Abstract
Iron-sulphur clusters are present in more than 200 different types of enzymes or proteins and constitute one of the most ancient, ubiquitous and structurally diverse classes of biological prosthetic groups. Hence the process of iron-sulphur biosynthesis is essential to almost all forms of life and is remarkably conserved in prokaryotic and eukaryotic organisms. Three distinct types of iron-sulphur cluster assembly machinery have been established in bacteria, termed the NIF, ISC and SUF systems, and in each case the overall mechanism involves cysteine desulphurase-mediated assembly of transient clusters on scaffold proteins and subsequent transfer of preformed clusters to apo proteins. A molecular level understanding of the complex processes of iron-sulphur cluster assembly and transfer is now beginning to emerge from the combination of in vivo and in vitro approaches. This review highlights recent developments in understanding the mechanism of iron-sulphur cluster assembly and transfer involving the ubiquitous U-type scaffold proteins and the potential roles of accessory proteins such as Nfu proteins and monothiol glutaredoxins in the assembly, storage or transfer of iron-sulphur clusters.
Keywords: iron-sulphur clusters, scaffolds, cysteine desulphurase, chaperones, glutaredoxins
Introduction
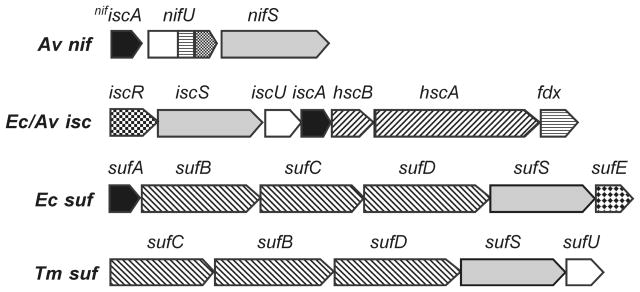
Clusters involving iron and inorganic sulphide that are attached to the polypeptide primarily via cysteinate iron ligation constitute one of the most ubiquitous and structurally and functionally diverse classes of biological prosthetic groups, for recent reviews see Refs [1]. In accord with the hypothesis that Fe-S clusters are one of the most ancient types of prosthetic group, the biosynthetic machineries for Fe-S cluster biogenesis are widely conserved in all three kingdoms of life, for recent reviews see Refs [2–5]. Following the pioneering work of Dennis Dean on the function of the nitrogen fixation (nif) genes, three distinct types of biosynthetic machinery, NIF, ISC and SUF have been emerged in bacteria, archaea and eukaryotic organelles, based initially on biochemical evidence and the organization of genes in bacterial operons [6;7], see Fig. 1. The NIF system is generally specific for the maturation of Fe-S proteins in nitrogen fixing organisms such as Azotobacter vinelandii (Av). The ISC system is the primary system for general Fe-S cluster biosynthesis in bacteria such as Escherichia coli and Av [6;8]. Moreover, along with a few additional components, the ISC system constitutes the eukaryotic mitochondrial machinery for Fe-S cluster biogenesis [2;3]. This is in accord with the hypothesis that the mitochondrial ISC system was inherited from the bacterial endosymbiotic ancestor of this organelle. The third bacterial assembly system, termed SUF, plays a similar general role as ISC in many bacteria, but is operative only under conditions of iron limitation or oxidative stress [7;9]. It is therefore not surprising that the bacterial SUF system also forms the basis of the Fe-S cluster biogenesis machinery in plant chloroplasts [2], an O2-producing organelle that was most likely inherited from the cyanobacterial ancestor of plastids. The SUF system also appears to be the sole system for Fe-S cluster biogenesis in archaea and cyanobacteria, as well as many gram-positive, pathogenic and thermophilic bacteria.
Figure 1. Organization of genes in selected bacterial nif, isc, and suf operons.
Av,Azotobacter vinelandii; Ec, Escherichia coli; Tm, Thermotoga maritima.
The central theme of all three Fe-S cluster assembly systems is cysteine desulphurase-mediated assembly of transient [Fe2S2]2+ and [Fe4S4]2+ clusters on scaffold proteins and subsequent transfer of preformed clusters to the apo forms of acceptor proteins [10;11]. All involve a cysteine desulphurase (NifS, IscS, SufS) and at least one type of potential scaffold protein capable of assembling both [Fe2S2]2+ and [Fe4S4]2+ clusters, i.e. U-type (NifU, IscU, SufU) [10;11] and/or A-type (NifIscA, IscA, SufA) [12;13]. Cysteine desulphurases are homodimeric, pyridoxal phosphate-dependent enzymes that catalyze the conversion of L-cysteine to L-alanine and a mobile enzyme-bound cysteine persulphide [14] which can transfer S0 directly, or via the SufE sulphurtransferase [9], to the active-site cysteines of scaffold proteins [15;16]. The nature of the immediate Fe donor for cluster assembly has yet to be fully resolved. Frataxin and the bacterial homolog, CyaY, are viable candidates for the Fe donor to U-type scaffold proteins based on in vivo and in vitro studies [3;17]. A role for A-type scaffold proteins as the Fe donor to U-type scaffold proteins, rather than an alternative scaffold protein for cluster assembly under specific cellular conditions or for maturation of a specific subset of Fe-S proteins [12;13;18], has also been proposed based on in vitro evidence for high affinity binding of a monomeric S = 3/2 ferric center that can reductively released by cysteine and used as the Fe source for cluster assembly on U-type scaffold proteins [19] and references therein. Consequently, although there is no disagreement concerning the role of U-type proteins as primary scaffold proteins for cysteine desulphurase-mediated assembly of Fe-S clusters, there is as yet no consensus concerning the role of the ubiquitous A-type proteins in Fe-S cluster biogenesis. In large part, this is because the effects of inactivation of the genes encoding NifIscA, IscA/Isa, or SufA proteins on cellular cluster assembly are generally minor and often manifest only under oxidative stress conditions, whereas deletion or disruption of the genes encoding for U-type scaffold genes are usually lethal [20]. The mechanism of cluster assembly on U-type scaffold proteins and subsequent cluster transfer to acceptor proteins are the focus of this review.
The nif, isc, and suf operons also encode for additional proteins or protein domains that play important roles in Fe-S cluster biosynthesis. In the NIF system, NifU is a modular protein, see Fig. 1, containing a N-terminal U-type scaffold domain, a central Fd-like domain containing a redox-active [Fe2S2]2+,+ cluster of unknown function, and a C-terminal domain with a CXXC motif that provided the basis for identifying a separate class of Fe-S cluster assembly proteins, known as Nfu proteins. The ISC system contains a regulatory protein, IscR, two heat-shock-cognate proteins, HscA and HscB, and a [Fe2S2] ferredoxin (Fdx). IscR serves a transcriptional repressor of the entire isc operon in its [Fe2S2]2+ cluster-bound form [21], and as a global regulator of numerous Fe-S proteins and an activator of both the isc and suf operons in response to oxidative stress [22]. HscA/HscB are essential molecular chaperones that are closely related to DnaK and DnaJ, molecular co-chaperones that have intrinsic ATPase activity and participate in non-specific protein folding and renaturation [6]. However, IscU appears to be the sole substrate for HscA/HscB [23], indicating a specific role Fe-S protein biogenesis using the ISC system. Fdx contains a redox-active [Fe2S2]2+,+ cluster and has an essential but ill-defined redox function in assembly of Fe-S cluster on the IscU scaffold protein. The SUF system, contains SufB, SufC and SufD which are essential for Fe-S cluster formation or repair in bacteria under oxidative stress and Fe limitation conditions [9]. SufC is an atypical cytoplasmic ABC-ATPase [24] which forms a soluble complex with SufB and SufD and has enhanced ATPase activity in the presence of SufB [25]. The function of the SufBCD complex is still unclear, but possibilities include providing energy for mobilizing Fe destined for Fe-S cluster biosynthesis or for the assembly or transfer of clusters produced by the SUF machinery. In addition, the recent demonstration of an interaction between SufB and SufE and for SufSE-mediated assembly of a [Fe4S4] cluster on SufB [26] has raised the possibility that SufB could function as a primary scaffold protein for [Fe4S4] cluster assembly in the bacterial SUF system.
Cluster assembly on U-type scaffold proteins
U-type scaffold proteins (N-terminal domain of NifU, IscU/Isu and SufU) constitute the primary site for Fe-S cluster assembly in the NIF and ISC systems and are components of many bacterial or archaeal SUF systems, see Fig. 1. All contain three conserved cysteines (Cys37, Cys63 and Cys 106 in Av IscU) that are required for cluster incorporation, see Fig. 2, but the primary sequences differ in three important respects [4]. SufUs have 18–20 residue insertion between the second and third cysteine residues and a lysine rather than a histidine immediately before the third cysteine and only IscUs have the LPPVK motif that is required for interaction with HscA, see Fig. 2. Structural information on U-type scaffold proteins is currently limited to NMR studies of the apo form T. maritima SufU [27] and the Zn-bound form of Haemophilus influenzae IscU [28] and a 2.3 Ǻ resolution crystal structure of the Zn-bound form of Streptococcus pyogenes SufU [29]. The initial NMR studies of apo T. maritima SufU indicated a molten globule-like state with reasonably well defined of secondary structure, but with fluxional tertiary structure and an undefined cluster binding site [27]. In contrast, the NMR structure of Zn-bound form of H. influenzae IscU shows flexibility in the N-terminus and C-terminal His-tag, but well ordered secondary and ternary structure for the core that includes a well-defined and solvent-exposed Zn binding site, involving the three conserved cysteine residues and the histidine immediately preceding the third cysteine residue. A similar structure was reported for the Zn-bound form for S. pyogenes SufU, except that the fourth ligand was a rigorously conserved aspartate (D39 in Av IscU) that is present in all IscU, NifU, SufU sequences, rather than the histidine which is not present in SufU. The structural studies suggest that Zn or Fe-S cluster binding is essential for well-ordered ternary structure and indicate a solvent-exposed Fe-S cluster binding site with the three conserved cysteine residues, the conserved aspartate one residue removed from the first cysteine, and the histidine (NifU and IscU) or lysine (SufU) immediately preceding the third cysteine as potential cluster ligands.
Figure 2. Primary sequence comparisons for U-type scaffold proteins.
Numbers refer to the conserved cysteines in the Azotobacter vinelandii IscU sequence. Fully conserved residues are shaded in black. The LPPVK sequences that are required for IscU interactions with HscA and the histidine (NifU and IscU) and lysine (SufU) residues in the cluster binding pocket are shaded in grey. Av,Azotobacter vinelandii; Ec, Escherichia coli; Hi, Haemophilus influenzae; At, Arabidopsis thaliana; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; Sp, Streptococcus pyogenes; Bs, Bacillus subtilis; Tm, Thermotoga maritima.
Spectroscopic characterization of the timecourse of in vitro cluster assembly on homodimeric Av IscU and the N-terminal domain of homodimeric Av NifU under steady-state conditions with catalytic IscS/NifS and excess Fe2+ and cysteine revealed sequential formation of stable forms containing one [Fe2S2]2+ cluster per dimer, two [Fe2S2]2+ clusters per dimer and one [Fe4S4]2+ cluster per dimer [11;30], see Fig. 3. The one [Fe2S2]2+ cluster-bound form is particularly stable and resistant to iron chelators such as EDTA and the two [Fe2S2]2+ cluster-bound form is clearly a viable intermediate on the pathway to assembling a [Fe4S4]2+ cluster. The stoichiometry and absence of significant [Fe2S2]2+ cluster degradation suggested a mechanism for [Fe4S4]2+ cluster formation at the subunit interface involving two-electron reductive coupling of two adjacent [Fe2S2]2+ clusters. However, formation of the [Fe4S4]2+ cluster occurred very slowly over period of several hours and the source of reducing equivalents was unclear. Subsequently, immediate and quantitative reductive coupling was observed using dithionite as the electron donor and partial reductive coupling using stoichiometric reduced Isc Fdx, suggesting a possible role as the physiological electron donor [31]. Our interpretation is that the Isc Fdx redox potential is poise to induce partial reductive coupling in order to make IscU a more versatile and environmentally responsive scaffold protein capable of providing clusters for the maturation of both [Fe2S2]2+ and [Fe4S4]2+ cluster-containing proteins. Although the reductive coupling is not a reversible electrochemical process, spectroscopic studies showed that O2 exposure converts the [Fe4S4]2+ cluster-bound form back to the one [Fe2S2]2+ cluster-bound form of IscU, see Fig. 3. Hence, the ability of IscU to accommodate either [Fe2S2]2+ or [Fe4S4]2+ clusters in response to cellular redox status and/or O2 levels may provide an effective way to populate appropriately cluster-loaded forms of IscU for maturation of different types of Fe-S proteins under different cellular conditions. In the absence of an exogenous one-electron donor, the two electrons required for slow reductive coupling are proposed to originate from disulphide formation involving two of the cysteines that are released via the cluster conversion, see Fig. 3.
Figure 3.
Working hypothesis for the mechanism of IscS-mediated [Fe2S2]2+ and [Fe4S4]2+ cluster assembly on IscU and transfer to the apo forms of acceptor proteins.
Cysteine mutagenesis results coupled with resonance Raman and Mossbauer properties of the one [Fe2S2]2+ cluster-bound form of IscU and the N-terminal domain of NifU indicate cluster ligation by all three of the conserved cysteine residues and one non-cysteinyl ligand [11]. Furthermore, structural and mutagenesis data implicate the rigorously conserved aspartate, (D39 in Av IscU) as the non-cysteinyl ligand. Substitution of this aspartate with alanine results in major changes in the resonance Raman spectrum of the [Fe2S2]2+ center in both the N-terminal domain of NifU [32] and in IscU (H. Gao and M. K. Johnson, unpublished results) and the crystal structure of Zn-bound S. pyogenes SufU places this residue in the cluster binding pocket [29]. However, whereas substitution of a cluster-ligating residue by alanine generally destabilizes Fe-S clusters, the D39A substitution stabilizes the cluster in IscU and the N-terminal domain of NifU as judged by the ability to purify the one [Fe2S2]2+ cluster-bound form with the cluster intact [10] and the dramatic decrease in the rates of cluster transfer to acceptor proteins [33]. Our working hypothesis based on the available structural and spectroscopic data is that the [Fe2S2]2+ cluster is stabilized in these variants by ligation of the histidine immediately preceding the third conserved cysteine in NifU and IscU. Hence the role of the aspartate ligand appears to be in labilizing the [Fe2S2]2+ cluster for transfer to an acceptor protein. A similar conclusion is likely for the [Fe4S4]2+ cluster assembled at the subunit interface of IscU, since the resonance Raman Fe-S stretching frequencies are best interpreted in terms of one non-cysteinyl ligand [11] and the D39A mutation perturbs the resonance Raman spectrum, stabilizes the cluster and inhibits cluster transfer to aconitase [34]. Our current working hypothesis for the organization, ligation and mechanism of assembly of Fe-S clusters on U-type scaffold proteins and the means of populating different cluster-loaded forms is schematically depicted in Fig. 3.
Cluster transfer from IscU
Genetic experiments have established that U-type scaffold proteins are involved in maturation of Fe-S proteins that require either [Fe2S2] or [Fe4S4] clusters for their biological activities [3;4;20]. In both cases, in vitro studies indicate that maturation involves intact cluster transfer from [Fe2S2]2+ or [Fe4S4]2+ cluster-loaded of the U-type scaffold protein to the apo-form of the acceptor protein. Intact [Fe2S2]2+ cluster transfer from IscU or SufU to apo IscFdx has been demonstrated by several research groups [35] and references therein, and the intact [Fe4S4]2+ cluster transfer from the N-terminal domain of NifU to apo nitrogenase Fe-protein and from IscU to apo aconitase has been demonstrated by the Johnson and Dean groups [30;34]. The crucial role of the conserved aspartate residue (Asp39 in Av IscU) in facilitating release of both types of cluster is evident by the dramatic decrease in the rates of cluster transfer when this residue is substituted with alanine [33;34]. Since cluster transfer reactions exhibit second order kinetics [35], we therefore propose that that they proceed by successive ligand exchange reactions after formation of a transient donor-acceptor complex, with the rate determining step involving the initial displacement of the coordinated aspartate by a cysteine residue on the acceptor protein.
Numerous genetic and biochemical studies have established an essential role for the HscA/HscB co-chaperones, and their eukaryotic homologs (Ssp1/Jac1 in yeast), in the efficient maturation of Fe-S proteins using the ISC assembly machinery [3–5]. However, their specific role in the biogenesis of Fe-S clusters was controversial with conflicting reports favoring a role in stabilizing clusters assembled on IscU, based on in vitro studies with the generic DnaK/DnaJ molecular chaperone system [36], or in facilitating cluster transfer from IscU to acceptor proteins based on in vivo 55Fe immunoprecipitation studies [37]. Our in vitro results using the IscU-specific HscA/HscB co-chaperone system provided direct evidence for a role in facilitating cluster transfer from IscU to acceptor proteins and showed for the first time that cluster transfer is an ATP-dependent reaction [35]. The presence of the HscA, HscB and MgATP, individually or together, was found to have no significant effect on the ability to assemble either [Fe2S2]2+ and [Fe4S4]2+ clusters on IscU in an IscS-mediated reaction. However, both HscA and HscB were found to bind independently to IscU in the vicinity of the [Fe2S2]2+ cluster, based on changes in the CD spectra of the [Fe2S2]2+ cluster. Moreover, HscA and HscB together were shown to enhance the rate of [Fe2S2]2+ cluster transfer from IscU to apo IscFdx approximately twenty-fold in an ATP dependent reaction, as judged by cluster transfer experiments monitored by CD and EPR. These results provide the basis for mechanistic proposals for coupling the HscA ATPase cycle with cluster release from [Fe2S2]-IscU [23;35]. Surprisingly, the presence of HscA/HscB/MgATP had no effect on the rate of [Fe4S4]2+ cluster transfer from IscU to apo aconitase [34], suggesting different conformations for the [Fe2S2]2+ or [Fe4S4]2+ cluster-bound forms of IscU, with only the former having the ability to interact productively with HscA.
Role of Nfu proteins in Fe-S cluster biogenesis
Nfu proteins constitute a ubiquitous class of proteins containing a redox-active CXXC motif that are not generally associated with specific bacterial operons and were originally identified as the C-terminal domain of NifU (Nfu indicates NifU-like protein). In systems containing U-type scaffold proteins, gene knockout studies indicate an important auxiliary role in Fe-S cluster biogenesis that is primarily evident under oxidative stress conditions [3;38;39]. In vitro studies have shown that that bacterial and mitochondrial Nfu proteins or domains can assemble [Fe4S4]2+ clusters, most likely ligated by the cysteines in the CXXC motif at the subunit interface, based on cluster stoichiometry, spectroscopic properties and cysteine mutagenesis studies [30;38;40]. Moreover, these clusters can be used for the assembly of [Fe4S4] centers in physiologically relevant acceptor proteins via intact cluster transfer. This was first demonstrated using NifU, when [Fe4S4]2+ clusters assembled on the C-terminal domain of Av NifU were found to be effective for activation of apo nitrogenase Fe protein [30]. More recently, we have investigated the properties of Av NfuA; a ubiquitous bacterial protein with an N-terminal domain that exhibits homology to an A-type scaffold protein, albeit without the three conserved cysteine residues, and a C-terminal domain Nfu-domain. Gene disruption experiments indicated that Av nfuA is required for optimal aconitase activity and is essential for growth under elevated oxygen concentrations [38]. Most interesting was the observation that the [Fe4S4]2+ cluster assembled on NfuA could be rapidly and quantitatively transferred to apo Av aconitase based on cluster transfer assays monitored by aconitase activity. The rate constant for cluster transfer was an order of magnitude greater than that determined using [Fe4S4]2+-IscU and occurred with 100% efficiency compared to ~33% efficiency for cluster transfer from [Fe4S4]2+-IscU. Since previous gene disruption studies have demonstrated an essential role for IscU in the general maturation of Fe-S proteins including aconitase [20], even under oxidative stress conditions, NfuA is proposed to be an intermediate [Fe4S4]2+ cluster carrier or chaperone for clusters assembled on IscU [38], see Fig. 3. However, in O2-rich organisms such as cyanobacteria and plant chloroplasts which utilize the SUF system without a U-type scaffold protein, the available evidence indicates that Nfu-type proteins function as de novo scaffold proteins in the maturation of both [Fe2S2]2+ and [Fe4S4]2+ cluster-containing proteins [41–43]
Role of monothiol glutaredoxins in Fe-S cluster biogenesis
Glutaredoxins (Grxs) are small proteins that normally function in the reduction of disulphide bridges or glutathionylated proteins. However, gene disruption studies in Saccharomyces cerevisiae showed that Grx5, a mitochondrial monothiol Grx with a CGFS active site, is involved with Fe-S cluster biosynthesis [37;44]. Yeast cells deleted for the grx5 gene were found to be more sensitive to oxidative stress, to accumulate free iron, and to have impaired mitochondrial Fe-S cluster biogenesis and respiratory growth [44]. Moreover, immunoprecipitation studies using 55Fe radiolabeled samples revealed build up of Fe on IscU implicating a role in mediating transfer of clusters preassembled on the IscU scaffold protein to acceptor proteins [37]. Monothiol Grxs with CGFS active sites are widespread among prokaryotes and eukaryotes [45], and the ability of prokaryotic, cytosolic and chloroplastic monothiol Grxs to rescue the defects of grx5 mutants in yeast suggests functional conservation among this class of proteins [46;47].
The most obvious role for monothiol Grxs in Fe-S cluster biogenesis lies in facilitating Fe-S cluster assembly or transfer by reducing internal disulphides or glutathionylated cysteines on scaffold proteins or apo forms of acceptor proteins. However, the recent discovery that a wide variety of recombinant monothiol Grxs can be purified containing [Fe2S2]2+ clusters under anaerobic conditions and that [Fe2S2]2+ clusters can be reconstituted on apo forms in the presence of glutathione [47;48], has raised the possibility that they may function in the assembly, storage, or transfer of [Fe2S2] clusters or as sensors of the cellular Fe-S cluster status. Moreover, the [Fe2S2] cluster-bound form of plant chloroplast monothiol Grx, GrxS14, was recently shown to be competent for rapid and efficient maturation of apo plant chloroplast ferredoxin via intact cluster transfer [47]. Consequently in chloroplasts, which utilize the SUF system without a U-type scaffold protein, monothiol Grxs may function as de novo scaffold proteins for the assembly and delivery of [Fe2S2]2+ clusters. However, in light of the yeast immunoprecipitation results [37], they are more likely to play a role as a delivery system for [Fe2S2]2+ clusters assembled on U-type scaffold proteins in systems containing U-type scaffold proteins, see Fig. 3. Experiments are currently in progress to test this hypothesis.
Although there is currently no high resolution structural data for a [Fe2S2]2+ cluster-bound monothiol Grx, spectroscopic and analytical studies indicate one [Fe2S2]2+ cluster with complete cysteinyl ligation per dimer [47;48]. Only the active site cysteine in the CGFS motif and glutathione are required for cluster assembly, which suggests that the cluster is coordinated at the subunit interface by one cysteine from each Grx and two glutathiones. A similar cluster ligation has been established by crystallography for the stable [Fe2S2]2+ cluster-bound forms of human Grx2 (SCSYS active site) [49] and plant GrxC1 (YCGYC active site) [50]. However, marked differences in spectroscopic properties and labilities of the clusters in the structurally characterized Grxs compared with monothiol Grxs indicate significant differences in the arrangement of ligands or the [Fe2S2]2+ cluster environment in monothiol Grxs [47]. Structural studies are in progress to address [Fe2S2]2+ cluster environment in monothiol Grxs.
Conclusions and Prospects
The combination of in vivo and in vitro data has demonstrated that U-type scaffold proteins serve as the primary scaffold proteins for the cysteine desulphurase-mediated assembly of [Fe2S2]2+ and [Fe4S4]2+ clusters that are used for maturation of Fe-S proteins by the ISC and NIF Fe-S cluster biogenesis systems. The roles of A-type, Nfu-type, and monothiol Grx-type scaffold proteins are less well defined, but the ability of Nfu-type proteins and monothiol Grxs to rapidly and efficiently transfer [Fe4S4] and [Fe2S2] clusters to acceptor proteins, respectively, suggests a role as cluster chaperones for the delivery of clusters assembled on U-type scaffold proteins. Nfu and monothiol Grx scaffold proteins may also serve as primary scaffolds in the SUF system, particularly in organisms or chloroplasts that lack U-type scaffold proteins, although here the situation is unclear due to lack of a well-defined role of the SufBCD complex and the possibility that SufB may serve as a primary scaffold protein. Determining the specific function(s) of each type of potential scaffold protein will require a detailed characterization of interprotein cluster transfer relationships among scaffold proteins and evaluation of their specificity for the maturation of a wide range of acceptor proteins using both in vivo and in vitro approaches. Ultimately the goal is a molecular level understanding of the mechanism of cluster assembly and transfer using the components of the NIF, ISC and SUF systems. Consequently there is pressing need for structural characterization of cluster-bound forms of scaffold proteins and kinetic studies of cluster assembly under turnover rather than steady state conditions. Although great progress has been made of the past decade in identifying the key proteins involved in Fe-S cluster biogenesis, unraveling the details of this complex process at the molecular level is only just beginning.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM62524). We are indebted to our collaborators, Dennis Dean, Vincent Huynh, Nicolas Rouhier, Jean-Pierre Jacquot and their coworkers, for supplying samples and plasmids and for numerous stimulating discussions.
References
- 1.Johnson MK, Smith AD. In: Iron-sulfur proteins. In Encyclopedia of Inorganic Chemistry. 2. King RB, editor. John Wiley & Sons; Chichester: 2005. pp. 2589–2619. [Google Scholar]
- 2.Balk J, Lobreaux S. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 2005;10:324–331. doi: 10.1016/j.tplants.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Lill R, Muhlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: Components and mechanisms. Annu Rev Cell Dev Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 5.Ayala-Castro C, Saini A, Outten FW. Fe-S Cluster Assembly Pathways in Bacteria. Microbiol Mol Biol Rev. 2008;72:110–125. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi Y, Nakamura M. Functional assignment of the ORF2-iscS-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J Biochem. 1999;126:917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 9.Fontecave M, Ollagnier-de-Choudens S, Py B, Barras F. Mechanisms of iron-sulfur cluster assembly: the SUF machinery. J Biol Inorg Chem. 2005;10:713–721. doi: 10.1007/s00775-005-0025-1. [DOI] [PubMed] [Google Scholar]
- 10.Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc Natl Acad Sci USA. 2000;97:599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agar JN, Krebs B, Frazzon J, Huynh BH, Dean DR, Johnson MK. IscU as a scaffold for iron-sulfur cluster biosynthesis: Sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry. 2000;39:7856–7862. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 12.Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK. IscA, an alternative scaffold for Fe-S cluster biosynthesis. Biochemistry. 2001;40:14069–14080. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- 13.Ollagnier-de-Choudens S, Mattioli T, Takahashi Y, Fontecave M. Iron-sulfur cluster assembly. Characterization of IscA and evidence for a specific functional complex with ferredoxin. J Biol Chem. 2001;276:22604–22607. doi: 10.1074/jbc.M102902200. [DOI] [PubMed] [Google Scholar]
- 14.Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- 15.Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. Sulfur transfer from IscS to IscU: The first step in iron-sulfur cluster biosynthesis. J Am Chem Soc. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- 16.Sendra M, Ollagnier-de-Choudens S, Lascoux D, Sanakis Y, Fontecave M. The SUF iron-sulfur cluster biosynthetic machinery: Sulfur transfer from the SUFS-SUFE complex to SUFA. FEBS Lett. 2007;581:1362–1368. doi: 10.1016/j.febslet.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 17.Layer G, Ollagnier-de-Choudens S, Sanakis Y, Fontecave M. Iron-sulfur cluster biosynthesis. Characterization of Eschericia coli CyaY as an iron donor for the assembly of [2Fe-2S] clusters in the scaffold IscU. J Biol Chem. 2006;281:16256–16263. doi: 10.1074/jbc.M513569200. [DOI] [PubMed] [Google Scholar]
- 18.Loiseau L, Gerez C, Bekker M, Ollagnier-de-Choudens S, Py B, Sanakis Y, Teixeira M, Fontecave M, Barras F. ErpA, an iron-sulfur (Fe-S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding H, Yang J, Coleman LC, Yeung S. Distinct iron binding property of two putative iron donors for the iron-sulfur cluster assembly: IscA and the bacterial frataxin ortholog CyaY under physiological and oxidative stress conditions. J Biol Chem. 2007;282:7997–8004. doi: 10.1074/jbc.M609665200. [DOI] [PubMed] [Google Scholar]
- 20.Johnson DC, Unciuleac MC, Dean DR. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J Bacteriol. 2006;188:7551–7561. doi: 10.1128/JB.00596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci USA. 2001;98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 23.Vickery LE, Cupp-Vickery JR. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit Rev Biochem Mol Biol. 2007;42:95–111. doi: 10.1080/10409230701322298. [DOI] [PubMed] [Google Scholar]
- 24.Kitaoka S, Wada K, Hasegawa Y, Minami Y, Fukuyama K, Takahashi Y. Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery. FEBS Lett. 2006;580:137–143. doi: 10.1016/j.febslet.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 25.Eccleston JF, Petrovic A, Davis CT, Rangachari K, Wilson RJ. The kinetic mechanism of the SufC ATPase: The cleavage step is accelerated by SufB. J Biol Chem. 2006;281:8371–8378. doi: 10.1074/jbc.M513455200. [DOI] [PubMed] [Google Scholar]
- 26.Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de-Choudens S, Lascoux D, Fontecave M, Outten FW. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J Biol Chem. 2007;282:13342–13350. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- 27.Bertini I, Cowan JA, del Bianco C, Luchinat C, Mansy SS. Thermotoga maritima IscU. Structural characterization and dynamics of a new class of metallochaperone. J Mol Biol. 2003;331:907–924. doi: 10.1016/s0022-2836(03)00768-x. [DOI] [PubMed] [Google Scholar]
- 28.Ramelot TA, Cort JR, Goldsmith-Fischman S, Kornhaber GJ, Xiao R, Shastry R, Acton TB, Honig B, Montelione GT, Kennedy MA. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J Mol Biol. 2004;344:567–583. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Oganesyan N, Shin DH, Jancarik J, Yokota H, Kim R, Kim SH. Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form. Proteins: Struct Func Bioinform. 2005;59:875–881. doi: 10.1002/prot.20421. [DOI] [PubMed] [Google Scholar]
- 30.Smith AD, Jameson GNL, Dos Santos PC, Agar JN, Naik S, Krebs C, Frazzon J, Dean DR, Huynh BH, Johnson MK. NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry. 2005;44:12955–12969. doi: 10.1021/bi051257i. [DOI] [PubMed] [Google Scholar]
- 31.Chandramouli K, Unciuleac MC, Naik S, Dean DR, Huynh BH, Johnson MK. Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry. 2007;46:6804–6811. doi: 10.1021/bi6026659. [DOI] [PubMed] [Google Scholar]
- 32.Agar JN, Dean DR, Johnson MK. Iron-sulfur cluster biosynthesis. In: Ljungdahl LG, Adams MWW, Barton LL, Ferry JG, Johnson MK, editors. Biochemistry and Physiology of Anaerobic Bacteria. Springer-Verlag; New York: 2003. pp. 46–66. [Google Scholar]
- 33.Wu SP, Wu G, Surerus KK, Cowan JA. Iron-sulfur cluster biosynthesis: Kinetic analysis of [2Fe-2S] cluster transfer from holo ISU to apo Fd: Role of redox chemistry and a conserved aspartate. Biochemistry. 2002;41:8876–8885. doi: 10.1021/bi0256781. [DOI] [PubMed] [Google Scholar]
- 34.Unciuleac MC, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry. 2007;46:6812–6821. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- 35.Chandramouli K, Johnson MK. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu SP, Mansy SS, Cowan JA. Iron-sulfur cluster biosynthesis. Molecular chaperone DnaK promotes IscU-bound [2Fe-2S] cluster stability amd inhibits cluster transfer activity. Biochemistry. 2005;44:4284–4293. doi: 10.1021/bi0483007. [DOI] [PubMed] [Google Scholar]
- 37.Muhlenhoff U, Gerber J, Richhardt N, Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Iscu1p. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandyopadhyay S, Naik S, O’Carroll IP, Huynh BH, Dean DR, Johnson MK, Dos Santos PC. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J Biol Chem. 2008;283:14092–14099. doi: 10.1074/jbc.M709161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angelini S, Gerez C, Ollagnier-de-Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;289:14084–14091. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- 40.Tong WH, Jameson GNL, Huynh BH, Rouault TA. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci USA. 2003;100:9762–9767. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yabe T, Morimoto K, Kikuchi S, Nishio K, Terashima I, Nakai M. The arabidopsis chloroplast NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for the biogenesis of ferredoxin and photosystem I. Plant Cell. 2004;16:993–1007. doi: 10.1105/tpc.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touraine B, Boutin JP, Marion-Poll A, Briat JF, Peltier G, Lobreaux S. Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulfur cluster assembly in Arabidopsis chloroplasts. Plant J. 2004;40:101–111. doi: 10.1111/j.1365-313X.2004.02189.x. [DOI] [PubMed] [Google Scholar]
- 43.Balasubramanian R, Shen G, Bryant DA, Golbeck JH. Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcussp. strain PCC 7002. J Bacteriol. 2006;188:3182–3191. doi: 10.1128/JB.188.9.3182-3191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrero E, de la Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64:1518–1530. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molina-Navarro MM, Casas C, Piedrafita L, Belli G, Herrero E. Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett. 2006;580:2273–2280. doi: 10.1016/j.febslet.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 47.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 49.Johansson C, Kavanagh KL, Gileadi O, Oppermann U. Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria. J Biol Chem. 2007;282:3077–3082. doi: 10.1074/jbc.M608179200. [DOI] [PubMed] [Google Scholar]
- 50.Rouhier N, Unno H, Bandyopadhyay S, Masip L, Kim SK, Hirasawa M, Gualberto JM, Lattard V, Kusunoki M, Knaff DB, Georgiou G, Hase T, Johnson MK, Jacquot JP. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci USA. 2007;104:7379–7384. doi: 10.1073/pnas.0702268104. [DOI] [PMC free article] [PubMed] [Google Scholar]