Abstract
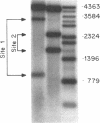
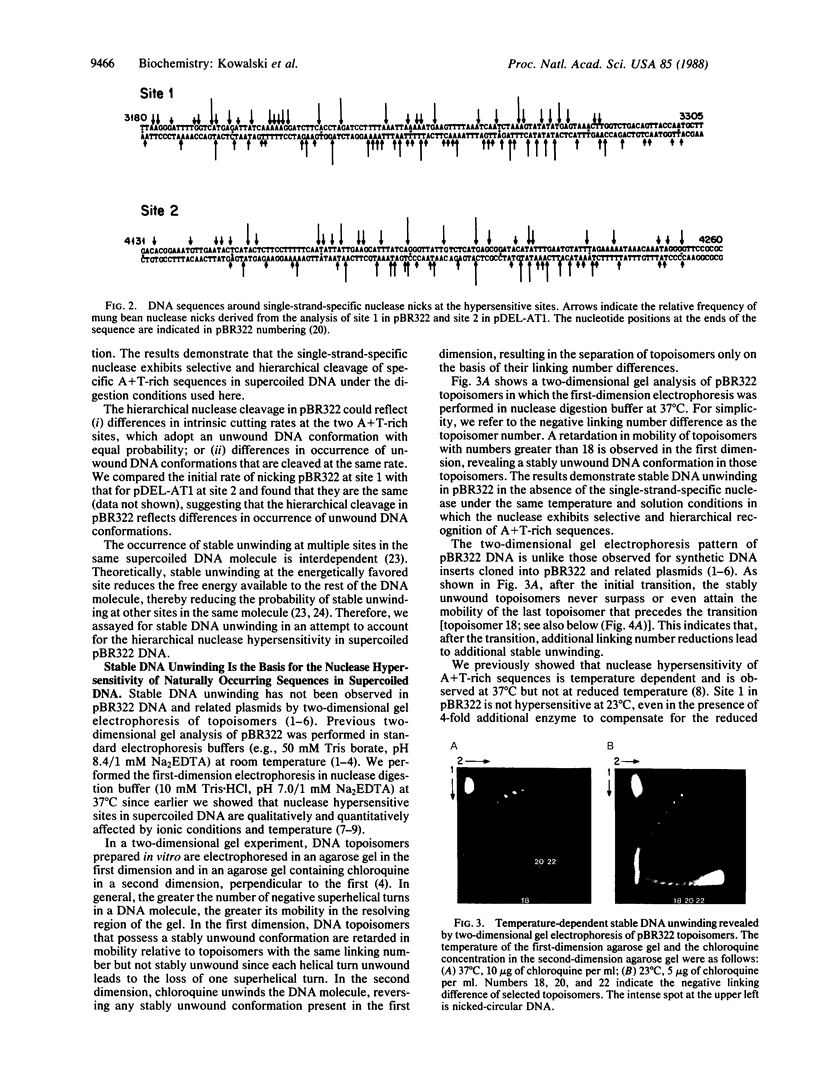
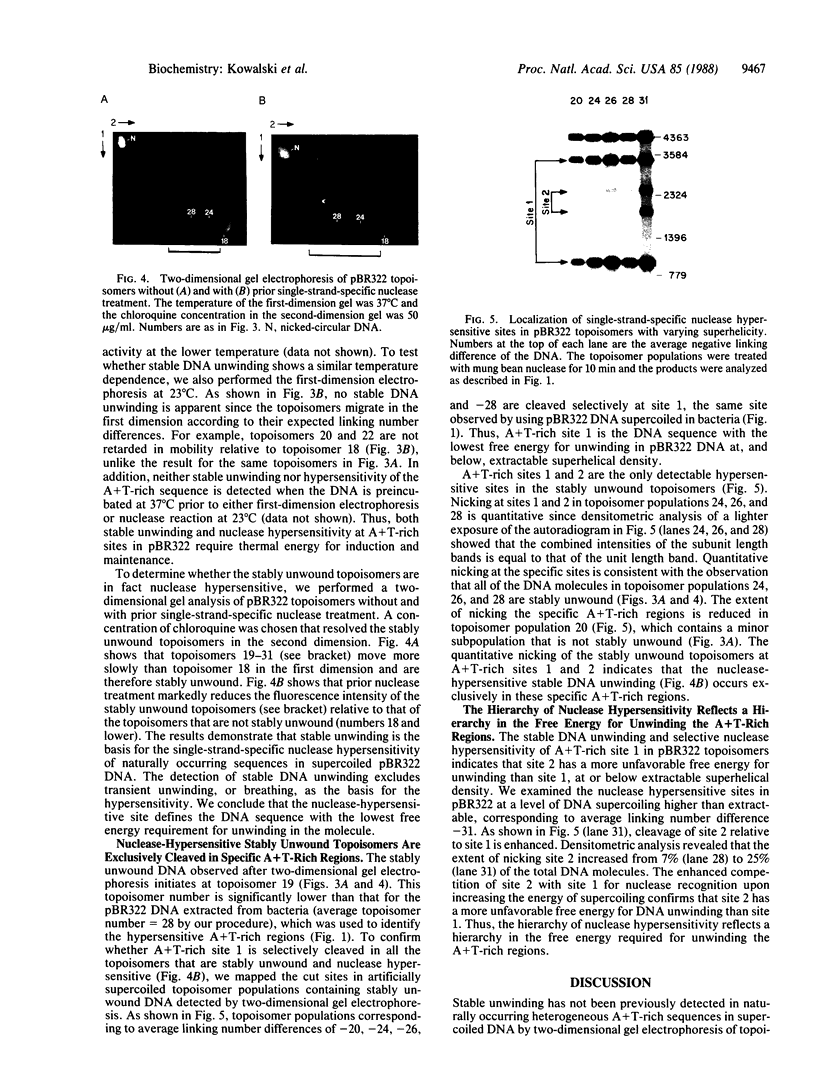
A long A+T-rich sequence in supercoiled pBR322 DNA is hypersensitive to single-strand-specific nucleases at 37 degrees C but not at reduced temperature. The basis for the nuclease hypersensitivity is stable DNA unwinding as revealed by (i) the same temperature dependence for hypersensitivity and for stable unwinding of plasmid topoisomers after two-dimensional gel electrophoresis, (ii) preferential nuclease digestion of stably unwound topoisomers, and (iii) quantitative nicking of stably unwound topoisomers in the A+T-rich region. Nuclease hypersensitivity of A+T-rich sequences is hierarchical, and either deletion of the primary site or a sufficient increase in the free energy of supercoiling leads to enhanced nicking at an alternative A+T-rich site. The hierarchy of nuclease hypersensitivity reflects a hierarchy in the free energy required for unwinding naturally occurring sequences in supercoiled DNA. This finding, along with the known hypersensitivity of replication origins and transcriptional regulatory regions, has important implications for using single-strand-specific nucleases in DNA structure-function studies.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Bond P. J., Chandrasekaran R. Visualization of an unwound DNA duplex. Nature. 1980 Oct 9;287(5782):561–563. doi: 10.1038/287561a0. [DOI] [PubMed] [Google Scholar]
- Benham C. J. Theoretical analysis of competitive conformational transitions in torsionally stressed DNA. J Mol Biol. 1981 Jul 25;150(1):43–68. doi: 10.1016/0022-2836(81)90324-7. [DOI] [PubMed] [Google Scholar]
- Benham C. J. Torsional stress and local denaturation in supercoiled DNA. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3870–3874. doi: 10.1073/pnas.76.8.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Ellison M. J., Ho P. S., Rich A. Competitive behavior of multiple, discrete B-Z transitions in supercoiled DNA. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6342–6346. doi: 10.1073/pnas.83.17.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D. Changes in site specificity of single-strand-specific endonucleases on supercoiled PM2 DNA with temperature and ionic environment. Nucleic Acids Res. 1984 Sep 25;12(18):7071–7086. doi: 10.1093/nar/12.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D., Kroeker W. D., Laskowski M., Sr Mung bean nuclease I. Physical, chemical, and catalytic properties. Biochemistry. 1976 Oct 5;15(20):4457–4463. doi: 10.1021/bi00665a019. [DOI] [PubMed] [Google Scholar]
- Kowalski D., Sanford J. P. Action of mung bean nuclease on supercoiled PM2 DNA. J Biol Chem. 1982 Jul 10;257(13):7820–7825. [PubMed] [Google Scholar]
- Lee F. S., Bauer W. R. Temperature dependence of the gel electrophoretic mobility of superhelical DNA. Nucleic Acids Res. 1985 Mar 11;13(5):1665–1682. doi: 10.1093/nar/13.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. A pH-dependent structural transition in the homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1985 Oct;3(2):327–338. doi: 10.1080/07391102.1985.10508420. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Sawadaishi K., Miura K., Ohtsuka E., Ueda T., Shinriki N., Ishizaki K. Structure- and sequence-specificity of ozone degradation of supercoiled plasmid DNA. Nucleic Acids Res. 1986 Feb 11;14(3):1159–1169. doi: 10.1093/nar/14.3.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheflin L. G., Kowalski D. Altered DNA conformations detected by mung bean nuclease occur in promoter and terminator regions of supercoiled pBR322 DNA. Nucleic Acids Res. 1985 Sep 11;13(17):6137–6154. doi: 10.1093/nar/13.17.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheflin L. G., Kowalski D. Mung bean nuclease cleavage of a dA + dT-rich sequence or an inverted repeat sequence in supercoiled PM2 DNA depends on ionic environment. Nucleic Acids Res. 1984 Sep 25;12(18):7087–7104. doi: 10.1093/nar/12.18.7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüber D., Bujard H. Organization of transcriptional signals in plasmids pBR322 and pACYC184. Proc Natl Acad Sci U S A. 1981 Jan;78(1):167–171. doi: 10.1073/pnas.78.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tricoli J. V., Kowalski D. Topoisomerase I from chicken erythrocytes: purification, characterization, and detection by a deoxyribonucleic acid binding assay. Biochemistry. 1983 Apr 12;22(8):2025–2031. doi: 10.1021/bi00277a045. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell. 1988 Feb 26;52(4):559–567. doi: 10.1016/0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. Yeast regulatory sequences preferentially adopt a non-B conformation in supercoiled DNA. Nucleic Acids Res. 1987 Jun 11;15(11):4467–4480. doi: 10.1093/nar/15.11.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Yagil G., Sussman J. L. Structural models for non-helical DNA. EMBO J. 1986 Jul;5(7):1719–1725. doi: 10.1002/j.1460-2075.1986.tb04416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]