Abstract
HCV/HIV coinfection has emerged as a major cause of morbidity and mortality due to liver disease. Interferon-based therapy response rates have been disappointingly low. Baseline HCV complexity and the relationship between complexity and viral kinetic parameters has not been well described in HCV/HIV subjects. A subset of patients enrolled in ACTG 5071 underwent sampling to evaluate viral kinetics and HCV complexity changes. Early kinetic parameters, baseline complexity, and treatment outcomes, including rapid (RVR), early (EVR), and sustained (SVR) viral response were evaluated. HCV monoinfected subjects were matched to HCV/HIV coinfected subjects.
RESULTS
Baseline complexity was determined in 108 HCV/HIV coinfected subjects and 13 HCV controls. Quasispecies complexity was 2.24 in HCV/HIV and 1.90 in monoinfected subjects (p=0.14). Lower baseline complexity was associated with EVR (p=0.04) and approached significance for SVR. In patients who underwent viral kinetic modeling, complexity decrease was associated with RVR (p= 0.03), and was independent of the correlation between first phase viral decline efficiency and RVR.
CONCLUSION
Baseline HCV complexity is an independent predictor of early viral response in HCV/HIV subjects. Complexity decrease occurs by 4 weeks of interferon-based therapy and is associated with RVR. These findings may enhance predictive modeling of treatment outcomes in HCV/HIV patients.
Keywords: HCV, HIV, RNA, Quasispecies, Complexity, Pegylated-interferon, Coinfection
INTRODUCTION
In the decade following widespread use of highly active antiretroviral therapy (HAART) among patients with human immunodeficiency virus infection (HIV), there has been a dramatic decrease in morbidity and mortality associated with traditional opportunistic infections, and a concomitant increase in liver disease associated with viral coinfections[1, 2]. Hepatitis C virus (HCV) infection is present in approximately 25% of HIV infected patients in the United States, and represents the most important and serious coinfection [3]. Both hepatitis B and hepatitis C appear to be associated with rapidly progressive liver injury and fibrosis[4, 5].
It is therefore not surprising that considerable effort has been devoted to development and performance of clinical trials in HCV/HIV coinfected subjects, in order to determine the optimal role and efficacy of agents that were proven useful in controlling/eradicating HCV monoinfection. Recently, three pivotal multicenter trials and several other single center trials have demonstrated that pegylated interferon alfa (PEG-IFN) plus ribavirin is the most effective current therapy in coinfected populations being treated for HCV infection[6-8]. However, there is a significant decrement in treatment intervention response compared to HCV monoinfected subjects. A meta-regression of the existing literature demonstrated that the overall sustained viral response rate (SVR) was 33% with a wide range between individual trials. Differences between trials were not attributable to the presence of cirrhosis, age, gender, race, or other factors[9]. Subsequently, it has been suggested that high baseline viral load and inadequate ribavirin dosing may contribute to the decreased treatment response observed in HCV/HIV coinfected persons[10].
HCV quasispecies represent a population of closely related viral genomes that are present in sera and tissues. This intrapatient variability is due to the error-prone nature the HCV RNA-dependent RNA polymerase. The dominant viral population may be static or evolving, due to the combination of viral replicative fitness and concurrent immune selection pressures which drive selection. We have previously reported that HCV quasispecies complexity and diversity were different in HCV/HIV coinfected subjects vs. patients with HCV alone[11]. The population variability appears to be closely related to clinical outcomes, both in acute and advanced liver disease [12-14]. In this paper, we examine the relationship between baseline HCV quasispecies, early viral selection following introduction of interferon-based therapy, and treatment outcomes.
METHODS
Study Population
The study population was derived from the parent clinical trial, ACTG 5071 which was conducted in 21 U.S. academic research centers between 2002 and 2004 [6]. Briefly, subjects with HCV/HIV coinfection were enrolled and randomized to receive either pegylated interferon alfa-2a 180 mcg SQ/week plus ribavirin (600–1000 mg/day) vs. standard interferon alfa-2a (induction dosing of 6 M.U. SQ tiw followed by 3 M.U. tiw) ± ribavirin (same regimen) per day for 48 weeks. ACTG 5091s was a substudy embedded in the primary trial which permitted enrollment of up to 20 HCV monoinfected subjects, prospectively matched to coinfected subjects for age (± 5 years), gender, race, HCV genotype, and presence/absence of cirrhosis on liver biopsy. The protocol allowed for up to two controls per selected coinfected case. Frequent serum samples were collected in the first 28 days following drug initiation for the purpose of viral kinetic modeling among coinfected and control subjects enrolled in the substudy. Subsequent samples with detectable virus collected at weeks 8, 12, 24, 48, and 72 weeks from start of therapy were available for analysis in most substudy patients. All study sites were approved to enroll patients by their respective Institutional Review Boards, and all subjects provided appropriate informed consent.
Quasispecies Analysis
Heteroduplex complexity assays (HCA) were utilized to evaluate quasispecies or viral complexity. This assay is a non-isotopic method for determination of quasispecies complexity which has previously been compared to clonal sequenced-based analysis.[15]First, HCV RNA was extracted from 140 ul of patient serum. The amplification was performed in the E2/NS1 coding domain using the following primers : external forward (EF), 5′-GGTGCTCACTGGGGAGTCCT-3′ position 1048–1067; external reverse (ER), 3′-CATTGCAGTTCAGGGCCGTGCTA-5′ position 1291 1269; internal forward (IF), 5′-TCCATGGTGGGGAACTGGGC-3′ position 1087–1106; and internal reverse (IR), 3′-TGCCAACTGCCGTTGGTGTT-5′ position 1262–1243, (primer positions corresponding to the Choo HCV sequence)[16]. The E2/NS1 region was selected for evaluation because it is the most variable and least functionally constrained region of the HCV genome, providing the greatest potential for detecting changes in complexity due to non-specific (e.g., interferon) external pressure. Other regions are more highly constrained due to functional restrictions. Since interferon/ribavirin antiviral effects are not selective for a particular region, the overall variability of quasispecies is easiest to identify in this domain. The HCA assay has been previously described[15]and results confirmed with sequenced-based analysis in a study of quasispecies complexity in patients with alcoholic liver disease. Briefly, polymerase chain reaction amplicons from the HCV RNA amplification were heat denatured leading to unwinding and separation of cDNA strands. Slow reannealing leads to the development of heteroduplexes when mixed, codominant populations are present. The assay has been validated for reproducibility and sensitivity using simultaneous cloning and sequence analysis, and can detect quasispecies with 1–2% nucleotide difference down to a mix proportion of 10% of the total population.[15] The samples are visualized on a non-denaturing gel, and the band number is confirmed visually and using a densitometry scanner set to an appropriate threshold sensitivity. Samples from baseline were evaluated for quasispecies complexity in all patients. In the viral kinetic substudy samples, baseline and week 4 quasispecies were assayed as well as additional timepoints (weeks 8, 12, 24, 48, and 72 weeks from start of therapy) for those with detectable virus.
Viral Kinetic Parameters
Non-linear mathematical modeling was performed to determine key early kinetic parameters. The methodology utilized is described in detail in a prior publication[10]. Briefly, frequent early blood sampling following initiation of therapy was utilized to calculate epsilon (ε), as well as lambda1 and lambda2, which represent the first and second phases of viral decline for each subject. Rapid viral response (RVR) was defined as an HCV viral load below the limit of detectability (<600 IU/mL) for the Cobas Amplicor assay (Roche Diagnostics, Nutley, NJ) four weeks after drug initiation. Early viral response (EVR) was defined as undetectable HCV RNA or a two log drop from baseline HCV RNA twelve weeks after the start of therapy. Finally, sustained viral response (SVR) was defined as undetectable HCV RNA 24 weeks after completion of therapy.
Statistical Analysis
Baseline characteristics were compared using parametric and non-parametric methods including Chi-square, Wilcoxon rank-sum, Kruskal-Wallis, Student’s t (using Satterthwaite correction for unequal variances), and ANOVA as appropriate for the data being evaluated. Linear regression modeling was used in the evaluation of factors associated with baseline quasispecies variability. The analysis was performed using Statistix 9.0 (Analytical Software, Tallahassee, FL). A two-tailed hypothesis was utilized for all testing and an alpha= 0.05 was considered significant.
RESULTS
Study Population
Baseline serum samples were available for 136 subjects with HCV/HIV coinfection. This included three subjects who were consented and screened, but not dosed in the parent study (ACTG 5071). In addition, fifteen HCV monoinfected controls that were prospectively matched to coinfected study participants were treated and studied, as described in the Methods section. Amplification of HCV RNA from in the E2/NS1 coding domain was successful in 121 subjects, including 108 HCV/HIV coinfected subjects, and 13 HCV monoinfected control subjects. Three coinfected subjects were screen failures. Though initial quasispecies data are available, no other demographic response information was provided and these were also excluded. Demographic profiles of the subjects with quasispecies and demographic data are shown in Table 1. Briefly, the mean age of the 105 HCV/HIV coinfected subjects who received treatment was 44.5 year (SE ± 0.45). Eighty-five were male (81%). Racial breakdown included 44% Caucasian, 33% black, non-Hispanic, 17% Hispanic, and 6% other racial groups. Mean CD4 cell count was 533 cells/mm3 (SE ± 26). Plasma HIV viral load was undetectable in 63%. Liver biopsy revealed Ishak fibrosis (E) scores of 4–6 (incomplete to complete cirrhosis) in 20.2% and advanced fibrosis/cirrhosis (Ishak 3-6) in 44.2%.
Table 1.
Demographics
| HCV/HIV (entire 5071 cohort) | HCV/HIV (5091s substudy) | HCV (5091s substudy) | |
|---|---|---|---|
| N | 105 | 11 | 13 |
| Age (years) | 44.5 | 46.5 | 47.2 |
| Gender (%Male) | 81 | 82 | 92 |
| Race (%) | |||
| Caucasian | 44 | 36.4 | 38 |
| Black, non-Hispanic | 33 | 36.4 | 62 |
| Other | 23 | 27.2 | 0 |
| HCV VL (log IU/ml) | 6.24 | 6.22 | 6.17 |
| HCV Genotype 1 (%) | 78 | 82 | 100 |
| CD4 (cells/mm3) | 533 | 563 | na |
| Cirrhotic (%) | 20.2 | 27.3 | 7.7 |
| RVR (%) | Not done | 0 | 15.4 |
| EVR (%) | 27.4 | 27.3 | 46.2 |
| SVR (%) | 14.2 | 18.2 | 30.8 |
The mean age of 13 evaluable HCV-monoinfected controls enrolled in ACTG 5091s was 47.2 years (SE ± 1.1). Twelve were male, 5 were Caucasian and the remainder were black, non-Hispanic. These HCV monoinfected controls nearly matched the subgroup of coinfected subjects in the substudy population, which were representative of the entire coinfected cohort, except for percentage of cirrhotics and blacks.
HCV Quasispecies Complexity
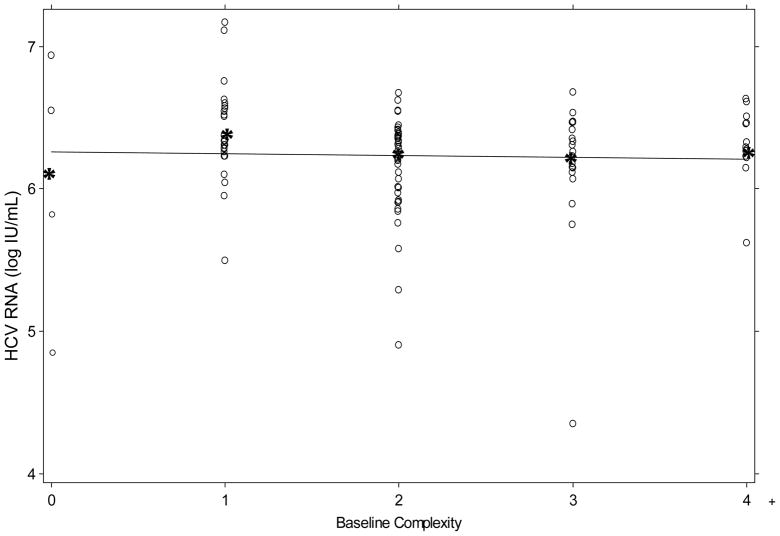
The baseline complexity of the entire coinfected cohort was 2.24 bands (SE ± 0.1) and for the HCV monoinfected controls was 1.9 (SE ± 0.3). This difference trended toward, but did not achieve statistical significance (t-test with Satterthwaite correction for unequal variances, p= 0.14). Linear regression modeling was performed to investigate factors associated with baseline quasispecies variability. Factors in the model included age, gender, race, CD4 count, degree of fibrosis, HIV RNA detectability, and HCV RNA titer/genotype. None of these factors alone or in combination were associated with the baseline quasispecies complexity. The lack of relationship between viral load and quasispecies complexity is illustrated in Figure 1.
Figure 1.
Relationship between HCV viral load and HCV RNA complexity as measured by quasispecies complexity assay (HCA) band number. Mean HCV RNA (IU/mL) titer represented by “*”. Solid line is best fit regression.
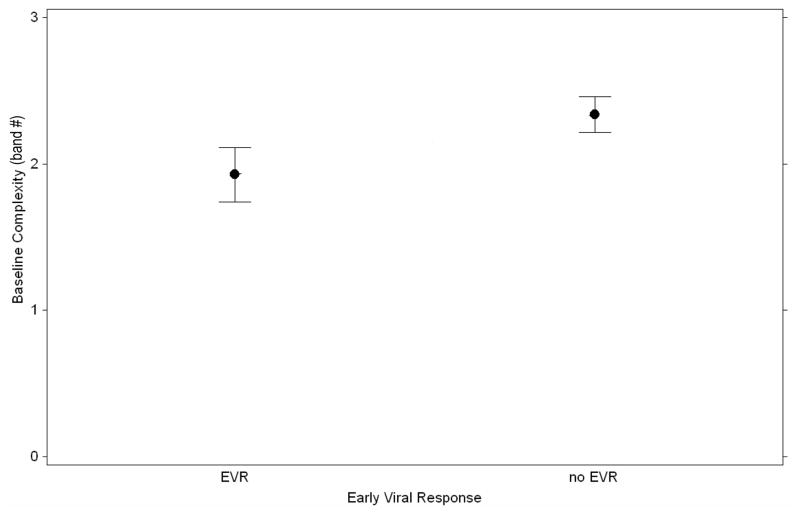
In contrast to the baseline comparison, early treatment response (EVR) was highly associated with reduced quasispecies complexity. Among both coinfected and monoinfected subjects who achieved EVR, the mean baseline quasispecies complexity was 1.90 vs. 2.37 for patients who did not achieve EVR (p= 0.04). (Figure 2) This trend held for the relationship between baseline complexity and sustained viral response (SVR) (1.88 vs. 2.26) but failed to reach statistical significance due to the greater variance associated with smaller sample size in the SVR group. The association between EVR and baseline complexity remained, even after exclusion of the control subjects (p=0.04) and the trends regarding SVR were comparable to those observed in the whole group.
Figure 2.
Mean and standard errors of HCV RNA complexity as measured by HCA band number vs. treatment response (p= 0.04).
Early Viral Kinetic Parameters and Quasispecies Complexity
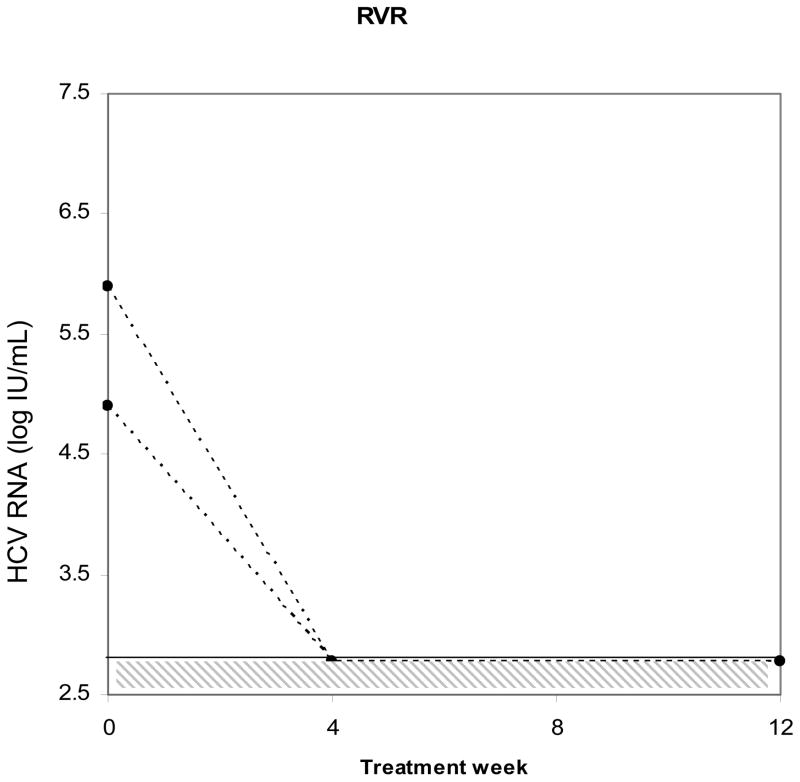
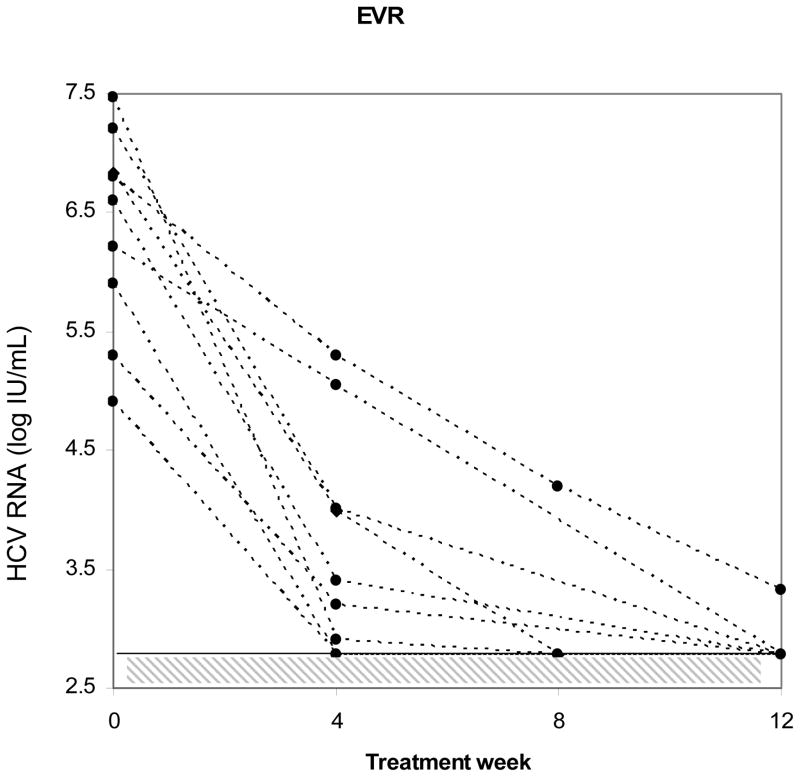
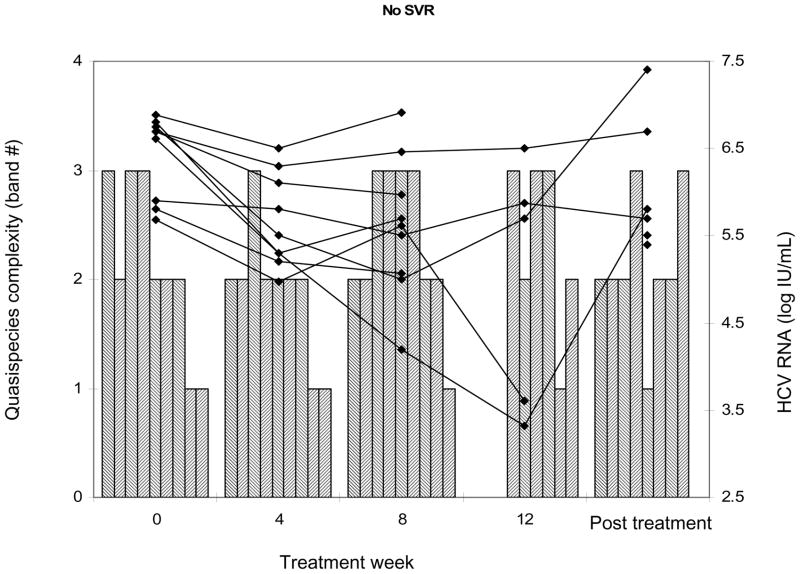
To more closely evaluate the relationship between HCV viral complexity and early viral kinetic parameters, we examined serial samples in a subset of the study population. Thirteen evaluable monoinfected controls were closely matched to 11 coinfected subjects for age, gender, race, HCV genotype, and histological stage and samples were serially collected at the same viral kinetic timepoints. Figure 3 illustrates individual patient response to initiation of interferon-based therapy (achievement of RVR, EVR, or SVR) on HCV RNA viral levels. In this subgroup, four week viral load data were available, which permitted evaluation of RVR. This parameter could not be evaluated in the larger study population, due to the lack of stored samples at week 4. As before, there was no statistical difference in baseline complexity between the subgroup of HCV/HIV coinfected and HCV monoinfected subjects. The relationship between complexity and viral kinetic parameters including (treatment efficacy), lambda1 and lambda2 was examined. Lower baseline complexity was strongly associated with RVR (p= 0.02). Furthermore, a decrease in complexity from baseline was also highly associated with RVR (−2 ± 0.48 vs. −0.15 ± 0.1; p= 0.03, Kruskal-Wallis ANOVA). This decrease was most closely correlated with the first phase decline slope (r= 0.6, p = 0.0042) lambda1. There was a lower degree of correlation between change in complexity and lambda2, the second phase decline slope (r= 0.39, p= 0.07). Epsilon (efficacy) was not associated with decreased HCV complexity. However, it was significantly associated with RVR among coinfected subjects (€ = 0.99 if RVR occurred vs. 0.76 if RVR absent; p= 0.004 by Wilcoxon rank sum). Figure 3, panel c illustrates the determination of quasispecies complexity and HCV viral levels in subjects who did not achieve SVR. The left axis shows HCA quasispecies complexity band counts as represented by bars, and the right axis shows HCV RNA viral levels as represented by solid lines. Not all timepoints are connected due to lack of available amplifiable samples. Among those subjects with no SVR, quasispecies complexity changes were noted after treatment when compared to earlier treatment timepoints.
Figure 3.
Individual subject response to initiation of interferon-based therapy. Panels a and b: Subjects who achieved RVR or EVR. Hashed area shows limit of detection of HCV RNA viral load assay. Panel c: Subjects who did not achieve SVR. Left axis shows HCA quasispecies complexity as represented by bars and right axis shows HCV RNA viral titers represented by solid lines. Some timepoints are missing due to unavailable amplifiable samples.
Genotype was also highly associated with a decrease in complexity. The mean decrease among subjects with genotype 2, independent of coinfection status, was 1 (SEM± 0.6) band during the first 4 weeks of therapy. Among genotype 1a subjects, the mean decrease was 0.23 bands (SEM± 0.15). There was no change in band complexity among three subjects with genotype 1b or 1 classification.
DISCUSSION
Treatment of HCV in the setting of HIV infection remains a clinical challenge. A recent meta-analysis suggested that only 33% of subjects treated achieve SVR[9]. However, high disease prevalence, and increased rates of fibrotic progression in coinfected patients supports attempts of active intervention among selected patients. The relative toxicities associated with PEG-IFN ± ribavirin regimens mandate that investigators continue to explore better predictors of treatment response that might permit earlier termination of treatment or that might provide guidance to the patient and clinician during the pre-treatment consent process. In this regard, we explored the role of quasispecies complexity as a marker of treatment response.
Quasispecies complexity in HCV/HIV coinfected subjects prior to treatment intervention has been a controversial topic, with data showing greater complexity compared to those with HCV monoinfection, as well as no difference from those with monoinfection or even less complexity [11]. Since quasispecies complexity may also be affected by duration of infection, viral load, histology, and whether the subject is a progressor vs. non-progressor in terms of liver disease, it is likely that the variability in reporting reflects differences in comparison groups randomly selected for cross-sectional evaluation [11, 17, 18]. Variation over time was clearly demonstrated in the longitudinal cohort described by Qin et al. [14]. Over a 10 year period, progressors narrowed their quasispecies display, while non-progressors did not. Thus, a cross-section study that included more patients with advanced liver disease would show lower complexity across the group than one heavily weighted by patients with milder liver disease.[14] However, another source of variability may be related to the methodology used to determine quasispecies complexity. In this study we utilized a well-validated non-isotopic assay. Other investigators have utilized isotopic heteroduplex assays and single-strand conformational polymorphism (SSCP) methodologies. There are limited data that permit direct comparison between assays, though our group compared HCA to SSCP and found that correlation between assays was present, but that SSCP reproducibility was low (data not shown). In this study, we noted a trend toward higher baseline complexity among subjects with HCV/HIV coinfection, but this failed to reach statistical significance, and may be reflective of the small sample size of our control population.
In contrast, we found that baseline complexity was highly associated with treatment outcomes, particularly with regard to EVR. Reasons for failure to achieve SVR appear to be significantly more complex than EVR because while virtually all patients that achieve SVR have EVR, the converse is not true. Several studies demonstrate that SVR rates among those who achieve EVR are more dependent upon drug adherence and total ribavirin dose than the initial ability to respond favorably to the therapeutic intervention [19]. The current regression analysis suggests that among both coinfected and monoinfected subjects, baseline HCV complexity is an independent factor in achieving EVR. Other investigators have noted an association between treatment response and baseline quasispecies complexity among HCV monoinfected subjects [20, 21, 22].There was no difference in quasispecies complexity as analyzed at baseline prior to treatment assignment in the entire cohort.(t-test; p= 0.76) Therefore, while treatment affected ability to achieve both EVR and SVR, this effect was independent of the quasispecies influence on the outcome overall. There were insufficient samples in the substudy to further separate the effect of treatment type.
Recently, rapid viral response (RVR) has emerged as a much more reliable predictor of SVR among monoinfected and coinfected subjects than EVR. However, there has been little effort to characterize the relationship between quasispecies changes following initiation of HCV therapy and the occurrence of RVR. Furthermore, the relationship between viral kinetic parameters and quasispecies selection has not been previously described. Herein, we demonstrated that a decrease in quasispecies complexity is highly associated with development of RVR. The narrowing of quasispecies is akin to prior observations in the setting of spontaneous HCV clearance following acute infection[13]. In our population, this finding was independent of viral load and supportive of the concept that the population of virus prior to treatment is made up of a mix of codominant species that have variable sensitivity to interferon. While a nearly infinite number of single nucleotide changes may be generated daily during the replication process, only a limited number of species have the ability to emerge as dominant or codominant. This pool represents the primary target for initial response following administration of interferon-based therapy.
The relationship between viral kinetic parameters and quasispecies clearance is also interesting. While the efficacy parameter ( ) is predictive of RVR, it appears to be independent of the quasispecies complexity. Since this parameter is calculated during the first 48 hours of viral decline, it reflects an overall change in viral load, which as noted above seems to be independent of baseline complexity which also factors into the development of RVR. Decreased HCV complexity is highly correlated with the slope of the first phase decline. These data further support the concept that both viral load and degree of complexity independently affect viral clearance, as reflected by both RVR and SVR. Identification of specific HCV signature motifs that are more or less prone to early clearance may improve our understanding of mechanisms of interferon resistance, though other groups have found it difficult to reliably identify such motifs[23, 24].
This study is limited by the small number of subjects for whom RVR could be documented. Unfortunately, early sampling was only performed on the subset of subjects within the ACTG 5091 substudy. The larger cohort did not save evaluable samples between baseline and week 12. However, newer studies being designed within the ACTG and elsewhere will provide samples at critical timepoints that will permit extension of our observations. Larger studies will also permit development of predictive algorithms which take into account baseline quasispecies complexity, adding to the array of tools already used in the treatment decision process (e.g. genotype, viral load, fibrosis). Though complexity assays are only available as research tools at this time, it is possible that they could achieve wider clinical application if their value is validated. Finally, there is growing evidence that early viral selection plays a critical role in treatment outcomes when small molecule antiviral agents are utilized in conjunction with PEG-IFN. There are limited data suggesting that such selection will need to be incorporated into viral kinetic models to ensure predictive accuracy as the field of HCV treatment continues to evolve.
In conclusion, HCV quasispecies complexity prior to treatment appears to be an important and independent predictor of both RVR and SVR and early quasispecies selection is critical in development of RVR. These findings have important implications for predictive modeling and for advancement of our understanding of HCV viral clearance in HCV/HIV coinfected patients.
Acknowledgments
The authors would like to thank Dr. Janet Andersen, ScD who provided assistance with data management and analysis.
This work was supported by grants from the NIAID AIDS Clinical Trials Group (AI25897, AI068636, and U01 AI38858) and by an award from NIDDK to KES (K24 DK070528), and Roche Laboratories provided the study drug and HCV RNAmonitoring kits.
Footnotes
Conflict of interest statements for all authors: Kenneth E. Sherman-Roche supports research funding for the institution, Susan D. Rouster-No conflict of interest, Sandra Stanford-No conflict of interest, Jason T. Blackard-No conflict of interest, Norah Shire-No conflict of interest, Margaret Koziel-No conflict of interest, Marion Peters-Consultant for Roche, clinical care options, Raymond T. Chung-Receives Roche research support
Bibliography
- 1.Weber R, Sabin CA, Friis-Moller N, et al. Liver-related deaths in persons infected with the human immunodeficiency virus: the D:A:D study. Arch Intern Med. 2006;166:1632–41. doi: 10.1001/archinte.166.15.1632. [DOI] [PubMed] [Google Scholar]
- 2.Bica I, McGovern B, Dhar R, et al. Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis. 2001;32:492–7. doi: 10.1086/318501. [DOI] [PubMed] [Google Scholar]
- 3.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C Virus prevalence among patients infected with Human Immunodeficiency Virus: a cross-sectional analysis of the US Adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34:831–7. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 4.Sulkowski MS, Mehta SH, Torbenson MS, et al. Rapid fibrosis progression among HIV/hepatitis C virus-co-infected adults. Aids. 2007;21:2209–16. doi: 10.1097/QAD.0b013e3282f10de9. [DOI] [PubMed] [Google Scholar]
- 5.Thio CL, Seaberg EC, Skolasky R, Jr, et al. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS) Lancet. 2002;360:1921–6. doi: 10.1016/s0140-6736(02)11913-1. [DOI] [PubMed] [Google Scholar]
- 6.Chung RT, Andersen J, Volberding P, et al. Peginterferon Alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351:451–9. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, et al. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–50. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 8.Carrat F, Bani-Sadr F, Pol S, et al. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. Jama. 2004;292:2839–48. doi: 10.1001/jama.292.23.2839. [DOI] [PubMed] [Google Scholar]
- 9.Shire NJ, Welge JA, Sherman KE. Response rates to pegylated interferon and ribavirin in HCV/HIV coinfection: a research synthesis. J Viral Hepat. 2007;14:239–48. doi: 10.1111/j.1365-2893.2006.00824.x. [DOI] [PubMed] [Google Scholar]
- 10.Sherman KE, Shire NJ, Rouster SD, et al. Viral kinetics in hepatitis C or hepatitis C/human immunodeficiency virus-infected patients. Gastroenterology. 2005;128:313–27. doi: 10.1053/j.gastro.2004.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherman KE, Andreatta C, O’Brien J, Gutierrez A, Harris R. Hepatitis C in human immunodeficiency virus-coinfected patients: increased variability in the hypervariable envelope coding domain. Hepatology. 1996;23:688–94. doi: 10.1002/hep.510230405. [DOI] [PubMed] [Google Scholar]
- 12.Farci P, Shimoda A, Coiana A, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–44. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 13.Farci P, Strazzera R, Alter HJ, et al. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci U S A. 2002;99:3081–6. doi: 10.1073/pnas.052712599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin H, Shire NJ, Keenan ED, et al. HCV quasispecies evolution: association with progression to end-stage liver disease in hemophiliacs infected with HCV or HCV/HIV. Blood. 2005;105:533–41. doi: 10.1182/blood-2004-04-1452. [DOI] [PubMed] [Google Scholar]
- 15.Sherman K, Rouster S, Mendenhall C, Thee D. Hepatitis C RNA quasispecies complexity in patients with alcoholic liver disease. Hepatology. 1999;30:265–70. doi: 10.1002/hep.510300131. [DOI] [PubMed] [Google Scholar]
- 16.Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–5. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neau D, Jouvencel AC, Legrand E, et al. Hepatitis C virus genetic variability in 52 human immunodeficiency virus-coinfected patients. J Med Virol. 2003;71:41–8. doi: 10.1002/jmv.10451. [DOI] [PubMed] [Google Scholar]
- 18.Laskus T, Wilkinson J, Karim R, et al. Hepatitis C virus quasispecies in HIV-infected women: role of injecting drug use and highly active antiretroviral therapy (HAART) Hepatology. 2007;46:359–70. doi: 10.1002/hep.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McHutchison JG, Manns M, Patel K, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1061–9. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 20.Moreau I, Levis J, Crosbie O, Kenny-Walsh E, Fanning LJ. Correlation between pre-treatment quasispecies complexity and treatment outcome in chronic HCV genotype 3a. Virol J. 2008;5:78. doi: 10.1186/1743-422X-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmeron J, Casado J, Rueda PM, et al. Quasispecies as predictive factor of rapid, early and sustained virological responses in chronic hepatitis C, genotype 1, treated with peginterferon-ribavirin. J Clin Virol. 2008;41:264–9. doi: 10.1016/j.jcv.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 22.Ueda E, Enomoto N, Sakamoto N, et al. Changes of HCV quasispecies during combination therapy with interferon and ribavirin. Hepatol Res. 2004;29:89–96. doi: 10.1016/j.hepres.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Chung RT, Monto A, Dienstag JL, Kaplan LM. Mutations in the NS5A region do not predict interferon-responsiveness in american patients infected with genotype 1b hepatitis C virus. J Med Virol. 1999;58:353–8. [PubMed] [Google Scholar]
- 24.McKechnie VM, Mills PR, McCruden EA. The NS5a gene of hepatitis C virus in patients treated with interferon-alpha. J Med Virol. 2000;60:367–78. [PubMed] [Google Scholar]