Abstract
Nanoparticle-based platforms for identification of chemical and biological agents offer substantial benefits to biomedical and environmental science. These platforms benefit from the availability of a wide variety of core materials as well as the unique physical and chemical properties of these nanoscale materials. This review surveys some of the emerging approaches in the field of nanoparticle based detection systems, highlighting the nanoparticle based screening methods for metal ions, proteins, nucleic acids, and biologically relevant small molecules.
Keywords: Nanoparticles, assembly, absorbance, surface plasmon band, fluorescence, protein, DNA, bacteria
1. Introduction
Detection of chemical and biological agents plays a pivotal role in medical, forensic, agricultural, and environmental sciences [1]. Sensitive methods that allow identification of biomarkers such as proteins and nucleic acids at early disease states provide the prospect of better health and more effective therapy. Technological platforms that provide sensors of high sensitivity, selectivity and stability are therefore in high demand.
Sensing systems consist of two functional components: recognition elements for binding with target analytes and a transduction process to signal the binding event. The efficiency of these two components are critically related to the outcome of the detection process in terms of the response time, signal-to-noise (S/N) characteristics, sensitivity, and selectivity of the system. Thus, the challenges in development of novel detection systems have been concerned with improving the recognition process as well as designing new signal transduction mechanisms. Nanomaterials provide novel systems for the pursuit of new recognition and transduction processes, as well as increasing the signal-to-noise ratio through miniaturization of the system components [2].
Nanoparticles (NPs) possess several distinctive physical and chemical attributes that make them promising synthetic scaffold for the creation of novel chemical and biological detection systems [3]. Indeed, in the last few years nanostructured materials, such as noble metal nanoparticles, quantum dots, and magnetic nanoparticles, have been employed in a broad spectrum of highly innovative approaches for assays of metal ions, small molecules and protein and nucleic acid biomarkers [4, 5, 6, 7]. In addition to the large surface-to-volume ratio that favors miniaturization, nanoparticles possess unique optical, electronic and magnetic properties depending on their core materials. Furthermore these properties of the nanomaterials depend on their size and shape, and vary with their surrounding chemical environment. Additionally, nanoparticles can be fashioned with a wide range of small organic ligands and large biomacromolecules by using tools and techniques of surface modification. Each of these capabilities has allowed researchers to design novel diagnostic systems that offer significant advantages in terms of sensitivity, selectivity, reliability and practicality. This review provides recent research advances involving the use of nanoparticles in the detection and diagnosis of analytes including metal ions, small molecules, nucleic acids, proteins, and microorganisms.
2. Optical Detection of Metal Ions and Small Molecules
2.1. Fluorescence-Based Detection Using Nanoparticles
Quantum dots (QDs) are characterized by several unique intrinsic optical properties [8]. The properties include broad absorption spectra with high extinction coefficient, along with a narrow emission with a full width at half maximum (FWHM) of 20-30 nm. The QD emission can also be readily tuned by changing the nanocrystal size, and is environmentally responsive. These intrinsic optical properties of QDs make them promising candidates for optical detection of various analytes. For example, detection of cyanide was achieved using 2-mercaptoethane sulfonate functionalized CdSe nanoparticles by monitoring the quenching of QD emission upon addition of CN− [9]. Quenching of QD emission was observed in the presence of CN− ions leading to a detection of μM concentration of CN−. Importantly, the presence of SO42−, NO3−, Cl−, Br− and acetate anions in the system did induce any quenching of QD emission In another example, acetylcholine (ACh) was used to quench the emission of calix[4]arene coated CdSe/ZnS QDs [10].
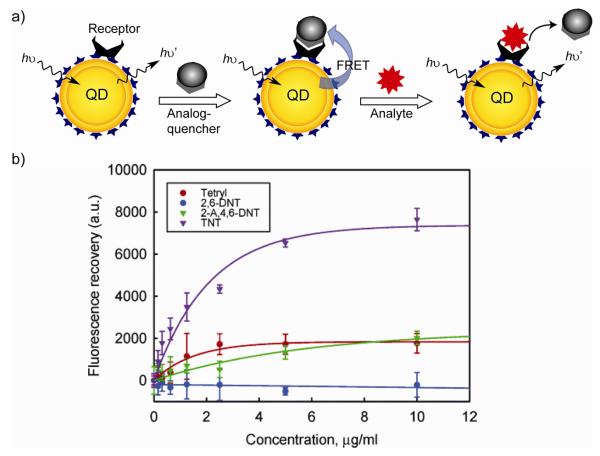
The efficiency of QDs as donors in FRET processes has been exploited in sensing of local pH, metal ions and small molecules. For pH sensing Snee and coworkers have utilized a conjugated system comprising of a pH-sensitive squaraine dye and QD [11]. The dye absorbance depends on the solution pH, correspondingly the FRET efficiency also become a function of environmental pH. Similarly, detecting system targeting the explosive 2,4,6-trinitrotoluene (TNT) was developed by Mattoussi et al. [12]. The QD was functionalized with a recognition element (single chain antibody fragment specifically selected against TNT). Preassembling a TNT analog, consisting of a dark quenching dye with the antibody binding site quenches the QD emission through FRET. When the assembly was exposed to TNT, it displaces the quencher, disrupting the energy transfer from the QDs to the quencher and recovering the QD emission.
Gold nanoparticles (AuNPs) have extraordinarily high molar extinction coefficients (1×109 M−1 cm−1 for d = 20 nm AuNP) as compared to common organic dyes (104-106 M−1 cm−1) [13,14]. Therefore, AuNPs can be treated as nonmolecular chromophores with excellent light collecting ability. Their exceptional quenching ability makes them suitable energy acceptors in the FRET based assays [15]. In one example, anionic tiopronin-coated AuNPs were used to efficiently quench the fluorescence of Poly-pyridyl complex [Ru(bpy)3]2+[16]. The fluorophore then can be dissociated from the nanoparticle surface by addition of electrolytes such as K+, Bu4N+, and Ca2+ salts. Similarly, selective detection of aminothiols was possible by the use of Nile red-adsorbed AuNPs [17]. Zhu, Li and collaborators have devised a Cu2+ sensor by employing bispyridyl perylene bridged AuNPs, where the initially quenched fluorescence of perylene is recovered by Cu2+ ions through formation of stronger pyridine-Cu2+ coordination [18]. Recently, a phosphorescent sensor for alkaline earth metal ions and transition metal ions have been devised based on lanthanum complexes of bipyridine-functionalized AuNPs [19].
2.2. Colorimetric Detection
A promising avenue for analyte detection arises from the unique size and shape dependent optical, magnetic and electronic properties of nanomaterials. For instance, spherical gold nanoparticles (AuNPs) exhibit a variety of colors in solution from brown to violet as the core size increases from 1 to 100 nm. Spherical AuNPs usually exhibit an intense absorption peak from 500 to 550 nm, corresponding to the surface plasmon band of nanometer scale noble metal nanoparticles [20]. This absorption arises from the collective oscillation of the valence electrons due to resonant excitation by the incident photons. Surface plasmon resonance (SPR) is absent in both small nanoparticles (d < 2 nm) and bulk materials and strongly reliant on the particle size. Not only to the nanoparticle size, the SPR is also sensitive to the surrounding environment such as ligand, solvent and temperature and most importantly SPR is strongly dependent on the proximity to other nanoparticles. Thus, clustering of AuNPs of appropriate sizes (d > 3.5 nm) evokes interparticle surface plasmon coupling, resulting in a significant red-to-blue shifting (to ca. 650 nm) and broadening of the SPR band that can be readily observed by the naked eye at nanomolar concentrations [21].
The colorimetric detection of alkali metal ions using NPs was achieved by the incorporation of chelating agents onto the gold nanoparticle surface. AuNPs were functionalized with 15-crown-5 moieties to detect the physiologically important potassium ions. The presence of K+ induces the aggregation of 15-crown-5 functionalized 18 nm AuNPs through the sandwich complex formation which results in a red-to-blue color change at μM to mM concentration of K+ [22]. This system has been extended by incorporating 12-crown-4 onto the AuNPs surface to detect sodium ions [23]. Likewise, phenanthroline-functionalized 4 nm AuNPs to detect Li+ and lactose-functionalized 16 nm AuNPs have been used to sense Ca2+ have also been constructed [24,25].
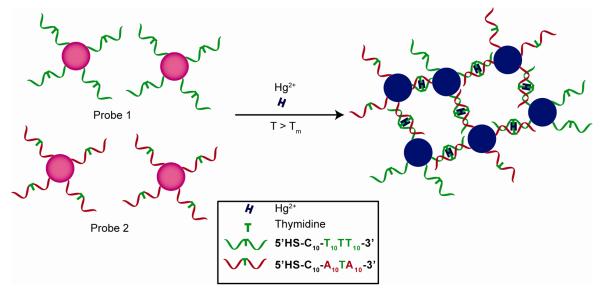
Heavy metal ions such as Pb2+, Cd2+, and Hg2+ are quite toxic, making their detection of great importance for environmental science. Hupp et al. have reported a simple heavy metal ion sensing system based on the aggregation of nanoparticles functionalized with appropriately designed ligands where the surface carboxylates act as chelating groups and the nanoparticle bridging is driven by heavy-metal ion chelation by Pb2+, Cd2+, or Hg2+ (≥400 μM) [26, 27]. A colorimetric sensor for Pb2+ was also developed by forming mixed monolayer-protected AuNPs carrying both carboxylate and 15-crown-5 functionalities [28]. In this system aggregates of AuNPs form due to hydrogen bonding interaction between carboxylic acid residues, and Pb2+ ions disrupt the hydrogen-bonded assembly by associating with crown ether moiety and generating an electrostatic repulsion between the AuNPs, resulting in. a blue to red color change. Similarly, AuNPs have been fabricated with cysteine and peptide-functionality to detect Cu2+ and Hg2+, respectively [29,30]. Recently, DNA-functionalized AuNPs have been employed for the detection of Hg2+ by Mirkin et.al. using thymidine- Hg2+-thymidine coordination chemistry [31].
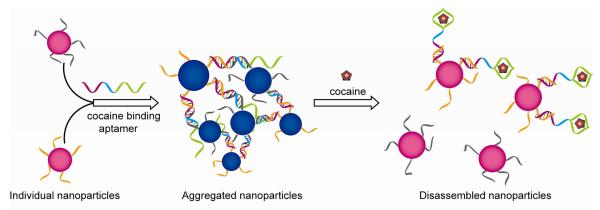
In ionic sensing, simultaneous addressing of selectivity and sensitivity issues are the key goals. Liu and Lu have provided an elegant example of selective and sensitive colorimetric Pb+2 biosensors implementing DNAzyme-directed assembly of AuNPs [32, 33, 34, 35, 36]. Initially the DNAzyme provides blue-colored assemblies of the DNA-functionalized AuNPs through Watson-Crick base pairing. The presence of Pb2+ in system activates the DNAzyme, which subsequently cleaves the substrate strand to dissemble the AuNPs resulting in a blue-to-red color change.
In anion sensing, Kubo et al. have reported a colorimetric sensing of oxoanions such as AcO−, HPO42−, and malonate in aqueous methanol solution by using isothiouronium group functionalized AuNPs [37]. Similarly, thioglucose-grafted AuNPs have been fabricated to sense fluoride anions in water [38].
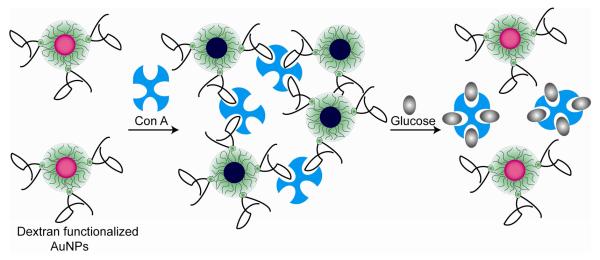
In neutral molecule sensing, Geddes et al. have demonstrated glucose sensing by using assemblies of concanavalin A (Con A) and high-molecular-weight dextran-coated AuNPs on the basis of a competitive colorimetric assay [39, 40]. Con A is a multivalent protein having four sugar binding sites at pH 7. Due to its multiple binding sites, it allows dextran-coated nanoparticles to assemble around its binding sites. This assembly affords cross-linked nanoparticles with broadened and red-shifted SPR of AuNPs. The presence of glucose in the system releases the individual dextran-coated AuNPs by competitively interacting with Con A, generating a blue-to-red shift with a dynamic sensing range of 1 – 40 mM. The incorporation of MUA-AuNPs into the polymer matrix of molecularly imprinted polymers (MIPs) has been used to provide colorimetric sensor for adrenaline [41]. The initially shrunken MIP gel placed between two glass slides in the absence of adrenaline affords the close proximity of AuNPs. However, selective rebinding of the target analytes with the MIP causes swelling of the MPI gel. This swelling process results in separation of the nanoparticles with a blue-shift in the SPR band of the immobilized AuNPs.
Aptamers are single-stranded oligonucleic acid-based binding molecules that can bind to a variety of targets with high affinity and specificity. An effective cocaine sensor was designed by Lu and coworkers, allowing quantification in the range of 50 to 500 μM [42, 43, 44].
3. Nanoparticle-Based Detection of Proteins
3.1. Fluorescence-Based Detection Using Nanoparticles
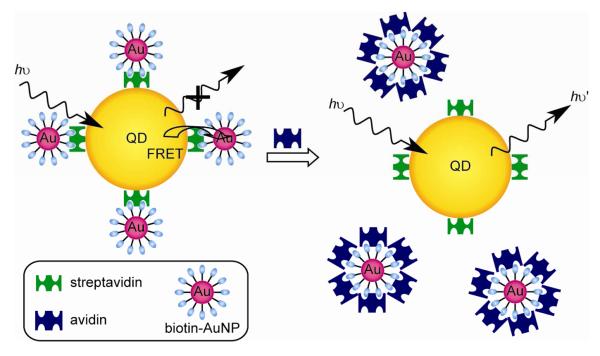
The identification of proteins offers direct applications in therapeutics, forensic analysis and environmental monitoring. QDs have shown potential for the detection of proteins. Nagasaki et al. reported a biotin-PEG/polyamine CdS quantum dot [45]. Due to the specific interaction of this QD with Texas Red-labeled streptavidin, an effective fluorescent energy transfer (FRET) takes place which is proportional to the concentration of the dye-labeled protein. It can thus be applied as a highly sensitive detection motif. In another system, Kim et al fabricated a FRET donor-acceptor couple using biotinylated AuNPs and streptavidin coated QDs [46]. In presence of avidin, the fluorescence of QDs is regenerated due to the interruption of streptavidin-biotin interaction (Fig. 7). The same concept was also used to detect glycoproteins [47].
Fig. 7.
Assay for the detection of avidin by using QD-AuNP conjugate.
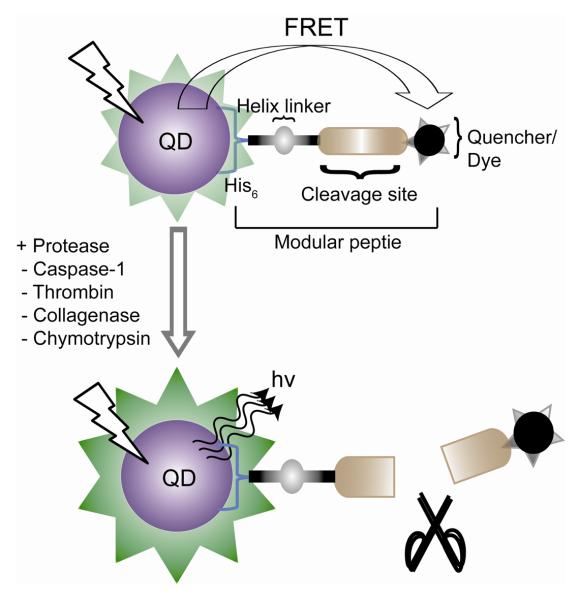
Luminiscent QD bioconjugates were applied in detecting the proteolytic activity of several enzymes by Mattoussi [48]. For this study, dyes labeled multifunctional modular peptides containing a substrate sequence were designed. The peptides were then self-assembled on dihydrolipoic acid-capped QD surface to get an efficient FRET from the QD to the proximal dye. Presence of proteases in the system cleaves the substrate strand, altering the FRET signature.
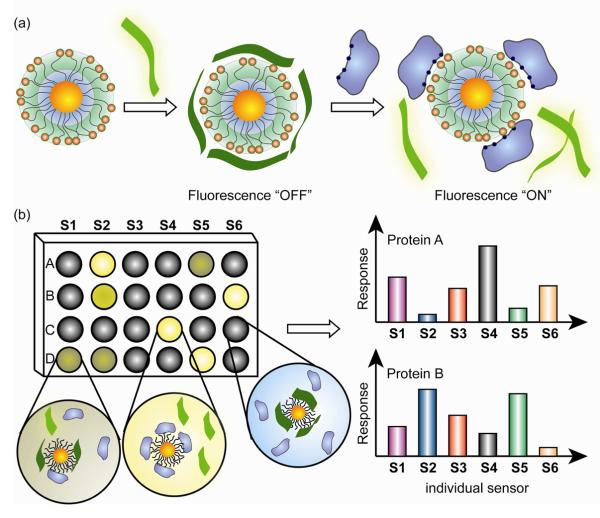
Another paradigm for detection of proteins by functionalized gold nanoparticles was introduced by Rotello and co-workers employing “chemical nose approach” [49]. This approach relies on array-based sensing using selective recognition elements. They fabricated a sensor array by using cationic AuNPs with various head groups and anionic poly(p-phenyleneethynylene) (PPE) fluorescent polymer that serves as the fluorescence transduction element. In this sensor design, the cationic nanoparticles significantly quench the intrinsic fluorescence of the PPE polymer. Competitive binding of analyte proteins releases the PPE polymer, resulting in a fluorescence recovery (Fig. 9). Linear discrimination analysis (LDA) was then used to identify unknowns from the training set.
Fig. 9.
Illustration of ‘chemical nose’ sensor array. (a) The competitive binding between protein and polymer-NP complexes leads to the fluorescence recovery. (b) Fingerprint response patterns for individual proteins.
3.2. Colorimetric Detection
AuNPs with diverse ligand functionalities provide one of the potential scaffolds in detecting proteins. For example, the aggregation induced by specific recognition of galactosefunctionalized AuNPs with agglutinin, a bivalent lectin, causes a visible color change that can serve as the colorimetric sensor for proteins [50].Other glyconanoparticles have also been used for detecting and quantifying several proteins such as Concanavalin A [51] and cholera toxin [52] by colorimetric detection methods.
The optical properties of the metallic NPs have been employed in amplifying the detection of proteins. Willner and coworkers have amplified the detection of aptamer-thrombin complexes in solution and glass surface as a result of catalytic enlargement of aptamer functionalized AuNPs [53]. The aptamer covalently attached to the glass surface binds first to the thrombin target. Then the aptamer-functionalized AuNPs was associated with the other thrombin binding site leading to a sandwich complex. The immobilized AuNPs are further enlarged in a growth solution containing HAuCl4, CTAB, and NADH, which enhances the surface plasmon coupling interaction of adjacent nanoparticles [54].
The specific interaction of antigen-coated nanoparticle and the antibodies provide another avenue for detection of proteins. Rosenzweig and co-worker developed an immunoassay procedure in which AuNPs coated with protein A were used to determine the level of anti-protein A in aqueous and serum solutions [55]. The presence of anti-protein A causes aggregation of the antigen coated nanoparticle resulting in a change in absorption at 620 nm and thus can be sensed in the solution.
3.3. Bio-bar-code Assay
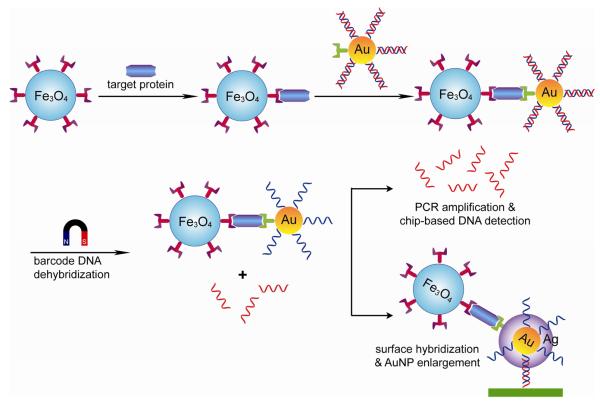
Mirkin's group has developed an AuNP-based bio-barcode assay to amplify the target analytes. This method provides highly multiplexed and ultrasensitive detection of proteins [56, 57]. The bio-barcode assay was first employed to analyze PSA, which is a biomarker protein for prostate and breast cancer [58]. The recognition element for protein is monoclonal antibody functionalized on magnetic microparticle. The other component is an AuNP coated with both polyclonal antibodies for the target protein and oligonucleotides hybridized to bar-code strands. In this method, the magnetic microparticles first bind to the target protein followed by sandwich structure formation with AuNPs. A magnetic field is then applied to separate the complexed target from the sample solution and the bar-codes were released in buffer chemically or by heating. The barcodes were identified with attomolar detection limit using chip-based sandwich hybridization with ss-DNA functionalized AuNP probes followed by silver amplification method. This approach was also applied to multiplexed detection of protein using different biobar-coded AuNP probes [57].
4. Detection of Nucleic Acids
4.1. Fluorescence-Based Detection Using Nanoparticles
As with organic fluorescent dyes, the emission of semiconductor QDs can be effectively quenched by AuNPs in appropriate vicinity. Melvin et al. have designed a fluorescent competitive assay for DNA detection by using QDs and AuNPs as the FRET donor-acceptor couple [59]. In their protocol, the CdSe QDs linked to a short DNA strand are hybridized with a complementary DNA strand linked to an AuNP, leading to quenched assemblies due to the surface-contact between QDs and AuNPs. When unlabelled complementary oligonucleotides are present, the AuNP-DNA is displaced from the QD-DNA, regenerating the QD fluorescence.
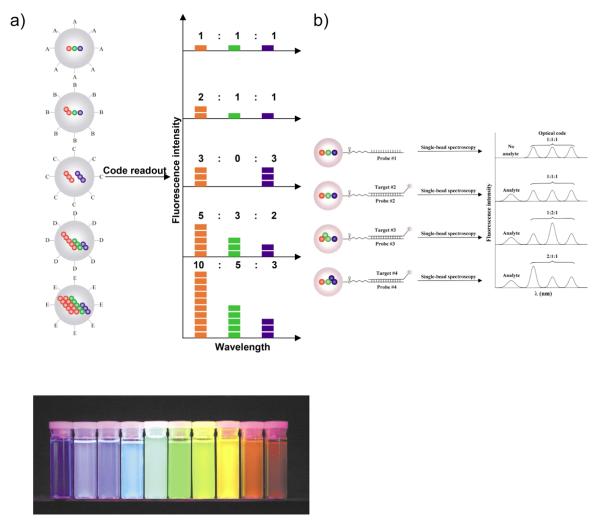
Nie and coworkers have developed a strategy for multiplexed DNA detection [60]. For this protocol, they labeled the target DNA with a fluorophore. Correspondingly, oligonucleotidefunctionalized polymeric microbeads were imbedded with QDs which are designed to emit at various specified wavelengths (other than target DNA). As shown in Fig. 11, microbeads with different ratios of QDs showed different emission intensities at different wavelengths. After binding with target DNA, single-bead spectroscopy was used to determine the presence and the identity of the target DNA. Later, Alivisatos and co-workers reported a DNA-QD conjugate, applicable to chip based DNA microarray for single-nucleotide polymorphism and multimarker detections [61]. These chip-based assays exhibited true-to-false signal ratios above 10 and the detection limit as low as 2 nM concentration of DNA-NP probes.
Fig. 11.
a) Fluorescence signature of microbeads with different ratios of QDs. b) DNA hybridization assay using microbeads. (adapted with permission from reference [60])
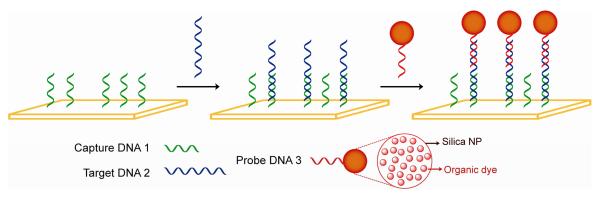
Tan's group developed highly fluorescent bioconjugated silica NPs as labels for chip-based sandwich DNA assays [62]. The silica NPs encapsulates large numbers of fluorophores inside a single NP which produces a strong fluorescence signal associated with each target recognition event without any preamplification. Moreover, the silica matrix provides a high photostability because of shielding effect to protect the fluorophores from environmental oxygen. The ultrasensitive DNA analysis assay showed a 0.8 fM detection limit using a bioconjugated NP-based sandwich assay and provided 100:7 discrimination between target DNA and one-base mismatched DNA sequences. They also utilized this bioconjugated NP-based bioassay for detection of pathogenic bacteria based on antibody-antigen interaction and recognition.
4.2. Colorimetric detection
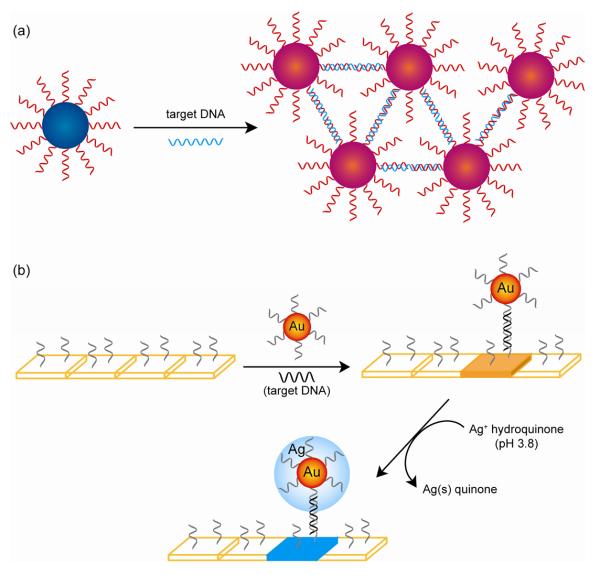
The potential of NPs as DNA detection agents was first described by Mirkin et al. using oligonucleotide-functionalized AuNPs and sequence-specific particle assembly events induced by target DNA [63, 64]. Since then, oligonucleotide-directed NP aggregation has been extensively used in the colorimetric detection of oligonucleotides [65, 66, 67, 68, 69, 70]. Generally, two ssDNA-modified NPs are used for the detection of oligonucleotides. The base sequences in ssDNA are complementary to both ends of the target oligonucleotides. As illustrated in Fig. 13a, the presence of target oligonucleotide causes the NP aggregation. Concomitantly a change in optical properties of the NPs was observed. Intense absorptivity of NPs as well as strong and highly specific base-pairing of DNA molecules facilitates the ultrasensitive optical detection of oligonuceotides. Generally, when large AuNPs (e.g. 50 nm or 100 nm) were employed a better sensitivity in detection was achieved. Interestingly, based on simple electrostatic interaction such as single-base-pair mismatches, citrate-stabilized AuNPs were able to distinguish ssDNA and dsDNA at the level of 100 fmol [71].
Fig. 13.
a) DNA mediated nanoparticle assembly. b) Chip-based system for DNA assay.
Another advantage offered by the colorimetric technique for DNA detection is the tunable selectivity due to the sharp melting transitions of NP-labeled DNA assemblies. This advantage was utilized in a chip-based system based on a sandwich assay [72]. This assay consists of an oligonucleotide-modified glass slide, a NP probe and target DNA, as illustrated in Fig. 13b. The immobilized DNA strand recognizes the DNA of interest and changes the melting profiles of the targets from an array substrate. This change gave the differentiation of an oligonucleotide sequence from targets with single nucleotide mismatches with a high selectivity.
4.3. SERS-Based Detection
SERS using AuNPs has been used to sense DNA. Mirkin et al. used AuNP probes labeled with Raman-active dyes and oligonucleotides to accomplish multiplexed detection of oligonucleotide targets [73]. Using a sandwich assay and silver enhancement, SERS signals were observed from the immobilized Raman dyes. This method was able to discriminate between single nucleotide polymorphisms in six different viruses. Raman tags have also been incorporated into DNAfunctionalized AuNPs for the detection of DNA using SERS [74].
4.4. Electrical and electrochemical detection
Electrochemical detection provides an alternative to optical approaches for the detection of DNA[75]. Using this strategy, DNA recognition events are transduced into electrical signals using NP mediators. Mirkin et al. have developed a DNA array detection method where the binding of oligonucleotide-functionalized AuNPs generates conductivity changes [76]. In their studies, target DNA has been detected at concentrations of 500 fmol with a point mutation selectivity factor of ~ 100,000:1.
The redox properties of NPs make them useful as electrochemical labels for the detection of oligonucleotides. Ozsoz et al. have shown that the incubation of a DNA-modified electrode with complementary DNA strands conjugated to NPs generated a gold oxide wave at +1.2 V [77]. A number of amplification strategies have been developed, including silver deposition [78] and the conjugation of electrochemically active groups onto NPs [79, 80]. In one approach ferrocene-capped AuNP/streptavidin conjugates were attached to a DNA detection probe of a “sandwich” DNA complex on the electrode. [78, 81]. Fan et al. likewise described a detection approach employing hybridization with AuNP-labeled reporter probe DNA and the subsequent treatment with [Ru(NH3)6]3+ complexes [82].In recent studies, Willner et al. reported the electrochemical detection of DNA using aggregation of AuNPs on electrodes coupled with intercalation of methylene blue into the DNA [83].The methylene blue dyes act as electrochemical indicator for the formation of double-stranded DNA and the AuNP assemblies facilitate the electrical contact of methylene blue.
4.5. QCM-based Detection and Bio-Bar-Code Assay
Quartz crystal microbalances (QCM), are piezoelectric devices that provide an ultrasensitive mass sensor. QCM based technique of DNA sensing have been widely used in biodiagnosis, due to the high sensitivity, economic effectiveness, and convenient operation of QCM instruments [84]. In practice, immobilization of thiol-terminated ssDNA onto the gold coated QCM followed by a hybridization step with target oligonucleotides leads to a detectable signal. As a consequence of large surface to volume ratio of AuNPs, the introduction of a layer of AuNPs between the gold film and the immobilized ssDNA significantly improves the detection capacity of the system [85]. In an alternative way, “sandwich” approaches can also drastically improve the detection limit of the system [86, 87, 88, 89, 90]. In the “sandwich” approach one end of target oligonucleotides hybridizes with the immobilized ssDNA molecules (recognition elements) while the other end hybridizes with ssDNA-modified AuNPs (signal amplifier). To further improve the sensitivity of the QCM approach, catalyzed deposition of gold onto the amplifier AuNPs has also been demonstrated [91], with a detection limit of ~1 fM
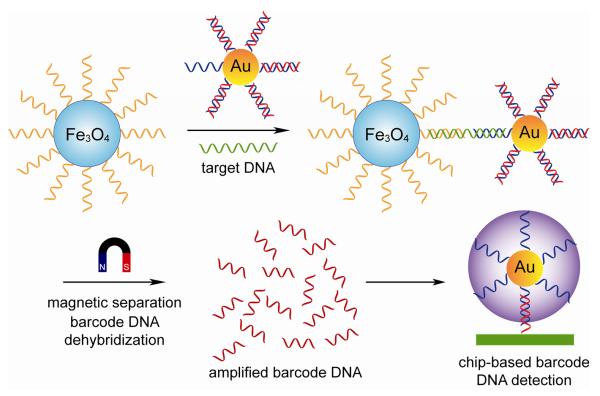
The principle of bio-bar-code amplification described for protein analyes, has been employed for DNA detection [92, 93]. As illustrated in Fig. 14, specific ssDNAs were first immobilized onto a magnetic microparticle surface. Consequently, sandwich assemblies were formed when target DNA hybridizes with both the magnetic particle probes and the bio-bar-coded AuNP probes. The magnetic separation of the sandwich complex followed by thermal dehybridization releases the free bar-code nucleotides, which was then subjected to analysis. This method has led to 500 zeptomolar sensitivity, which is comparable to many PCR-based approaches without the need for enzymatic amplification [92]. Additionally, multiplexed DNA detection is well suited with this system by using a mixture of different biobarcoded NP probes [93].
Fig. 14.
NP-based bio-bar-code assay of DNA
5. Detection of Microorganisms Using Nanoparticles
5.1. Fluorescence-Based Detection Using Nanoparticles
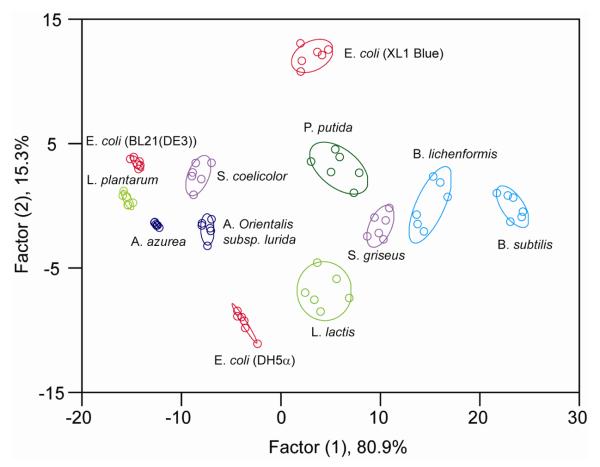
The efficient detection of pathogenic microorganisms is of great importance in food, medical, forensic, and environmental sciences [94]. AuNP-conjugated polymer systems were used to detect pathogens [95]. Three cationic AuNPs and one anionic PPE carrying carboxylate and oligo(ethylene glycol) arms were combined to generate non-covalent complexes. In the presence of bacteria, the initially quenched fluorescent polymers recover their fluorescence. The sensor array has been used to identify 12 microorganisms, which contain both Gram-positive (e.g. A. azurea, B. subtilis) and Gram-negative (e.g. E. coli, P. putida) species. As shown in Fig. 15, LDA discerns not only the species, but also the strains of the bacteria. The outstanding performance of this system is attributed to the exceptional quenching ability of AuNPs as well as the ‘molecular wire’ effect of PPE polymer [96].
Fig. 15.
Canonical score plot for the fluorescence response patterns of three AuNP-conjugated polymer constructs in the presence of bacteria processed with LDA. The first two factors consist of 96.2% variance and the 95% confidence ellipse for the individual bacteria are depicted.
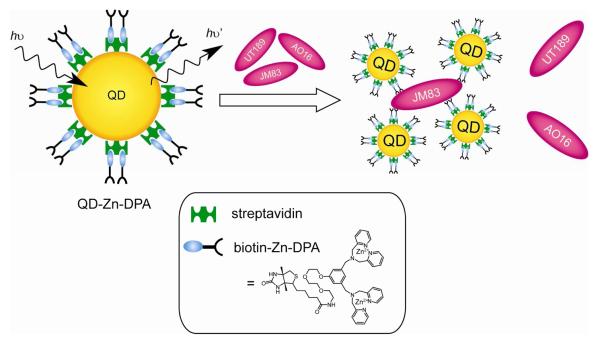
The use of QDs as a fluorescence labeling system in microorganism detection has been successfully demonstrated [97]. Fluorescent CdSe NPs were conjugated with wheat germ agglutinin (WGA), which can respond to gram-positive bacteria [98]. In the presence of the bacteria, the QD-WGA conjugate can bind to sialic acid and N-acetylglucosaminyl residues on the bacterial cell walls. QDs can also be conjugated with antibodies to detect specific pathogenic microorganisms such as Escherichia coli, Salmonella typhimurium, Cryptosporidium parvum, Giardia lamblia, and oral bacteria [99, 100, 101, 102]. QD-antibody systems have been shown to exhibit superior photostability and multiplexing capability, compared with traditional organic dyes. In addition, QDs conjugated with zinc-dipicolylamine (Zn-DPA) coordination complexes can selectively bind to a Escherichia coli mutant that lacks an O-antigen element, allowing optical detection in a living mouse leg infection model (Fig. 16) [103].
Fig. 16.
Schematic illustrations of QD-Zn-DPA and its selective detection to Gram-negative E. coli JM83.
6. Conclusion and Future Prospects
NPs present a versatile synthetic scaffold for the creation of detection systems for analyzing chemical and biological targets. NPs provide a suitable platform for the incorporation of various receptors, allowing the binding of target analytes with appropriate affinity and selectivity. Moreover, the environment-sensitive optoelectronic properties of NPs can be harnessed to realize the transduction of the binding events. Thus, functionalized NPs can act as both molecular receptor and signal transducer, simplifying system design.
For many sensor applications NPs exhibit distinctive attributes that can increase the sensitivity and selectivity of assays relative to conventional diagnostic techniques. Advanced nanodiagnostic techniques have also opened a promising avenue to provide rapid, low-cost, easy and multiplexed identification of biomarkers (e.g. proteins and genes) in the clinic. However, to meet the demand of clinical diagnostics for the development of personalized medicine, continuous efforts for optimization of these parameters are necessary. In particular, efforts are required for the development of efficient sensors with the ability to detect analytes in complex biological fluids like blood, urine, serum etc. A crucial factor in designing highly efficient sensors is the modulation of nanoparticle surface functionality for selective capture of target analytes. For this purpose, nanoparticle surfaces have been engineered with suitable functionality to utilize the highly selective recognition events like formation of double-stranded DNA, antibody-antigen, and aptamer-analyte interactions. These systems are useful, but have limitations as regards sensing of disease states. This is mainly due to the need of a tremendous amount of pertinent recognition elements for the multianalyte detection. This issue is being addressed in two parallel fashions. In one case, miniaturization of the sensor system allows more specific binders to be used in a given device. The other possible direction is the use of a differential sensor array approach. As in this case selectivity is required rather than specificity, a limited number of individual sensors may screen unlimited number of different target analytes. To this end, although it is clear that the NPs provide a powerful and evolving toolkit for designing ultrasensitive detection methods, much work needs to be done for transition of these settings towards the real world applications.
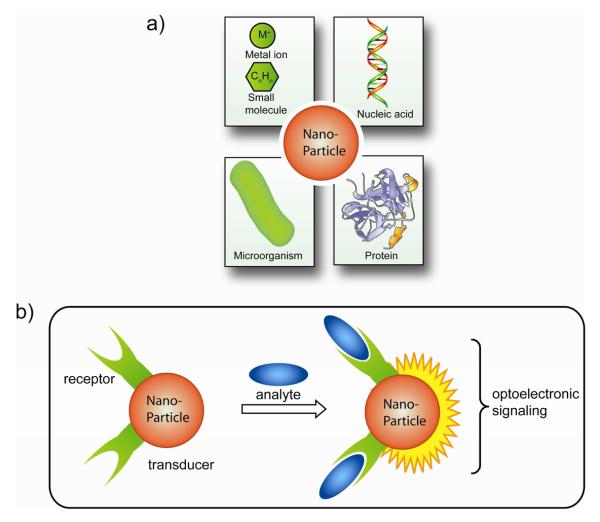
Fig. 1.
a) Examples of targets for nanoparticle based detection. b) Schematic depiction of a representative nanoparticle based detection system.
Fig. 2.
a) Schematic depictions of TNT sensor constructs. b) Specificity of the QD-based TNT sensor investigated in the presence of three TNT analogues (adapted with permission from reference [12]).
Fig. 3.
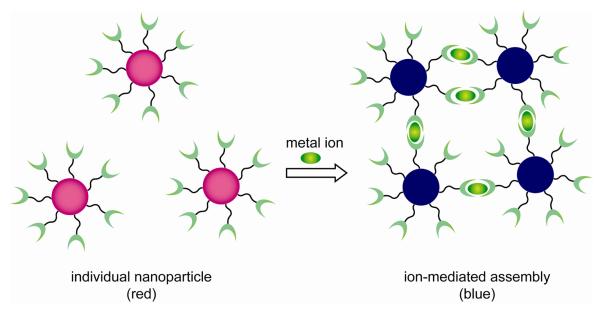
Schematic representation of metal ion-induced nanoparticle assembly.
Fig. 4.
Hg2+ detection using DNA functionalized nanoparticle and relying on the thymidineHg2+-thymidine coordination chemistry.
Fig. 5.
Glucose sensing using dextran functionalized nanoparticles.
Fig. 6.
Aptamer mediated cocaine detection.
Fig. 8.
QD-Peptide sensor for proteases.
Fig. 10.
Nanoparticle based bio-bar-code assay for proteins.
Fig. 12.
Bioconjugated silica NPs as labels for chip-based sandwich DNA assays.
7. Acknowledgement
This research was supported by the NIH grant (GM077173).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8. References
- 1.Diamond D. Principles of Chemical and Biological Sensors. John Wiley & Sons, Inc.; New York, NY: 1998. pp. 1–18. [Google Scholar]
- 2.Sheehan PE, Whitman LJ. Detection limits for nanoscale biosensors. Nano Lett. 2005;5:803–807. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 3.Rosi N, Mirkin CA. Nanostructures in Biodiagnostics. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 4.Alivisatos P. The use of nanocrystals in biological detection. Nat. Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 5.Niemeyer CM. Nanoparticles, proteins, and nucleic acids: Biotechnology meets materials science. Angew. Chem. Int. Edit. 2001;40:4128–4158. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 6.West JL, Halas NJ. Applications of nanotechnology to biotechnology - Commentary. Curr. Opin. Biotechnol. 2000;11:215–217. doi: 10.1016/s0958-1669(00)00082-3. [DOI] [PubMed] [Google Scholar]
- 7.Parak WJ, Gerion D, Pellegrino T, Zanchet D, Micheel C, Williams SC, Boudreau R, Le Gros MA, Larabell CA, Alivisatos AP. Biological applications of colloidal nanocrystals. Nanotechnology. 2003;14:R15–R27. [Google Scholar]
- 8.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 9.Jin WJ, Fernandez-Arguelles MT, Costa-Fernandez JM, Pereiro R, Sanz-Medel A. Photoactivated luminescent CdSe quantum dots as sensitive cyanide probes in aqueous solutions. Chem. Commun. 2005:883–885. doi: 10.1039/b414858d. [DOI] [PubMed] [Google Scholar]
- 10.Jin T, Fujii F, Sakata H, Tamura M, Kinjo M. Amphiphilic p-sulfonatocalix[4]arene-coated CdSe/ZnS quantum dots for the optical detection of the neurotransmitter acetylcholine. Chem. Commun. 2005:4300–4302. doi: 10.1039/b506608e. [DOI] [PubMed] [Google Scholar]
- 11.Snee PT, Somers RC, Nair G, Zimmer JP, Bawendi MG, Nocera DG. A ratiometric CdSe/ZnS nanocrystal pH sensor. J. Am. Chem. Soc. 2006;128:13320–13321. doi: 10.1021/ja0618999. [DOI] [PubMed] [Google Scholar]
- 12.Goldman ER, Medintz IL, Whitley JL, Hayhurst A, Clapp AR, Uyeda HT, Deschamps JR, Lassman ME, Mattoussi H. A hybrid quantum dot-antibody fragment fluorescence resonance energy transfer-based TNT sensor. J. Am. Chem. Soc. 2005;127:6744–6751. doi: 10.1021/ja043677l. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Atwater M, Wang J, Huo Q. Extinction Coefficient of Gold Nanoparticles with Different Sizes and Different Capping Ligands. Colloid Surf. B: Biointerfaces. 2006;58:3–7. doi: 10.1016/j.colsurfb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Jain PK, El-Sayed IH, El-Sayed MA. Au Nanoparticles Target Cancer. Nano Today. 2007;2:18–29. [Google Scholar]
- 15.Sapsford KE, Berti L, Medintz IL. Materials for Fluorescence Resonance Energy Transfer Analysis: Beyond Traditional Donor-Acceptor Combinations. Angew. Chem. Int. Ed. 2006;45:4562–4589. doi: 10.1002/anie.200503873. [DOI] [PubMed] [Google Scholar]
- 16.Huang T, Murray RW. Quenching of [Ru(bpy)3]2+ Fluorescence by Binding to Au Nanoparticles. Langmuir. 2002;18:7077–7081. [Google Scholar]
- 17.Chen S-J, Chang H-T. Nile red-adsorbed gold nanoparticles for selective determination of thiols based on energy transfer and aggregation. Anal. Chem. 2004;76:3727–3734. doi: 10.1021/ac049787s. [DOI] [PubMed] [Google Scholar]
- 18.He XR, Liu HB, Li YL, Wang S, Li YJ, Wang N, Xiao JC, Xu XH, Zhu DB. Gold nanoparticle-based fluorometric and colorimetric sensing of copper(II) ions. Adv. Mat. 2005;17:2811–2815. [Google Scholar]
- 19.Ipe BI, Yoosaf K, Thomas KG. Functionalized Gold Nanoparticles as Phosphorescent Nanomaterials and Sensors. J. Am. Chem. Soc. 2006;128:1907–1913. doi: 10.1021/ja054347j. [DOI] [PubMed] [Google Scholar]
- 20.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. Calculated Absorption and Scattering Properties of Gold Nanoparticles of Different Size, Shape, and Composition: Applications in Biological Imaging and Biomedicine. J. Phys. Chem. B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 21.Su K-H, Wei Q-H, Zhang X, Mock JJ, Smith DR, Schultz S. Interparticle coupling effects on plasmon resonances of nanogold particles. Nano Lett. 2003;3:1087–1090. [Google Scholar]
- 22.Lin S-Y, Liu S-W, Lin C-M, Chen C.-h. Recognition of Potassium Ion in Water by 15-Crown-5 Functionalized Gold Nanoparticles. Anal. Chem. 2002;74:330–335. doi: 10.1021/ac0156316. [DOI] [PubMed] [Google Scholar]
- 23.Lin S-Y, Chen C-H, Lin M-C, Hsu H-F. A Cooperative Effect of Bifuctionalized Nanaoparticles on Recognition: Sensing Alkali Ions by Crown and Carboxylate Moieties in Aqueous Media. Anal. Chem. 2005;77:4821–4828. doi: 10.1021/ac050443r. [DOI] [PubMed] [Google Scholar]
- 24.Obare SO, Hollowell RE, Murphy CJ. Sensing Strategy for Lithium Ion Based on Gold Nanoparticles. Langmuir. 2002;18:10407–10410. [Google Scholar]
- 25.Reynolds AJ, Haines AH, russell DA. Gold Glyconanoparticles for Mimics and Measurement of Metal Ion-Mediated Carbohydrate - Carbohydrate Interactions. Langmuir. 2006;22:1156–1163. doi: 10.1021/la052261y. [DOI] [PubMed] [Google Scholar]
- 26.Kim YJ, Johnson RC, Hupp JT. Gold nanoparticle-based sensing of “spectroscopically silent” heavy metal ions. Nano Lett. 2001;1:165–167. [Google Scholar]
- 27.Huang CC, Chang HT. Parameters for selective colorimetric sensing of mercury (II) in aqueous solutions using mercaptopropionic acid-modified gold nanoparticles. Chem. Commun. 2007:1215–1217. doi: 10.1039/b615383f. [DOI] [PubMed] [Google Scholar]
- 28.Lin S-Y, Wu S-H, Chen C.-h. A simple strategy for prompt visual sensing by gold nanoparticles: general applications of interparticle hydrogen bonds. Angew. Chem. Int. Ed. 2006;45:4948–4951. doi: 10.1002/anie.200600771. [DOI] [PubMed] [Google Scholar]
- 29.Yang WR, Gooding JJ, He ZC, Li Q, Chen GN. Fast colorimetric detection of copper ions using L-cysteine functionalized gold nanoparticles. J. Nanosci.Nanotechnol. 2007;7:712–716. [PubMed] [Google Scholar]
- 30.Si S, Kotal A, Mandal TK. One-dimensional assembly of peptide-functionalized gold nanoparticles: An approach toward mercury ion sensing. J. Phys. Chem. C. 2007;111:1248–1255. [Google Scholar]
- 31.Lee J-S, Han MS, Mirkin CA. Colorimetric detection of mercuric ion (Hg2+) in aqueous media using DNA-functionalized gold nanoparticles. Angew. Chem. Int. Ed. 2007;46:4093–4096. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Lu Y. A Colorimetric Lead Biosensor Using DNAzyme-Directed Assembly of Gold Nanoparticles. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 33.Liu JW, Lu Y. Optimization of a Pb2+-directed gold nanoparticle/DNAzyme assembly and its application as a colorimetric biosensor for Pb2+ Chem. Mat. 2004;16:3231–3238. [Google Scholar]
- 34.Liu JW, Lu Y. Accelerated color change of gold nanoparticles assembled by DNAzymes for simple and fast colorimetric Pb2+ detection. J. Am. Chem. Soc. 2004;126:12298–12305. doi: 10.1021/ja046628h. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Lu Y. Stimuli-responsive disassembly of nanoparticle aggregates for light-up colorimetric sensing. J. Am. Chem. Soc. 2005;127:12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Lu Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. 2006;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 37.Kubo Y. Isothiouronium-modified gold nanoparticles capable of colorimetric sensing of oxoanions in aqueous MeOH solution. Tetrahedron Lett. 2005;46:4369–4372. [Google Scholar]
- 38.Watanabe S, Seguchi H, Yoshida K, Kifune K, Tadaki T, Shiozaki H. Colorimetric Detection of Fluoride Ion in An Aqueous Solution Using a Thioglucose-Capped Gold Nanoparticle. Tetrahedron Lett. 2005;46:8827–8829. [Google Scholar]
- 39.Aslan K, Lakowicz JR, Geddes CD. Nanogold-plasmon-resonance-based glucose sensing. Analytical Biochemistry. 2004;330:145–155. doi: 10.1016/j.ab.2004.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aslan K, Lakowicz JR, Geddes CD. Nanogold plasmon resonance-based glucose sensing. 2. Wavelength-ratiometric resonance light scattering. Anal. Chem. 2005;77:2007–2014. doi: 10.1021/ac0484880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsui J, Akamatsu K, Nishiguchi S, Miyoshi D, Nawafune H, Tamaki K, Sugimoto N. Composite of Au nanoparticles and molecularly imprinted polymer as a sensing material. Anal. Chem. 2004;76:1310–1315. doi: 10.1021/ac034788q. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Lu Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. 2006;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Lu Y. Smart nanomaterials responsive to multiple chemical stimuli with controllable cooperativity. Adv. Mat. 2006;18:1667–1671. [Google Scholar]
- 44.Liu J, Mazumdar D, Lu Y. A simple and sensitive ‘dipstick’ test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew. Chem. Int. Ed. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- 45.Nagasaki Y, Ishii T, Sunaga Y, Watanabe Y, Otsuka H, Kataoka K. Novel molecular recognition via fluorescent resonance energy transfer using a biotin-PEG/polyamine stabilized CdS quantum dot. Langmuir. 2004;20:6396–6400. doi: 10.1021/la036034c. [DOI] [PubMed] [Google Scholar]
- 46.Oh E, Hong M-Y, Lee D, Nam S-H, Yoon HC, Kim H-S. Inhibition Assay of Biomolecules Based on Fluorescence Resonance Energy Transfer (FRET) Between Quantum Dots and Gold Nanoparticles. J. Am. Chem. Soc. 2005;127:3270–3271. doi: 10.1021/ja0433323. [DOI] [PubMed] [Google Scholar]
- 47.Oh E, Lee D, Kim YP, Cha SY, Oh DB, Kang HA, Kim J, Kim HS. Nanoparticle-based energy transfer for rapid and simple detection of protein glycosylation. Angew. Chem. Int. Ed. 2006;45:7959–7963. doi: 10.1002/anie.200601948. [DOI] [PubMed] [Google Scholar]
- 48.Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Uyeda HT, Chang EL, Deschamps JR, Dawson PE, Mattoussi H. Proteolytic activity monitored by fluorescence resonance energy transfer through quantum-dot-peptide conjugates. Nat. Mater. 2006;5:581–589. doi: 10.1038/nmat1676. [DOI] [PubMed] [Google Scholar]
- 49.You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Detection and identification of proteins using nanoparticle-fluorescent polymer ‘chemical nose’ sensors. Nat. Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 50.Otsuka H, Akiyama Y, Nagasaki Y, Kataoka K. Quantitative and Reversible Lectin-Induced Association of Gold Nanoparticles Modified with α-Lactosyl-ω-mercaptopoly(ethylene glycol) J. Am. Chem. Soc. 2001;123:8226–8230. doi: 10.1021/ja010437m. [DOI] [PubMed] [Google Scholar]
- 51.Tsai CS, Yu TB, Chen CT. Gold nanoparticle-based competitive colorimetric assay for detection of protein-protein interactions. Chem. Commun. 2005:4273–4275. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- 52.Schofield CL, Field RA, Russell DA. Glyconanoparticles for the colorimetric detection of cholera toxin. Anal. Chem. 2007;79:1356–1361. doi: 10.1021/ac061462j. [DOI] [PubMed] [Google Scholar]
- 53.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. Aptamer-Functionalized Au Nanoparticles for the Amplified Optical Detection of Thrombin. J. Am. Chem. Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 54.Xiao Y, Pavlov V, Levine S, Niazov T, Markovitch G, Willner I. Catalytic growth of Au nanoparticles by NAD(P)H cofactors: optical sensors for NAD(P)+-dependent biocatalyzed transformations. Angew. Chem. Int. Ed. 2004;43:4519–4522. doi: 10.1002/anie.200460608. [DOI] [PubMed] [Google Scholar]
- 55.Thanh NTK, Rosenzweig Z. Development of an aggregation-based immunoassay for anti-protein A using gold nanoparticles. Anal. Chem. 2002;74:1624–1628. doi: 10.1021/ac011127p. [DOI] [PubMed] [Google Scholar]
- 56.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, Klein WL, Mirkin CA. Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew. Chem. Int. Ed. 2006;45:3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 58.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 59.Dyadyusha L, Yin H, Jaiswal S, Brown T, Baumberg JJ, Booy FP, Melvin T. Quenching of CdSe Quantum Dot Emission, a New Approach for Biosensing. Chem. Commun. 2005:3201–3203. doi: 10.1039/b500664c. [DOI] [PubMed] [Google Scholar]
- 60.Han MY, Gao XH, Su JZ, Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 61.Gerion D, Chen FQ, Kannan B, Fu AH, Parak WJ, Chen DJ, Majumdar A, Alivisatos AP. Room-temperature single-nucleotide polymorphism and multiallele DNA detection using fluorescent nanocrystals and microarrays. Anal. Chem. 2003;75:4766–4772. doi: 10.1021/ac034482j. [DOI] [PubMed] [Google Scholar]
- 62.Zhao XJ, Tapec-Dytioco R, Tan WH. Ultrasensitive DNA detection using highly fluorescent bioconjugated nanoparticles. J. Am. Chem. Soc. 2003;125:11474–11475. doi: 10.1021/ja0358854. [DOI] [PubMed] [Google Scholar]
- 63.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 64.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 65.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 1998;120:1959–1964. [Google Scholar]
- 66.Reynolds RA, Mirkin CA, Letsinger RL. Homogeneous, Nanoparticle-Based Quantitative Colorimetric Detection of Oligonucleotides. J. Am. Chem. Soc. 2000;122:3795–3796. [Google Scholar]
- 67.Cao YC, Jin RC, Thaxton S, Mirkin CA. A two-color-change, nanoparticle-based method for DNA detection. Talanta. 2005;67:449–455. doi: 10.1016/j.talanta.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 68.Thaxton CS, Georganopoulou DG, Mirkin CA. Gold nanoparticle probes for the detection of nucleic acid targets. Clin. Chim. Acta. 2006;363:120–126. doi: 10.1016/j.cccn.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 69.Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat. Biotechnol. 2004;22:883–887. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakrabarti R, Klibanov AM. Nanocrystals modified with peptide nucleic acids (PNAs) for selective self-assembly and DNA detection. J. Am. Chem. Soc. 2003;125:12531–12540. doi: 10.1021/ja035399g. [DOI] [PubMed] [Google Scholar]
- 71.Li H, Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 73.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman Spectroscopic Fingerprints for DNA and RNA Detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 74.Sun L, Yu C, Irudayaraj J. Surface-enhanced Raman scattering based nonfluorescent probe for multiplex DNA detection, Anal. Chem. 2007;79:3981–3988. doi: 10.1021/ac070078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Castaneda MT, Alegret S, Merkoci A. Electrochemical sensing of DNA using gold nanoparticles. Electroanalysis. 2007;19:743–753. [Google Scholar]
- 76.Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 77.Ozsoz M, Erdem A, Kerman K, Ozkan D, Tugrul B, Topcuoglu N, Ekren H, Taylan M. Electrochemical genosensor based on colloidal gold nanoparticles for the detection of factor V leiden mutation using disposable pencil graphite electrodes. Anal. Chem. 2003;75:2181–2187. doi: 10.1021/ac026212r. [DOI] [PubMed] [Google Scholar]
- 78.Cai H, Wang YQ, He PG, Fang YH. Electrochemical detection of DNA hybridization based on silver-enhanced gold nanoparticle label. Anal. Chim. Acta. 2002;469:165–172. [Google Scholar]
- 79.Wang J, Li JH, Baca AJ, Hu JB, Zhou FM, Yan W, Pang DW. Amplified voltammetric detection of DNA hybridization via oxidation of ferrocene caps on gold nanoparticle/streptavidin conjugates. Anal. Chem. 2003;75:3941–3945. doi: 10.1021/ac0344079. [DOI] [PubMed] [Google Scholar]
- 80.Baca AJ, Zhou FM, Wang J, Hu JB, Li JH, Wang JX, Chikneyan ZS. Attachment of ferrocene-capped gold nanoparticle-streptavidin conjugates onto electrode surfaces covered with biotinylated biomolecules for enhanced voltammetric analysis. Electroanalysis. 2004;16:73–80. [Google Scholar]
- 81.Wang JX, Zhu X, Tu QY, Guo Q, Zarui CS, Momand J, Sun XZ, Zhou FM. Capture of p53 by electrodes modified with consensus DNA duplexes and amplified voltammetric detection using ferrocene-capped gold nanoparticle/streptavidin conjugates. Anal. Chem. 2008;80:769–774. doi: 10.1021/ac0714112. [DOI] [PubMed] [Google Scholar]
- 82.Zhang J, Song SP, Zhang LY, Wang LH, Wu HP, Pan D, Fan C. Sequence-specific detection of femtomolar DNA via a chronocoulometric DNA sensor (CDS): Effects of nanoparticle-mediated amplification and nanoscale control of DNA assembly at electrodes. J. Am. Chem. Soc. 2006;128:8575–8580. doi: 10.1021/ja061521a. [DOI] [PubMed] [Google Scholar]
- 83.Li D, Yan Y, Wieckowska A, Willner I. Amplified electrochemical detection of DNA through the aggregation of Au nanoparticles on electrodes and the incorporation of methylene blue into the DNA-croslinked structure. Chem. Commun. 2007:3544–3546. doi: 10.1039/b704731b. [DOI] [PubMed] [Google Scholar]
- 84.Marx KA. Quartz crystal microbalance: A useful tool for studying thin polymer films and complex biomolecular systems at the solution-surface interface. Biomacromolecules. 2003;4:1099–1120. doi: 10.1021/bm020116i. [DOI] [PubMed] [Google Scholar]
- 85.Lin L, Zhao HQ, Li JR, Tang JA, Duan MX, Jiang L. Study on colloidal Au-enhanced DNA sensing by quartz crystal microbalance. Biochem. Biophys. Res. Commun. 2000;274:817–820. doi: 10.1006/bbrc.2000.3233. [DOI] [PubMed] [Google Scholar]
- 86.Zhou X-C, O'Shea SJ, Li SFY. Amplified Microgavimetric Gene Sensor Using Au Nanoparticle Modified Oligonucleotides. Chem. Commun. 2000:953–954. [Google Scholar]
- 87.Patolsky F, Ranjit KT, Lichtenstein A, Willner I. Dendritic amplification of DNA analysis by oliogonucleotide-functionalized Au-nanoparticles. Chem. Commun. 2000:1025–1026. [Google Scholar]
- 88.Willner I, Patolsky F, Weizmann Y, Willner B. Amplified detection of single-base mismatches in DNA using microgravimetric quartz-crystal-microbalance transduction. Talanta. 2002;56:847–856. doi: 10.1016/s0039-9140(01)00658-0. [DOI] [PubMed] [Google Scholar]
- 89.Nie LB, Yang Y, Li S, He NY. Enhanced DNA detection based on the amplification of gold nanoparticles using quartz crystal microbalance. Nanotechnology. 2007;18:305501. [Google Scholar]
- 90.Liu T, Tang J, Jiang L. Sensitivity enhancement of DNA sensors by nanogold surface modification. Biochem. Biophys. Res. Commun. 2002;295:14–16. doi: 10.1016/s0006-291x(02)00628-9. [DOI] [PubMed] [Google Scholar]
- 91.Weizmann Y, Patolsky F, Willner I. Amplified detection of DNA and analysis of single-base mismatches by the catalyzed deposition of gold on Au-nanoparticles. Analyst. 2001;126:1502–1504. doi: 10.1039/b106613g. [DOI] [PubMed] [Google Scholar]
- 92.Nam JM, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J. Am. Chem. Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 93.Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew. Chem. Int. Ed. 2006;45:3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 94.Deisingh AK, Thompson M. Detection of infectious and toxigenic bacteria. Analyst. 2002;127:567–581. doi: 10.1039/b109895k. [DOI] [PubMed] [Google Scholar]
- 95.Phillips RL, Miranda OR, You CC, Rotello VM, Bunz UHF. Rapid and efficient identification of bacteria using gold-nanoparticle - Poly(para-phenyleneethynylene) constructs. Angew. Chem. Int. Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 96.Thomas SW, Joly GD, Swager TM. Chemical sensors based on amplifying fluorescent conjugated polymers. Chem. Rev. 2007;107:1339–1386. doi: 10.1021/cr0501339. [DOI] [PubMed] [Google Scholar]
- 97.Liu WT. Nanoparticles and their biological and environmental applications. J. Biosci. Bioeng. 2006;102:1–7. doi: 10.1263/jbb.102.1. [DOI] [PubMed] [Google Scholar]
- 98.Kloepfer JA, Mielke RE, Wong MS, Nealson KH, Stucky G, Nadeau JL. Quantum dots as strain- and metabolism-specific microbiological labels. Appl. Environ. Microbiol. 2003;69:4205–4213. doi: 10.1128/AEM.69.7.4205-4213.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Su XL, Li YB. Quantum dot biolabeling coupled with immunomagnetic separation for detection of Escherichia coli O157 : H7. Anal. Chem. 2004;76:4806–4810. doi: 10.1021/ac049442+. [DOI] [PubMed] [Google Scholar]
- 100.Zhu L, Ang S, Liu WT. Quantum dots as a novel immunofluorescent detection system for Cryptosporidium parvum and Giardia lamblia. Appl. Environ. Microbiol. 2004;70:597–598. doi: 10.1128/AEM.70.1.597-598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang LJ, Li YB. Simultaneous detection of Escherichia coli O157 : H7 and Salmonella Typhimurium using quantum dots as fluorescence labels. Analyst. 2006;131:394–401. doi: 10.1039/b510888h. [DOI] [PubMed] [Google Scholar]
- 102.Chalmers NI, Palmer RJ, Du-Thumm L, Sullivan R, Shi WY, Kolenbrander PE. Use of quantum dot luminescent probes to achieve single-cell resolution of human oral bacteria in biofilms. Appl. Environ. Microbiol. 2007;73:630–636. doi: 10.1128/AEM.02164-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leevy WM, Lambert TN, Johnson JR, Morris J, Smith BD. Quantum dot probes for bacteria distinguish Escherichia coli mutants and permit in vivo imaging. Chem. Commun. 2008:2331–2333. doi: 10.1039/b803590c. [DOI] [PMC free article] [PubMed] [Google Scholar]