Summary
Simulation-based assessment is a popular and frequently necessary approach to evaluation of statistical procedures. Sometimes overlooked is the ability to take advantage of underlying mathematical relations and we focus on this aspect. We show how to take advantage of large-sample theory when conducting a simulation using the analysis of genomic data as a motivating example. The approach uses convergence results to provide an approximation to smaller-sample results, results that are available only by simulation. We consider evaluating and comparing a variety of ranking-based methods for identifying the most highly associated SNPs in a genome-wide association study, derive integral equation representations of the pre-posterior distribution of percentiles produced by three ranking methods, and provide examples comparing performance. These results are of interest in their own right and set the framework for a more extensive set of comparisons.
Keywords: Efficient simulation, ranking procedures, SNP identification
1 Introduction
Hans van Houwelingen’s impact on statistical theory and practice is both broad and deep. His many contributions are detailed elsewhere in this volume and we mention only a few. Hans has made signature methodological contributions to non- and semi-parametric empirical Bayes, to large sample theory in several contexts, to cross-validation and other sample reuse methods. His applied collaborations are extensive and, as with his methods work, his impact is “zeer groot.”
Simulation-based assessment is a popular and frequently necessary approach to evaluation of statistical procedures. Approaches range from the most straightforward, but generally inefficient (prepare a straightforward program and just blast off) to more sophisticated approaches using importance sampling (to enable generation of the appropriate stochastic sequences or to improve efficiency), rejection sampling or Markov Chain Monte-Carlo (to enable generation of the appropriate stochastic sequences). Carlin and Louis (2009) provide details on these and others. Sometimes overlooked is the ability to take advantage of underlying mathematical relations and we focus on this aspect. We show how to take advantage of large-sample convergence of empirical distributions to develop mathematical relations that provide an excellent approximation to the results of a large-scale simulation. When components of these relations are available analytically, the approximations can replace simulations. If some components are themselves available only by simulation, then in some cases the mathematical relations can be used to improve efficiency. This use of two types of CPUs (one biologic and one in silico) most definitely represents a strategic approach that Hans would take. We trust that this article, at least to a small degree, captures the spirit of his innovation, creativity and eclectic use of statistical theory and simulation-based evaluations.
Hans has made important contributions to genomics analysis, in particular with regard to statistical methodology for dimension reduction in association studies (Goeman et al., 2006; Heidema et al., 2007; Uh et al., 2007), and efficient statistical methods for linkage and association studies that comprehensively employ all information available (Houwing-Duistermaat et al., 2007; Lebrec et al., 2008). We motivate our approach and provide proof of concept in the setting of Genome-Wide Association studies (GWAs). Specifically, we consider evaluating and comparing performance of ranking-based methods that can be used to identify the most highly associated SNPs in a GWAs.
1.1 Genome-wide association studies
A single-nucleotide polymorphism (SNP) is a locus in the DNA where some appreciable variation in the nucleotide sequence between the members of a species occurs. In humans for example, each person has one pair of each of the autosomes (chromosomes 1–22), with one copy inherited from each parent. While almost all loci have identical nucleotides in each DNA strand for each person, about every 500 base pair positions some differences between people exist. Interestingly, the vast majority of all SNPs are bi-allelic, that is, only 2 out of the 4 nucleotides occur in the population. For a locus with variation to be defined as a SNP, the frequency with which the minor allele occurs in the population is usually required to be above 1%. Since there are substantial differences in the genetic background between ethnic groups or geographically separated populations, the actualminor allele frequencies (MAFs) between sub-populations can differ significantly. The bi-allelic SNPs are typically encoded as nucleotide pairs within a person’s matched chromosome pairs (e. g. (CC,CT,TT)), or specifically for SNPs typed in genomic arrays, as a generic factor with three levels (e. g. (AA,AB,BB)). In the statistical modeling of SNP data, the SNPs are typically encoded as the number of variant alleles (0, 1, or 2), where the variant allele (as opposed to the common allele) is defined as the allele that occurs less frequent in the population. In a “homogeneous” population (and under certain assumptions such as random mating), the frequencies of the nucleotide pairs that define a SNP is governed by the Hardy-Weinberg equilibrium, producing prevalences of (p2, 2pq, q2), with p being the major and q = 1 − p the minor allele frequency. A typical GWAs assesses association between candidate genotypes and a disorder or a quantitative trait (the phenotype), often via Z-scores, testing the hypothesis of no association between the genotype (AA, AB, BB) and the phenotype. Then, these association measures are ranked and the most highly associated SNPs identified. See Ruczinski et al. (2009) for additional details. To detect the genetic causes of complex human disease, genome-wide association studies have become the method of choice for many researchers around the globe. Consistent with this is the sheer abundance of GWAs papers that have been published in the last couple of years.
1.2 Issues in analyzing GWAs data
The predominant strategy for analyzing GWAs data is to carry out SNP specific (marginal) tests such as the Cochran-Armitage trend test, sort the results from the smallest to the largest p-value, and declare significance based on a Bonferroni correction to limit the family-wise error rate at a fixed level (typically, 5%). The benefit of this approach is its straightforward and reproducible implementation, and has led to the identification of important genes for many diseases (Manolio et al., 2008, provide an overview). However, both from scientific and statistical perspectives, this approach is sub-optimal for various reasons.
Enforcing a tight control on the family-wise error rate, i.e. the probability of declaring at least one SNP significant that is not associated with the phenotype, comes at the expense of the truly associated SNPs which do not achieve ”genome wide” significance. It appears that this strategy works in strong opposition to the aims of a GWAs, typically declared to be hypothesis generating and discovery oriented. This stringent type I error control is particularly worrisome in the investigation of truly complex diseases such as asthma or schizophrenia, since many SNPs, each typically with a very modest effect size, contribute to the disease risk. Unless the sample size is huge, none of the SNPs might reach overall significance after multiple comparisons corrections. From a Bayesian perspective, a Bonferroni adjustment with very small α can be justified if one has a strong prior belief that all the nulls are true (Westfall et al., 1997), however, the opposite is the case in GWAs. Moreover, controlling the family wise error rate via Bonferroni completely ignores type II errors, and employs a uniform threshold for significance that ignores power. Using such a constant threshold in a frequentist setting leads to an inconsistent procedure, and viewed from a decision theory perspective, leads to an inadmissible procedure (Wakefield, 2007, 2008a,b).
Employing the family wise error rate is certainly reasonable if the confirmation of a list of candidate SNPs believed to be associated with the phenotype is the objective, for example in sequencing or fine mapping of genomic regions. However, for many complex diseases the a priori information of the genetic base is very limited, and GWAs are usually carried out as truly hypothesis generating. Thus, the objective is to generate a list of SNPs for further investigation about their role in the genetic underpinning of the disease, or their correlation with truly disease causing variants via fine mapping. The generation of such a SNP list obviously relies on proper ranking methods, and should take the conditional power of each variant into account. In this manuscript, we focus on such ranking procedures.
We focus SNP selection based on ranks with no direct attention to controlling type I error, which usually is the procedure of choice (see for example Manolio et al., 2008). We consider ranks based on hypothesis test Z-scores and those based on Bayes optimal ranks (Lin et al., 2006). Ultimately, we are interested in simulation-based comparison of the performance of these with the current approaches. However, a GWAs can include more than one million candidate SNPs (and the number is growing), so computational demands in a simulation-based approach can be substantial. However, fortunately, as we show when the number of candidate SNPs is large, asymptotic consistency of empirical distributions can be used to speed up the assessment. These results will enable future complete assessments.
1.3 Organization
The scope of this paper is to lay the foundation and to compare different ranking procedures in a setting where only one of many variables is associated with an outcome of interest. We motivate this problem within the context of SNP association studies, which will be our main future application. Thus, we use a spiked SNP to demonstrate the differences in ranking procedures, that is, a single genetic locus with simulated non-zero effect size. In section 2 we outline candidate ranking methods including Z-score bases and Bayesian optimal approaches. See Lin et al. (2006) for full details. Sections 2.3 and 3 derive distributions of percentile ranks for a spiked SNP when K is large. Section 4 reports on comparisons of candidate ranking methods for mathematically evaluable models. Section 5 presents some cautions and section 6 sums up.
2 Ranking Methods
A variety of ranking methods are in use (see Lin et al., 2006). We focus on ranks/percentiles based on hypothesis test Z-scores and two Bayes optimal approaches. We rank (K +1) SNPs, where k = 1, …, K are “null” SNPs (with null or moderate association with the phenotype), and k = 0 is the spiked SNP, a SNP with a stochastically larger association than for the null SNPs.
2.1 The z-score approach
For each SNP, compute a Z-score which is used to test the hypothesis of no association, producing (Z0, Z1, …, ZK). Rank these and divide the ranks by K + 2 (one more that the number of SNPs), producing P̈ and P̈k. In a GWAs, Z-scores are often produced by a trend test. In the appendix, we outline the score test approach in Ruczinski et al. (2009) that also accommodates uncertain genotype calls.
2.2 Bayes optimal approaches
As detailed by Carlin and Louis (2009) and many other authors, Bayesian analysis requires a full probability model consisting of a prior distribution for parameters and a data likelihood (sampling distribution), conditional on parameters. Observed data map the prior to the posterior distribution and the posterior is used to make inferences, sometimes guided by a loss function (e.g, decision theory). In applications, especially with a large K, empirical Bayes approaches are very attractive (Schwender and Ickstadt, 2008).
Lin et al. (2006) show that Bayesian ranking/percentiling procedures structured by a ranking-relevant loss function can outperform those produced by ranking Z-scores or MLEs. We report on two ranking procedures, one that is “general purpose” and one targeted at identifying a specific number of SNPs. We assume that θ is the true magnitude of the genotype/phenotype association and construct a model so that it is also the conditional mean function for observed data. For clarity, we assume that other parameters in the sampling model are known or well-estimated, that the sampling distribution is Gaussian and so the maximum likelihood estimate θ̂ of θ (subsequently denoted by Yk) is sufficient for computing the posterior distribution, and that we are interested in detecting only the large, rather than both the large and small θk. These assumptions allow focus on taking advantage of the large K situation rather than on the computational details of Bayesian analysis.
For the null SNPs we have, k = 1, …, K;
| (1) |
We will use in the sequel that,
| (2) |
the prior distribution. The expectation is with respect to fk(Yk | H), the pre-posterior predictive distribution of Yk.
In a likelihood-based analysis, inputs Yk(=θ̂k) and σ2 are the MLE and its associated variance (inverse information). However, in many contexts a score test (see the appendix) is used. As we show in this section, a parameter estimate and its variance can be extracted from a score test.
Representation of ranks
For Bayes optimal ranks use the following representation for the K null SNPs in the context of ranking a total of (K + 1) SNPs. The percentile for the spiked SNP is computed along with those for the null SNPs. The Bayesian approach uses as input the computation that would be made, if one were to know the θk without error. Specifically, for k = 0, 1, …, K, the ranks are;
The smallest θ has rank 1. Inferences depend on the posterior distribution of the Rk(θ) as induced by the posterior distribution for θ and this depends on the prior and on the (Yk, ).
P̂k (general purpose percentiles)
First, compute the posterior expected ranks,
Then, rank the R̄k to get R̂k and P̂k.
(threshold-specific percentiles)
Select 0 < γ < 1, and let
Then,
The (defined as the ranks divided by K +2) are ordered according to the posterior probability of θk being among the top (1 − γ) fraction of the θs. Therefore, the SNPs with are the optimal selection for the top (1 − γ)(K + 1) and to optimally identify the top N SNPs, set γ = 1−N/(K + 1). Of course, the posterior probabilities will indicate the degree of confidence in this selection; that other SNPs may have just missed the cut, and that generally there is considerable uncertainty in this selection. Furthermore, as shown in Lin et al. (2006), unless , different γ can produce different orderings.
Extracting θ̂k and from a trend test
We need to map the trend test Z-score numerator and denominator into an estimate of the implicit θ (the log odds ratio for risk) and an estimate of its conditional variance. Using the model in section 2.1 and equation (21) in the appendix, we have for θ near 0 (omitting the subscript k),
| (3) |
So,
| (4) |
We also need the relation,
Therefore, for a given sample size (n), the power of a test or the performance of a ranking procedure depends on the MAF and on the fraction of cases in the sample (π).
2.3 Taking advantage of large K
The ranking method specific, cumulative distribution of the percentile for the spiked SNP provides a complete description of performance. A fully simulation-based approach to estimating this cdf would proceed by generating data from the assumed scenario for all (K + 1) SNPs, computing the percentiles and saving the percentile for the spiked SNP. After a sufficient number of simulation replications, the empirical distribution function (edf) of these percentiles can be used as the estimate and edfs from competing methods can be compared. For clarity, in the sequel we drop and so “Yk” stands for “(Yk, ).” Specifically, using PK(Y0) to denote the percentile for the spiked SNP as a function of all data, (Y0, Y1, …, YK), we need to compute pr(PK(Y0) > s), s ∈ [0, 1] and compare these tail functions among ranking methods (Z-score based, Bayes) for a set of scenarios. We use the tail function because we are looking for the stochastically large distributions.
Our goal is to compute pr(PK(Y0) > s), but consider computing pr(P∞(Y0) > s). If K is sufficiently large, evaluation of the latter provides a good approximation to the former. This is easily seen in the case wherein all SNPs, including the spiked SNP, come from the same distribution. In this case, pr(P∞(Y0) > s) = 1− s and for finite K, the distribution of the percentile for the spiked SNP is discrete uniform on the points {1/(K + 2), …, (K + 1)/(K + 2)} and so,
| (5) |
The foregoing holds for a fixed s. For s = sK such that (1 − sK)K ≈ N, then both limits in equation (5) go to 0 and in this situation the K = ∞ representation provides a lower bound to performance in identifying a the small number of SNPs in a large K context.
In the following sections we approximate performance for large K by developing mathematical results for pr(P∞(Y0) > s).
2.4 Distributions for the null SNPs and the spiked SNP
In real association studies, there is usually a fair amount of information available that can be employed in the specification of the prior distribution H. While the vast majority of all SNPs will not be associated (in a scientifically meaningful way) with a particular phenotype, the number of truly associated SNPs and the respective effect sizes can differ substantially between various outcomes of interest (see for example the results published by the Wellcome Trust Case Control Consortium 2007 and Manolio et al. 2008). Moreover, a certain degree of controversy exists in the community about two distinct genetic models which can be described as “common disease - common variant” and “common disease - rare variant”. The former, assuming that a few common SNPs explain a substantial proportion of the phenotype variability in a population, is the basis of the GWAs. However, in particular for some psychiatric diseases such as schizophrenia and bipolar disorder, this paradigm has been challenged (McClellan et al., 2007). The “common disease - rare variant” models asserts that for certain heritable traits only one rare variant of large effect explains the phenotype in a particular family, but different rare variants with large effects (which may or may not be in the same gene) are present in other families (Walsh et al., 2008). Thus, according to this view, the disease status of the subjects in a population could largely be explained by a collection of individually rare variants, with large effects.
For the null SNPs we assume that the working and actual priors both equal to H. Therefore, using standard Bayesian computations, the pre-posterior expectation of the posterior distribution is the prior H and it can be used as the posterior ensemble posterior distribution for the null SNPs. It is the K = ∞ approximation to the finite K distribution. To see that this produces a good approximation for all but the most extreme percentiles, consider simulating K “null” SNPs (these are not necessarily unassociated with phenotype, but have generally low association) and a small number (we focus on 1) highly associated, “spiked” SNPs. If K is very large, for each simulation replication the empirical distribution function (edf) of the K simulated values used for ranking will be very close to the theoretical (the generating) distribution because this edf is the average of iid realizations with expectation the distribution of interest. So, for a fixed percentile, the average converges almost surely and uniformly to the distribution of interest and for K large, the edf is, for all practical purposes, equal to the induced distribution. For example, in the Z-score approach, if all K SNPs are truly null, then the edf of the Z-scores is essentially N(0, 1) for each replication; more generally the edf is essentially equal to the distribution induced by the sampling model.
The ensemble posterior for the null SNPs is
| (6) |
The summands in (6) are iid bounded random variables with mean H1 (see equation (2)). So, the average converges almost surely and uniformly to H1. For very large K, H̄1(t | Y) ≈ H1(t). Note that if for θ, H1 is a point mass at 0, then so will be H̄1.
The foregoing assumes that H (and so H1) is known, but if K is large, an empirical Bayes or Bayes empirical Bayes approach produces the same result so long as the true H is in the allowable space of estimated priors. For empirical Bayes, it is sufficient that the parametric family includes H or that a (smoothed) non-parametric approach is used. For Bayes empirical Bayes, it is sufficient that H is in the support of the hyper-prior. In this case, for all but the extreme percentiles, the ensemble posterior will be virtually identical to H. For example, if H1 is Gaussian and the assumed prior is also Gaussian with (μ, τ2) estimated from the data, the result holds as it does for the (smoothed) non-parametric maximum likelihood approach.
By taking advantage of this near-constancy of the edf, the stochastic positioning of the spiked SNP among the “null” SNPs can be related either analytically or with minimal computation to this distribution, thereby efficiently computing the edf for its percentile. Evaluation proceeds by one-time generation of K realizations for the null SNPs and then repeated generation of realizations for the spiked SNP(s). Ranks and percentiles are computed by comparing these values to the relevant edf induced by the null SNPs. In some cases, for example when the null SNPs have mean 0 and Z-scores are used, the induced distribution is N(0, 1) and no simulation of the null SNPs is needed. Similarly, the spiked SNP may have a mathematically tractable distribution (for example, Gaussian) and a full mathematical treatment is possible. In any case, to approximate performance for a large K, at least for percentiles that are not too extreme a one-time simulation (generally generating realizations for substantially more than K SNPs) is needed for the null SNPs to produce the comparison distribution and repeated simulation for the spiked. However, see section 5 for a warning about situations wherein there is little to no saving in a simulation. Taking advantage of this near-constancy of the relevant edf for the K SNPs produces considerable efficiency gains and insights for the Z-score approach, the Bayesian approach and other candidate approaches to ranking.
2.5 The spiked SNP
For the spiked SNP (k = 0), we denote a generic prior by G rather than H. Further, we use A for the actual prior and W for the working (assumed) prior. As for the null SNPs, we assume a Gaussian sampling model and denote the sampling variance by (it can be a random variable),
| (7) |
with actual prior A and marginal distribution fA(y0). That is,
The working prior W can be different from the actual prior A, producing,
The Actual and the Working priors (A and W) may be different and one or both may equal H. We can set dW = dθ0 for a “frequentist” analysis. Setting A to a point mass at a single θ-value, allows assessment of the frequentist performance of the Bayesian analysis that assumes θ ~ W.
Conditioning on Y0 generates the working posterior distribution,
with pre-posterior expectation, taken with respect to the actual prior
| (8) |
If W = A, GW in equation (8) equals A and if A = H it equals H. For a frequentist (θ-specific) analysis of a Bayesian working model, let A be degenerate at some θ0.
“Frequentist” Bayes computations
We present in the context of the spiked SNP, but similar relations hold for the null SNPs. A “frequentist” (likelihood-based) analysis results from use of a flat, improper working prior, dW(θ) = dθ0. The posterior is the normalized likelihood (the likelihood is integrable for the Gaussian sampling distribution). That is . The marginal posterior for θ0 is produced by mixing over the distribution of and the pre-posterior distribution by integrating with respect to the marginal distribution of Y0.
3 Pre-posterior Distributions
We are interested in comparing performance of P̈k P̂k, and . For each, we want the pre-posterior tail distribution and to this end also need the pre-posterior distribution of Z-scores and of Bayesian posterior distributions computed from a working model.
3.1 The z-score approach
We first consider the situation wherein all “null” SNPs have θ ≡ 0 and the more general case.
All competing SNPs are null
All Z-scores are N(0,1) and so for large K their edf is N(0,1) as is H̄. In equations (15) and (19), μh = 0, . For the spiked SNP, set τ a = 0 (a point mass at μa) and compute the Z-score based on . Here μa = θ and is the variance in (3). This Z-score has mean ξ = μa/σa and local variance 1 (for small μa).
“Null” SNPs come from a distribution
Assume that the θ for the null SNPs come from H. Conditional on (θ, σ), Z ~ N(m, 1) with m = θ/σ. H induces a distribution on m, call it H̃ (recall that the prior delivers both the θk and the ). So,
| (9) |
In general, H needs to be evaluated numerically, but a few special cases are available.
Case I: If and θk iid N(μ, τ2) then
| (10) |
Case II: If σ is inverse gamma, H will be a t-distribution.
3.2 Pre-posterior distribution of P̂(Y)
Here we want the posterior percentile of the spiked SNP relative to all others with the percentile defined as the expected number of competing SNPs that the spiked SNP beats. With θk denoting a typical null SNP, and using P(Y0) for P∞(Y0), we have,
| (11) |
With GW(θ0 | Y0) the working posterior for the spiked SNP, consider a randomly drawn θk from H (we use H rather than H1 in the sequel). Assume that H has a density, then:
| (12) |
For each Y0, this is a value ∈ [0, 1] and we want its distribution. In general, it won’t have a closed form, but we can simulate Y0 from fA(y0) and get an empirical estimate without simulating the K null SNPs.
Theorem 1
| (13) |
Note that when W = A = H (the spiked SNP is also null), equation (13) equals 1 − H(H−1(s)) = 1 − s, the right tail of the U(0, 1) distribution.
Proof
We need to “locate” the random variable that has distribution GW(·| Y0) amongst the random variables that follow H and it is convenient to map all distributions to the [0, 1] interval. Do so by using H to compute the probability integral transform, i.e., H(·). Random variables that have distribution H are transformed to the U(0, 1) distribution and the transformed GW(·| Y0) random variable has a distribution on [0, 1], but is not necessarily uniformly distributed. Using equation 12, the conditional distribution (given Y0) of the percentile is GW(H−1(s) | Y0). Take the pre-posterior expectation to obtain (13).
Equation (13) can be evaluated numerically or in some situations (e.g., both Ḡ and H are Gaussian) in “closed” form. In any case, H and ḠW(·| A) need to be computed only once, a considerable computational savings. Since H may be time-consuming to invert, for graphs it will be convenient to let s = H(t) and plot (H(t),G(t)) with t ranging over a suitably broad interval and evaluated on a sufficiently fine mesh.
The expected percentile is,
| (14) |
and if W = A = H (spiked is also null)
Gaussian W, A and H
For the Gaussian sampling distribution with a variance that does not depend on , we can obtain closed form when all of (W,A,H) are also Gaussian, but not necessarily the same Gaussian. In this case, all marginal and conditional distributions are Gaussian. If θ ~ N(μ, τ2) and [Y | θ] ~ N(θ, σ2), then [θ | Y] ~ N(μ + (1 − B)(Y − μ), (1 − B) σ2) with B = σ2/(σ2 + τ2) and the marginal distribution of Y is N(μ, σ2 + τ2) = N(μ, σ2/B) = N(μ, τ2/(1 − B)).
We use subscripts w, a, h to indicate each distribution and find,
| (15) |
Generally, , producing
Now, H−1(s) = τhΦ−1(s) + μh and so from (13),
| (16) |
For a frequentist evaluation of the (W,H) combination, set τa = 0 (Ba = 1) (a point mass at μa).
3.3 Pre-posterior distribution of P* (Y0)
Theorem 2
Let tγ= H−1(γ). Then,
| (17) |
Proof
To compute P*(Y0), we need to find pr(θ0 > tγ| Y0) (for the spiked SNP) and do the same for all of the null SNPs. Then, rank the probability for the spiked SNP relative to all the null SNPs. Thus, we need to compare GW(tγ| Y0) = GW(H−1(γ) | Y0) to the collection of the K posteriors for the null SNPs. Equivalently, we need to count as follows for finite K (omitting dependence on W),
| (18) |
where the H(tγ| Yk) are iid according to the distribution induced by FH(y). To see that (18) is the correct accounting, note that we want to rank the right-hand tail probabilities and when H > GW, the right-hand tail is smaller.
We have that pr(H(t | Yk) > u) = S(u | t) so, S(H(t | Yk) | t) ~ U(0, 1) and,
Equation 17 follows directly.
Note that if W = A = H, then S(GH(tγ| Y0) | tγ) ~ U(0, 1) and so P*(Y0) ~ U(0, 1).
Gaussian W, A and H
We use the notation as in section 3.2 and first find S. Under H (dropping the subscript h until the end) and letting Zu = Φ−1(u),
For this to be > u, we need
But,
So,
| (19) |
Use (15) and (19) to evaluate (17).
In this all-Gaussian situation, if W = H and , then the (apparently) optimal P*(Y0) percentiles (those based on the working model) don’t depend on γ and are equal to the P̂(Y).
Evaluation
To compute (17) we need to find S and then sample Y0 from FA(y). Getting S(u | t) may require simulation for a target tγ and over a grid of u-values, but it is a one-time computation for each scenario. The same holds for sampling Y0 from FA. For efficient evaluation of a variety of scenarios, importance sampling can be used.
4 Performance Comparisons
We provide performance evaluation of the Z-score approach based on P̈ and P̈k to detecting a spiked SNP, comparisons of fully Bayesian ranking via (P̂, P̂k) to the Z-score approach, and a comparison of “frequentist-Bayes” based on a flat prior and (P̂, P̂k) to (P̈, P̈k). These provide a proof of concept and a basis for additional studies that include and investigate performance of candidate approaches in with data-based priors.
4.1 Performance of z-score-based percentiles
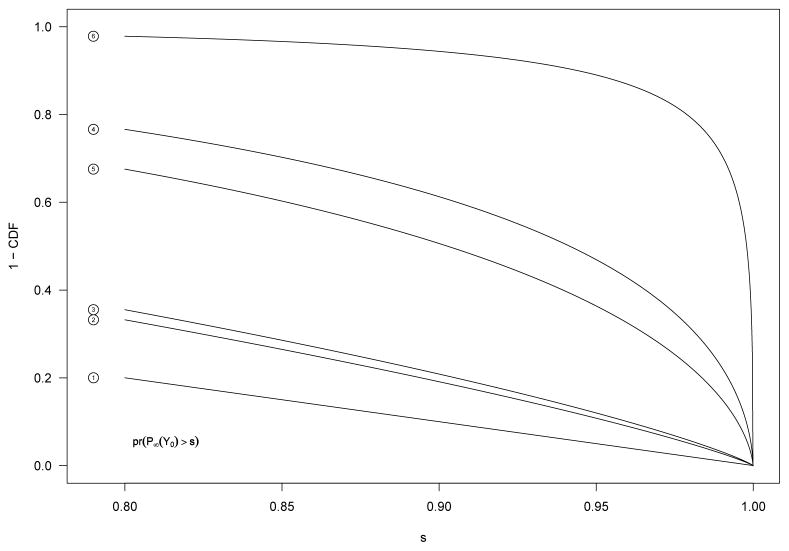
We compute the performance for Z-score based percentiles when θk ≡ 0 for various values of θ. We assume that for the null SNPs, H̄ = N(0, 1), and that the pre-posterior sampling distribution for the spiked SNP is N(θ/σ, 1) or equivalently N(θ, σ2). Table 1 contains scenarios for which the tail functions (from s = 0.80 to 1.00) are plotted in Figure 1. In this example, the probabilities that the percentile for the spiked SNP exceeds that for a null SNP (E(P̈)) are 0.50, 0.61, 0.63, 0.82, and 0.98, respectively.
Table 1.
Scenarios used for the evaluation and comparison of the performances of the ranking procedures described in the text. The performance evaluations illustrated in the manuscript figures are based on these parameters.
| Scenario | OR | θ | 1/σ | mean |
|---|---|---|---|---|
| 1 | 1.00 | 0 | 5.44 | 0 |
| 2 | 1.10 | 0.095 | 4.28 | 0.41 |
| 3 | 1.25 | 0.223 | 2.11 | 0.47 |
| 4 | 1.50 | 0.405 | 3.87 | 1.57 |
| 5 | 1.75 | 0.560 | 2.32 | 1.30 |
| 6 | 2.00 | 0.693 | 4.15 | 2.88 |
Fig. 1.
The tail distribution functions for P̈ (with rankings based on the ordinary z-statistics) for the six scenarios as outlined in Table 1. The probabilities that the percentile for the spiked SNP exceeds that for a null SNP (E(P̈)) are 0.50, 0.61, 0.63, 0.82, and 0.98, respectively.
4.2 Comparisons for Gaussian θ with a common sampling variance
For these comparisons, generically θ ~N(μ, τ2), [Y | θ] ~ N(θ, σ2) with the appropriate subscripts on (μ, τ2, σ2) to denote null SNPs or the spiked SNP. For the null SNPs, μ = 0 and we study performance as a function of μ for the spiked SNP.
The Z-score approach
The generic Z-score is Z = Y/σ2 and so the marginal distribution is N(μ/σ2, 1/B) (recall that τ2 = σ2(1 − B)/B). This distribution is equivalent to using in comparisons. For the null SNPs, μ = 0.
Fully Bayes
Using the fully Bayes approach, the marginal posterior is the prior and so is .
Comparisons
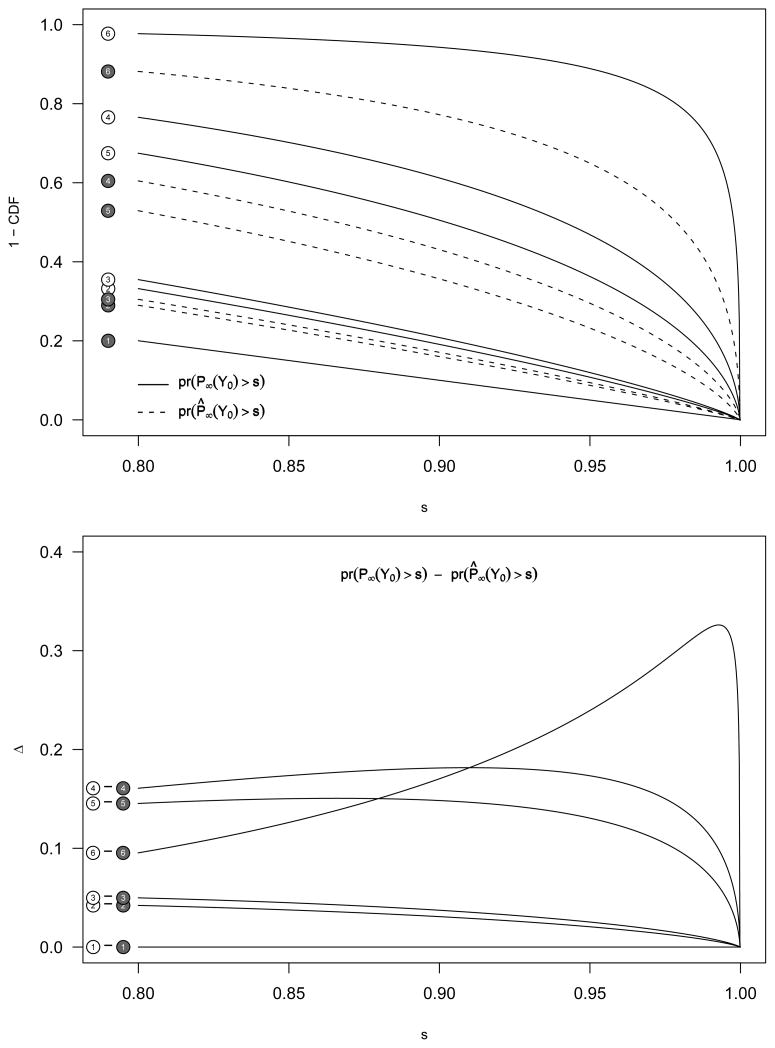
The pre-posterior distributions for the Z-score and fully Bayesian approaches are Gaussian with the same mean. Therefore, comparison of P̈k with P̂k depends on the variance. The variance for the Bayesian approach is (1 − B) times that for the Z-score approach, and so the Bayesian approach will out-perform the Z approach in that for μ > 0 the distribution of P̂ will be stochastically larger than that of P̈. This is no surprise, since P̂ takes advantage of the correct Bayesian structure. We illustrate this point in Figure 2.
Fig. 2.
The tail distribution functions for P̈ (z-score based, solid line) and P̂k (Bayes, dashed line) based on the six scenarios as outlined in Table 1. In this example, we assumed τ2 = 1, and the working prior (W) being equal to the actual prior (A). Thus, if the prior is approximately correct, the Bayesian ranking procedure will produce better results than the Z-score based ranking procedure.
Frequentist analysis
We investigate performance of the foregoing working analyses for fixed values of θ. We assume that θ1 = θ2 = … = θK = 0 and study performance for a fixed θ for the spiked SNP.
Z-score
Z = Y/σ2 and so the marginal distribution is N(θ/σ2, 1) This distribution is equivalent to using N(θ, σ2) in comparisons. For the null SNPs, θ = 0.
Bayes
Using the fully Bayes approach as the working model, the pre-posterior for a fixed θ is,
For the null SNPs, θ = μ = 0 and the mean is 0.
Comparison
The variance for the Bayesian approach is (1 − B)(2 − B) times that for the Z-score approach. However, the mean for the Bayesian approach is biased away from θ by B(μ − θ). Therefore, the “frequentist performance” (performance for specific θ-values) will be better than the Z-score approach for θ near μ and less good for θ far from μ. Of course, “on average” over the prior for θ, Bayes performance dominates.
4.3 Z-score and frequentist/Bayes with constant sampling variance
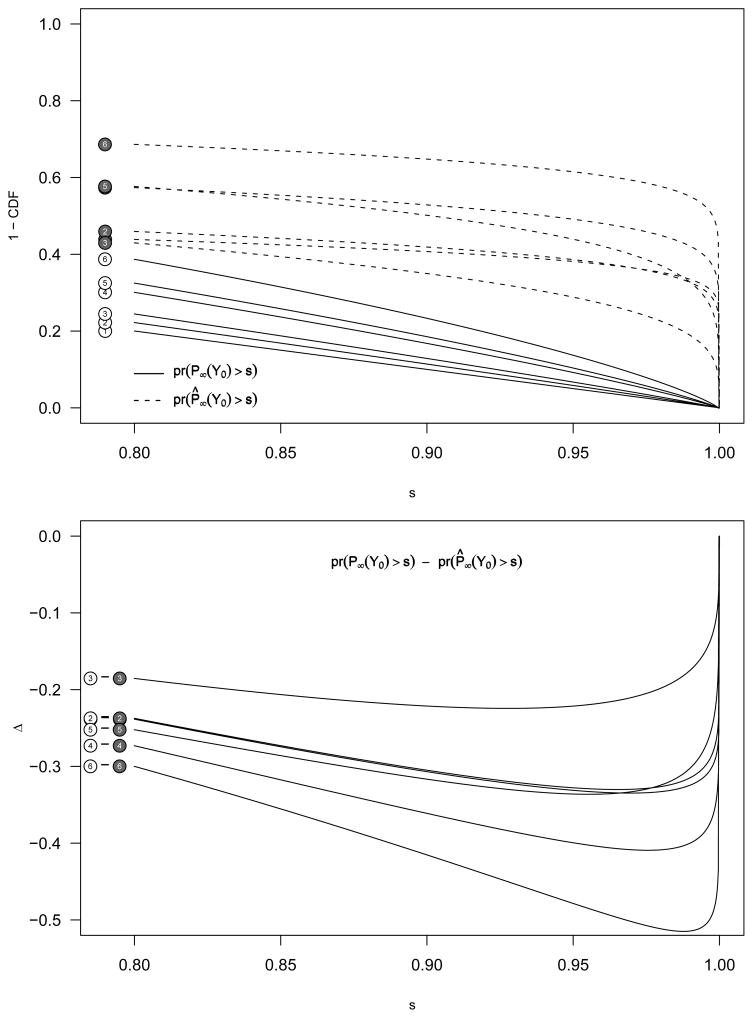
Here, the working model for the Bayesian analysis is flat-prior, producing [θ | Y] = N(Y, σ2). Conditioning on θ and mixing on Y produces N(θ, 2σ2) and is inferior to the Z-score approach which produces N(θ, σ2). Mixing on both Y and θ produces , with a variance (1+B) times that for the Z-score approach. Thus, frequentist/Bayes is never better than the Z-score approach, providing another example where use of an incorrect working model fails to achieve the Bayes advantage. We illustrate this point in Figure 3.
Fig. 3.
The tail distribution functions for P̈ (z-score based, solid line) and P̂k (“frequentist” Bayes, dashed line) based on the six scenarios as outlined in Table 1. If the truth is a fixed θ (Table 1) and the Bayesian analyst uses a very incorrect prior (τ2 = ∞ rather than τ2 = 0), there is a loss in performance compared to the frequentist approach using Zscores: the null SNPs produce a N(0, 2σ2) distribution, and the spiked a N(θ, 2σ2) marginal posterior.
5 Trade-offs when Simulations are Needed
The K = ∞ mathematical representations provide an excellent approximation to performance for large K so long as s is not too close to 1. However, simulation will still be needed if, for example H or other distributions are not mathematically evaluable and the number of draws (R) from the distribution being estimated needs to be specified. If one wants to have a good approximation for an extreme percentile, R may be sufficiently large that there is only a modest savings in computations and a standard simulation. As is the case in most statistical contexts, “there is no free lunch.” However, as shown below, there may be a reduced-price lunch.
This need for a large R will is most apparent if one wants to estimate the far right-hand tail of the distribution of the percentile for the spiked SNP. Accuracy for this requires an accurate estimate of the right-hand tail of the relevant distribution for the null SNPs. To see this, consider estimating an extreme percentile of a U(0,1) distribution. This case is sufficient for the ranking context because the probability integral transform for the null SNPs can be used to produce the distribution. In a simulation of size R, the variance of the empirical sth percentile is approximately s(1 −s)/R and the squared-coefficient of variation for the right-tail is s/{(1 − s)R} ≈ 1/{(1 − s)R} for s near 1. To control relative variation we require that this expression be no larger than V, producing, R = 1/{(1−s)V }. Now, consider the goal of identifying the top N SNPs so (1−s) = N/(K+1) and R = (K +1)/(NV). With V = 10−4, a relative standard deviation of 0.01, and the goal of identifying the top N = 100 SNPs, R = 100(K + 1) and the computational equivalent of 100 simulations, each of size (K + 1) are needed. This is still a considerable savings over a full simulation, but a smaller V or smaller N will generate the need for an even larger initial simulation.
6 Discussion
Simulation is a very effective tool for assessing the properties of statistical procedures. However, in many situations use of large-sample statistical properties can facilitate evaluations. In this context, we have shown how to use large-sample convergence of empirical distribution functions to speed up assessments of ranking procedures in some large sample size situations. This approach facilitates rapid evaluation and comparison of approaches for a broad array of scenarios and we provide a few examples as a proof of concept. More extensive comparisons and inclusion of the P* approach will build on the current results. In structuring the assessments, we have developed integral representations of the stochastic distribution for the percentile rank of a single “spiked” SNP in competition with a large number of “null” SNPs. In some settings the integral representations will require some numerical evaluation, but these are one-time operations. Generalizations include developing representations for the maximum percentile of more than one spiked SNP.
The large K context is ideal for the use of flexible, empirical priors including the non-parametric maximum likelihood (another area in which Hans has worked) or a smooth version of it. Use of such priors should be very efficient and robust and our integral representations will facilitate rapid assessments.
Though an attractive approach, the approximations are not a panacea. For extreme percentiles they provide a lower bound for the tail of the distribution of the spiked SNP and this bound can be very close to zero. Furthermore, as shown in section 5, if simulations are needed to evaluate components of the mathematical expressions, they may not need to be very large. Hans for your many contributions to statistics and to science, to your educational and mentoring success, and to your continued contributions we close with “dank u zeer.”
Acknowledgments
Support provided by grants R01 DK061662, from the U. S. NIH, National Institute of Diabetes, Digestive and Kidney Diseases, R01 HL090577 from the U. S. NIH, National Heart, Lung, and Blood Institute, and a CTSA grant to the Johns Hopkins Medical Institutions.
Appendix: Trend Test Z-Scores
We outline the score test approach from Ruczinski et al. (2009) that accommodates uncertain (“fuzzy”) genotype calls. We consider only the dominant model and so use g ∈ {0, 1} to indicate the true genotype and in the trend test use scores dg = g. With X the binary disease status indicator, define the risk relation, πg = pr(X = 1 | g) = H(μ + θdg) and for a specific SNP (index suppressed) let:
Xi = the disease status indicator for the e ith participant, i = 1, …, n.
gi = the true genotype.
Gi = the random variable taking on values gi (0 or 1).
wi = pr(Gi = 1), the working probabilities. If the genotype is known without error, then wi = gi.
w = the n-dimensional vector of the wi.
To test the hypothesis of no genotype/phenotype association, H0: π0 = π1 = π equivalently, H0: θ = 0, use the statistic:
| (20) |
with and nVar(w) = Σi(wi − w̄)2 = w̄(1 − w̄) − Σi wi(1 − wi).
Note that under H0, Z(w) ~ N(0, 1), showing that the null distribution is correct irrespective of the working probabilities w. Using a local alternative (θ = θ0 close to 0), and a logistic form for π we obtain:
with
| (21) |
Squaring the Z-statistic produces a one degree of freedom, chi-square statistic with non-centrality λ = m2(θ0,w)/2.
Footnotes
No conflict of interest: The authors have declared no conflict of interest.
References
- Carlin BP, Louis TA. Bayesian Methods for Data Analysis. 3. Chapman and Hall/CRC Press; Boca Raton, FL: 2009. [Google Scholar]
- Goeman JJ, van de Geer SA, van Houwelingen HC. Testing against a high dimensional alternative. Journal of the Royal Statistical Society: Series B. 2006;68:477–493. [Google Scholar]
- Heidema AG, Feskens EJM, Doevendans PAFM, Ruven HJT, van Houwelingen HC, Mariman ECM, Boer JMA. Analysis of multiple snps in genetic association studies: comparison of three multi-locus methods to prioritize and select SNPs. Genetic Epidemiology. 2007;31:910–921. doi: 10.1002/gepi.20251. [DOI] [PubMed] [Google Scholar]
- Houwing-Duistermaat JJ, Uh HW, van Houwelingen HC. A new score statistic to test for association given linkage in affected sibling pair-control designs. BMC Proceedings. 2007;1(Suppl 1):S39. doi: 10.1186/1753-6561-1-s1-s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrec JJP, Putter H, Houwing-Duistermaat JJ, van Houwelingen HC. Influence of genotyping error in linkage mapping for complex traits–an analytic study. BMC Genetics. 2008;9:57. doi: 10.1186/1471-2156-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Louis TA, Paddock SM, Ridgeway G. Loss function based ranking in two-stage, hierarchical models. Bayesian Analysis. 2006;1:915–946. doi: 10.1214/06-BA130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Brooks LD, Collins FS. A hapmap harvest of insights into the genetics of common disease. Journal of Clinical Investigation. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan JM, Susser E, King MC. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- Ruczinski I, Li Q, Carvalho B, Fallin M, Irizarry RA, Louis TA. Association tests that accommodate genotyping errors. Johns Hopkins University, Dept of Biostatistics Working Papers; 2009. Working Paper 181. [Google Scholar]
- Schwender H, Ickstadt K. Empirical Bayes analysis of single nucleotide polymorphisms. BMC Bioinformatics. 2008;9:144. doi: 10.1186/1471-2105-9-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uh HW, Mertens BJ, van der Wijk HJ, Putter H, van Houwelingen HC, Houwing-Duistermaat JJ. Model selection based on logistic regression in a highly correlated candidate gene region. BMC Proceedings. 2007;1(Suppl 1):S114. doi: 10.1186/1753-6561-1-s1-s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet ic Epidemiology. 2008a;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- Wakefield J. Reporting and interpretation in genome-wide association studies. International Journal of Epidemiology. 2008b;37:641–653. doi: 10.1093/ije/dym257. [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Johnson WO, Utts JM. A Bayesian perspective on the Bonferroni adjustment. Biometrika. 1997;84:419–427. [Google Scholar]