. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem J. 2008;413:505–516. doi: 10.1042/BJ20071392. [DOI] [PubMed] [Google Scholar]