Abstract
TNF and type I interferons (IFNs) are induced by microbial stimuli and mediate innate immune responses. They are also involved in pathogenesis of chronic inflammatory diseases, such as rheumatoid arthritis and systemic lupus erythematosus. Activated macrophages are an important driving force of inflammatory reactions and one of the major producers of TNF in innate immunity and chronic inflammation. Despite the fact that cells at sites of damage are continuously exposed to both cytokines, little is known about mechanisms regulating TNF and type I IFN interactions during inflammation. In this review, we will discuss the role of an IFN-β-mediated autocrine loop in regulation of gene expression program induced by TNF in myeloid cells.
The innate immune system is evolutionarily conserved and represents the first line of defense against pathogens. The mechanisms of recognition by this system are diverse and involve three strategies: recognition of ‘microbial non-self’, recognition of ‘missing self’ and recognition of ‘induced or altered self’. First strategy is the most universal in the animal kingdom and based on detection of conserved pathogen-associated molecular patterns (PAMPs) that are unique for microbes and are not expressed by the host. PAMPs are recognized by several structurally and functionally distinct classes of pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) [1]. Engagement of PRRs leads to production of proinflammatory cytokines, such as IL-1, IL-6, IL-12 and TNF, chemokines and interferons (IFNs). This attracts various cells to the site of damage and creates an inflammatory reaction, resulting in elimination of pathogens and dead tissues, followed by healing. Pathogen clearance and inflammation are tightly controlled and self-limited processes. TNF and type I IFNs are key effectors of innate immune responses. They are also involved in pathogenesis of chronic inflammatory diseases, such as rheumatoid arthritis and lupus [2, 3].
Virtually all nucleated cells express type I IFN receptors and TNF receptor 1, and thus can sense both cytokines. In this review, we focus on macrophages, one of the main players of the innate immune system. Activated macrophages are known to be major producers of TNF in acute and chronic inflammatory reactions. However, most studies in the TNF field were focused on activation of cell types other than macrophages, such as fibroblasts and endothelial cells. Macrophages also can secrete low amounts of type I IFN in response to microbial stimuli. There is evidence in the literature that TNF has some antiviral activities and is able to induce type I IFN in certain cell types [4]. The early events of TNF signaling mediated by nuclear factor κB (NF-κB) and mitogen-activated protein kinases (MAPKs) have been studied extensively, but little is known about the regulation of cell responses after continuous exposure to this cytokine. Recent work highlighted the importance of mechanisms and autocrine loops that sustain and regulate activity of signaling pathways and transcription factors after the initial and typically transient response to an extracellular ligand [5–8]. In this review, we will discuss a new autocrine loop induced by TNF, as well as role of TNF and IFN signaling in regulation of inflammatory gene expression.
Interferon-β Induction by TNF
IFNs were discovered more than 50 years ago and were named based on their ability to interfere with viral replication in a cell. Three classes of IFNs have been identified and classified according to the structure of their receptors. Type I IFNs are encoded by various genes including IFN-α (composed of a family of 13 genes in human and 14 genes in mice), -β, -ε, -κ, -ω and -δ. Type II IFN corresponds to the single IFN-γ that binds the IFN-γ receptor complex. It is encoded by single gene structurally unrelated to the type I IFNs and induced mainly in cells of the immune system such as T cells and natural killer cells and important for protection from nonviral pathogens. Type III IFNs include recently identified IFN-λ1 (IL-29), -λ2 (IL-28A), and -λ3 (IL-28B). Type III IFNs signal through receptors containing IFNLR1 (IL-28Rα) and IL-10Rβ, and are induced in virally infected cells by mechanisms similar to those that activate IFN-α and -β genes [9]. In this review, we will mainly focus on TNF's relationship with type I IFNs.
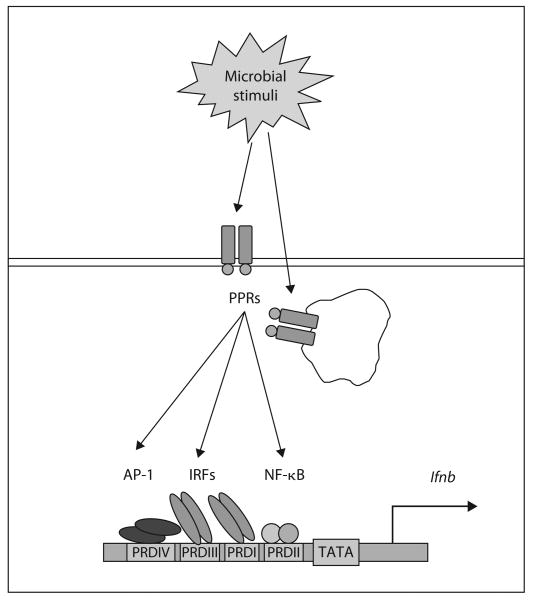
Type I IFNs are produced mainly in response to microbial stimuli (fig. 1), following the engagement of two types of PPRs: cytosolic, which include retinoic acid-inducible gene I, melanoma differentiation-associated gene 5 and DNA-dependent activator of IFN-regulatory factor (IRF), or transmembrane TLRs [9, 10]. All these pathways culminate in activation of NF-κB, activator protein 1 (AP-1) and IRFs, transcriptional factors that are driving the expression of type I IFN genes, and other mediators of innate immune reactions. For example, lipopolysaccharide (TLR4 ligand) and nucleic acids (ligands for TLR3, TLR7, TLR8, and TLR9) strongly induce type I IFN prowduction through activation of IRFs [9]. TLR3 and TLR4 activate IRF3, and TLR7–9 activate IRF7 [6, 9, 11–13].
Fig. 1.
Type I IFN induction by microbial stimuli. The ifnb1 promoter region is shown schematically. Microbial stimuli interact with extra- or intracellular PPRs and lead to the activation of NF-κB, AP-1 and IRFs transcriptional factors which bind to specific PRDs on the promoter region of IFN-β gene and driving the gene expression. The promoter contains four PRDs. PRD I and III are the binding sites for IRFs, PRD II binds NF-κB and PRD IV binds AP-1.
The promoter region of the IFN-β gene contains four regulatory cis elements – the positive regulatory domains (PRDs) I, II, III and IV [9]. PRD I and III are the binding sites for IRFs and PRD II and IV bind NF-κB and AP-1, respectively. After infection of cells with viruses, all of these transcription factors become activated, and together with the high-mobility group protein HMG-I(Y) bind cooperatively to the IFN-β promoter to form an enhanceosome [9]. In the steady state, the transcriptional start site of the promoter is covered by a nucleosome. Enhanceosome formation recruits histone acetyl transferases to acetylate histones H3 and H4 in the nucleosome, following the recruitment of a nucleosome modification complex, which displaces it and allows IFN-β gene transcription [9]. IRFs are required for the recruitment of chromatin modifiers and nucleosome alteration. TLR-induced IFN production triggers an autocrine loop by binding to its cognate receptor (a heterodimer of IFNAR1 and IFNAR2) and activating the IFN-stimulated gene factor 3 complex (a heterotrimer of STAT1, STAT2 and IRF9), which promotes expression of downstream IFN-induced and STAT-dependent genes [5, 9, 14]. An important function of TLR-induced signaling is a massive systemic production of type I IFNs in order to induce an antiviral state and protect the host, but it also can contribute to endotoxin lethality and autoimmune diseases [15, 16].
Many proinflammatory cytokines are powerful activators of NF-κB and AP-1 [1, 9, 16]. However, we have only limited information on induction of an IFN-mediated autocrine loop by endogenous inflammatory factors. Recently, our group discovered that in myeloid cells TNF induces expression of low but sustained levels of IFN-β mRNA and protein [17]. This induction occurred early after TNF induction and is mediated by IRF1. IRF1 was originally discovered as a transcriptional activator of IFN-β in virus-infected fibroblasts [18, 19]. However, after discovery of IRF3 and IRF7 as master regulators of type I IFN induction by PRRs the focus of research on IRF1 shifted to investigation of its role in mediating IFN-γ responses and in T and natural killer cell development and function [19]. In macrophages, IRF1 regulates induction of inducible nitric oxide synthase and IL-12p35, and has been implicated in the pathogenesis of autoimmune diseases [19, 20]. IRF1 is expressed in many cell types in basal amounts and its expression is further increased by various stimuli, including viral infection, IFNs and cytokines [19, 21]. Resting macrophages express IRF1 that is partially localized to the nucleus [Yarilina, unpubl. obs.], and TNF treatment increases IRF1 gene expression in a direct fashion [7, 17]. Recent work reveals that mechanisms of IRF1-mediated ifnb1 expression depend on the cell type and ligand [21, 22]. This work demonstrated a ‘licensing’ process whereby interaction of IRF1 with TLR9-activated MyD88 increases the rate of IRF1 nuclear translocation and transcriptional activation of target genes relative to that observed with IFN-γ-induced IRF1 that has not been licensed by MyD88. The mechanisms and molecular basis for licensing and/or activation of IRF1 function remain unknown, although posttranslational modification and/or activation of IRF1 was originally suggested in 1991, and recent work suggests potential phosphorylation [21, 22]. TNF does not utilize MyD88 for signaling or enhance IRF1 nuclear translocation [Yarilina, unpubl. obs.]. Also, production of IFN-β after TNF stimulation was several orders of magnitude lower than after TLR stimulation. Thus, similar to IFN-γ TNF increases expression of ‘unlicensed’ IRF1. As unlicensed IRF1 is a much weaker activator of gene expression [21], the gene-activating function of IRF1 induced by endogenous factors and cytokines such as IFN-γ and TNF will be much more dependent on synergistic interactions with additional signals and transcription factors such as NF-κB.
TNF Induces Sustained IFN Response Gene Expression
Similar to previous work categorizing TLR-induced genes into different response categories [5, 6], we were able to separate TNF-induced genes into three groups based on the kinetics of expression and requirement for the new protein synthesis [17]. Recently, a similar three-group pattern was described for TNF-activated mouse fibroblasts and bone marrow-derived macrophages, but classification was based on the differences in the mRNA stability between the genes in each group [7].
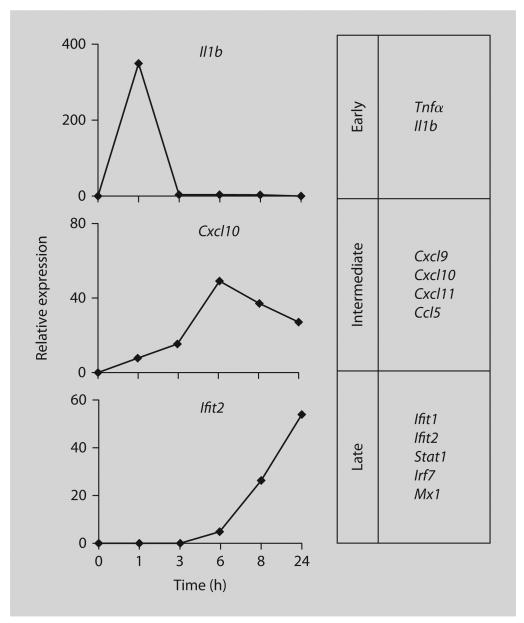
The first group includes well-known TNF-induced ‘primary response’ genes including NF-κB targets Tnf and Il1b1, whose expression increased rapidly and transiently after incubation with TNF and did not require new protein synthesis (fig. 2, upper section).
Fig. 2.
Kinetics of TNF-induced gene expression. Real-time PCR analysis of expression of indicated genes in mouse bone marrow-derived macrophage treated with mTNF for indicated periods of time, relative to untreated cells at the same time point (set at 1). Genes were divided into three groups according to the kinetics of their response to TNF.
A second group of ‘intermediate response’ genes that included the T helper type 1 chemokines Cxcl9, Cxcl10, Cxcl11 and Ccl5 increased in expression with slower kinetics, reaching maximal expression 3–8 h after TNF stimulation (fig. 2, middle section). Expression of these genes is known to be regulated by NF-κB-, STAT1- and IRF1-dependent pathways. At earlier time points, gene expression was independent of new protein synthesis, thus fitting into the delayed primary response category. However, sustained expression of these genes at later time points was dependent on autocrine IFN-β production, demonstrating a transition to the secondary response category.
The third group of genes (‘late response’ genes; fig. 2, lower section) was induced with delayed kinetics (induction first detected at about 6 h after TNF stimulation) in a manner entirely dependent on IFN-β and IRF-1, and included classical IFN response genes important for antiviral responses (Mx1, Isg54, Isg56), and for augmented subsequent responses to microbial products or inflammatory cytokines (Irf7, Ikke, Stat1) [5, 9, 11, 14, 23]. Consistent with a process involving autocrine type I IFN action, TNF induced activation of STAT proteins: in TNF-stimulated macrophages, both STAT1 and STAT2 were phosphorylated on the activating tyrosine residues (701 and 689, respectively). TNF also increased phosphorylation on STAT1 serine 727 residue that enhances its transcriptional activity. Thus, TNF induces a cascade of gene expression with an increasing need for IFN-β synthesis to sustain ‘intermediate’ gene expression after initial TNF stimulation and to induce the set of antiviral genes. The TNF-activated IRF1-IFN-Jak-STAT signaling pathway contributes to the proinflammatory functions of TNF, and provides evidence that induction of IFN-mediated autocrine loops is not limited to PRRs.
Synergy between TNF-Induced Nuclear Factor-κB and Interferon-β
Direct early events of cytokine signal transduction are relatively well studied. However, the mechanisms of signal propagation over time are not clear, and the importance of autocrine loops in regulating cellular responses has gained increasing attention [5, 6]. As was noted before, the modest increase in chemokine gene expression observed early after TNF stimulation was not dependent on autocrine IFN-β, but demonstrated an increased requirement for IFN-β signaling over the time. Notably, both IRF1 and IFN-β expression fit the profile of ‘intermediate response’ genes that are basally expressed, and then are further induced by TNF in a manner dependent on IFNAR signaling, supporting existence of a positive feedback loop [22]. Disruption of NF-κB signaling prevented induction of ‘intermediate response’ genes by TNF, and, in contrast to ‘late response’ genes, low amounts of IFN-β were not sufficient to rescue gene activation. Furthermore, simultaneous addition of TNF and low concentrations of IFN-β (1 U/ml) at early time points (prior to production of endogenous IFN-β) induced higher expression of ‘intermediate response’ genes. Thus, the temporal expression of ‘intermediate’ genes required a direct TNF-induced signal (activated NF-κB) as well as indirect signaling through an IFN-β-mediated autocrine loop.
Synergistic actions of IFNs and members of TNF family have been studied in the context of viral infection [4, 24]. It has been shown in several animal models that anti-HBV effects depend on the synergy between TNF and both type I and II IFNs, and another TNF family member – lymphotoxin cooperates with IFN-β to arrest the replication of HCMV in fibroblasts [25]. Adenoviral (Ad) vectors are used as a model to study immune response to viruses. Ad gene transfer is one of the most efficient techniques available for the in vivo gene transduction. However, the successful use of Ad vectors in gene therapy is limited by their rapid elimination from the circulation. The host response to Ad vectors is dependent on both type I IFNs and TNF [26]. Neutralization of autocrine TNF attenuated adenovirus-induced expression of IFN-β and IFN-dependent genes in human macrophages, providing additional insights into the mechanism of TNF-related clearance of Ad vectors via the synergy with type I IFN. Another important biological role of low concentrations of type I IFNs is ‘priming’ cells to produce more IFN upon subsequent stimulation. Low amounts of IFN-β induced by TNF enhanced type I IFN expression in human macrophages stimulated with TLR7/8 and TLR9 ligands, which are known as weak IFN inducers in macrophages. This mechanism may be involved in antiviral responses mediated by TNF.
Another significant outcome of synergy between TNF and IFN is strong induction of proinflammatory chemokines. Despite the modest induction of type I IFN by TNF compared to TLR ligands, the production of chemokines eventually reached similar amounts, and remained elevated for at least 2 days after TNF stimulation, supporting the functional importance of late TNF-induced gene induction in macrophages.
TNF induces a similar pattern of chemokine and IFN-mediated gene expression in vivo, when injected into mice intraperitoneally [17]. We also observed increased expression of type I IFN and IFN-induced genes in joints of mice expressing transgenic TNF [Yarilina, unpubl. obs.]. These mice develop spontaneous arthritis. These findings can explain an elevated expression of type I IFNs and type I IFN response genes in synovial macrophages from patients with rheumatoid arthritis, a sterile inflammatory condition that is driven by TNF (table 1). This phenomenon is known as ‘IFN signature’ and was described by several groups [3, 23, 27]. We recently found that ex-vivo blocking of endogenous TNF by soluble receptor etanercept prevented increase in type I IFN-dependent genes and chemokines in macrophages isolated from synovial fluids of patients with rheumatoid arthritis [Kalliolias, unpubl. obs.].
Table 1. Expression of IFN response genes in rheumatoid arthritis (RA) synovial macrophages.
| Description | Gene symbol | Fold increase RA vs. control | Mean signal intensity in RA |
|---|---|---|---|
| IFN-α-inducible protein 27 | IFI27 | 12.88 | 1,092 |
| Guanylate-binding protein 1, IFN-inducible, 67 kDa | GBP1 | 7.2 | 527 |
| IFN-induced transmembrane protein 1 (9–27) | IFITM1 | 5.9 | 906.6 |
| Signal transducer and activator of transcription 1, 91 kDa | STAT1 | 3.3 | 943 |
| Eukaryotic translation initiation factor 2-α kinase 2 | EIF2AK2 | 2.5 | 1,500 |
| IFN-induced protein with tetratricopeptide repeats 3 | IFIT3 | 2.2 | 1,001 |
| Chemokine (C-X-C motif) ligand 9 | CXCL9 | 80.5 | 33 |
| Indoleamine-pyrrole 2,3 dioxygenase | INDO | 6.8 | 499 |
| Chemokine (C-X-C motif) ligand 10 | CXCL10 | 6.2 | 282 |
| Chemokine (C-X-C motif) ligand 11 | CXCL11 | 5.7 | 81 |
| IRF1 | IRF1 | 3.2 | 983 |
| TNF (TNF superfamily, member 2) | TNF | 3.95 | 702 |
| INF-α41 | IFNA4 | 2.1 | 4.75 |
| INF-β1, fibroblast1 | IFNB1 | 2.8 | 11.75 |
Listed genes were elevated 2-fold or more in RA synovial macrophages (n = 5) relative to control macrophages isolated from peripheral blood of healthy donors (samples were pooled into 3 sets, 5 donors each), p < 0.05 (Welch's t test or Welch's analysis of variance with the Benjamini-Hochberg correction for false discovery rate multiple testing).
Expression levels were low. Difference was not statistically significant.
TNF and Toll-Like Receptors
It is interesting to compare activation of type I IFN-mediated autocrine loops by PRRs (such as TLR3 and TLR4) that respond to microbial pathogens with that activated by TNF. Activation of TRIF-dependent signaling pathways by TLR3 and TLR4 immediately results in TBK1-IKKε-mediated phosphorylation and activation of IRF3 (or IRF7 in plasmacytoid DCs or primed cells) with a rapid and massive production of IFNs and expression of IFN response genes [11–14]. This type of robust response has the advantages of efficient mobilization of host defenses against infection, and large amounts of IFNs act systemically to broadly induce an antiviral state [5, 9]. However, production of large amounts of systemic IFN can be toxic and contributes to endotoxin lethality [15], and in a chronic setting can predispose to autoimmunity [16]. In contrast, TNF uses unlicensed IRF1 to induce production of small amounts of IFN-β, and only minimal amounts of IFN-α mRNA and protein that act locally and require synergistic interactions with additional TNF-induced signals for effective induction of inflammatory genes. Thus, the toxicity associated with systemic IFN production is avoided, while sustained and synergistic induction of gene expression primes cells for strong responses to infectious pathogens if needed.
Conclusions
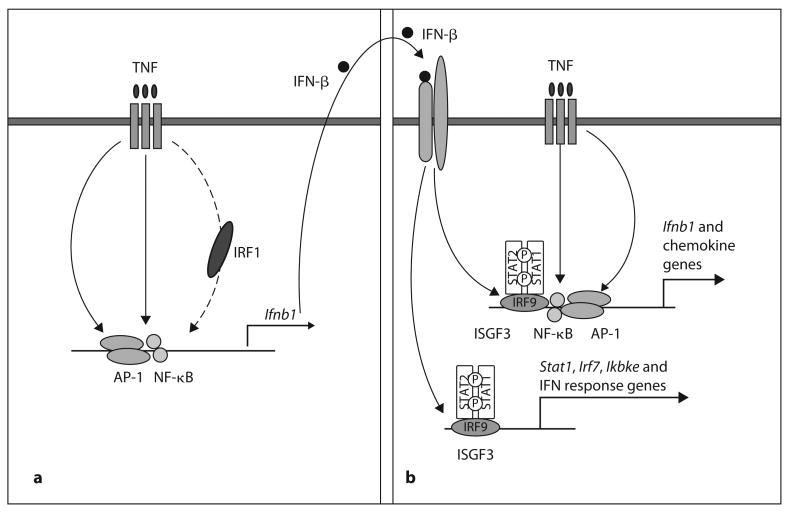
Our results suggest a model whereby TNF-induced gene expression is sustained and amplified by sequential induction of IRF1, IFN-β and STAT1 (fig. 3). Acute stimulation with TNF activates NF-κB and MAPK pathways, which lead to rapid expression of inflammatory genes, and to increased expression of IRF1. IRF1 works together with NF-κB and AP-1 to drive production of low amounts of IFN-β. IFN-β, in turn, activates Jak-STAT signaling that synergizes with other TNF-induced signals to sustain inflammatory chemokine and IFN-β expression, induce slow and delayed accumulation of mRNAs encoded by canonical IFN response genes, increase expression of signaling components such as IKKε, IRF7 and STAT1 that are known to further amplify activation of genes by low concentrations of IFNs and shift the balance of macrophage responses in an inflammatory direction [5, 14, 23, 28]. Regulation of sequential gene expression by autocrine loops induced by initial stimuli is well known in the IFN field [5, 6]. However, our study was focused on a very limited array of specific genes, related to type I IFN, induced by TNF. TNF induces many more target genes and pathways, and this activation is regulated on multiple levels and by different mechanisms. For example, microarray analysis of genes increased by TNF in mouse cells revealed hundreds of genes induced by TNF with similar kinetic patterns, but this study demonstrated that the kinetics of gene expression was dependent on mRNA stability [7].
Fig. 3.
Model for induction of a gene activation program by TNF. Stimulation with TNF (a) activates NF-κB and MAPK pathways leading to rapid expression of inflammatory genes and to increasing expression of IRF1. IRF1 works together with NF-κB and AP-1 to drive production of low amounts of IFN-β. IFN-β, in turn, activates STAT signaling (b) that synergizes with other TNF-induced signals to sustain inflammatory chemokine expression and Ifnb1 expression, and to induce canonical IFN response genes and increase expression of signaling components such as IKKε, IRF7 and STAT1. ISGF3 = IFN-stimulated gene factor 3.
Work from several laboratories demonstrated that low type I IFN signaling observed under physiological conditions in the absence of infection [29] is important for a robust response to microbial pathogens. The factors that regulate basal IFN production and signaling in the absence of pathogens are not well understood, but include ITAM-coupled immunoreceptors including Fc receptors [28, 30]. Our findings implicate TNF as a new endogenous inducer of type I IFN production and signaling. It is unlikely that the low amounts of IFN induced by TNF contribute significantly to overall IFN production in the setting of an infection where multiple PRRs are engaged. However, TNF-induced local production of IFN-β may regulate inflammation of noninfectious origin such as after tissue damage, and in chronic sterile TNF-dominated inflammation, such as the joint lining in rheumatoid arthritis [23, 27].
Acknowledgments
This work was supported by NIH grants AR050401, AR46713 and AI46712 to L.B.I.
References
- 1.Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 3.McInnes IB, Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol. 2007;7:429–442. doi: 10.1038/nri2094. [DOI] [PubMed] [Google Scholar]
- 4.Benedict CA. Viruses and the TNF-related cytokines, an evolving battle. Cytokine Growth Factor Rev. 2003;14:349–357. doi: 10.1016/s1359-6101(03)00030-3. [DOI] [PubMed] [Google Scholar]
- 5.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 7.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda K, Takaoka A, Taniguchi T. Type I interferon (corrected) gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci USA. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 13.Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A, Honda K, Taniguchi T. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem Biophys Res Commun. 2003;306:860–866. doi: 10.1016/s0006-291x(03)01049-0. [DOI] [PubMed] [Google Scholar]
- 14.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 15.Karaghiosoff M, Steinborn R, Kovarik P, Kriegshauser G, Baccarini M, Donabauer B, Reichart U, Kolbe T, Bogdan C, Leanderson T, et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 16.Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007:543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]
- 17.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 19.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Guan X, Tamura T, Ozato K, Ma X. Synergistic activation of interleukin-12 p35 gene transcription by interferon regulatory factor-1 and interferon consensus sequence-binding protein. J Biol Chem. 2004;279:55609–55617. doi: 10.1074/jbc.M406565200. [DOI] [PubMed] [Google Scholar]
- 21.Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, Takayanagi H, Ohba Y, Taniguchi T, Honda K. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc Natl Acad Sci USA. 2006;103:15136–15141. doi: 10.1073/pnas.0607181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz F, Heit A, Guggemoos S, Krug A, Mages J, Schiemann M, Adler H, Drexler I, Haas T, Lang R, et al. Interferon-regulatory-factor 1 controls Toll-like receptor 9-mediated IFN-beta production in myeloid dendritic cells. Eur J Immunol. 2007;37:315–327. doi: 10.1002/eji.200636767. [DOI] [PubMed] [Google Scholar]
- 23.Hu X, Herrero C, Li WP, Antoniv TT, Falck-Pedersen E, Koch AE, Woods JM, Haines GK, Ivashkiv LB. Sensitization of IFN-gamma Jak-STAT signaling during macrophage activation. Nat Immunol. 2002;3:859–866. doi: 10.1038/ni828. [DOI] [PubMed] [Google Scholar]
- 24.Mestan J, Digel W, Mittnacht S, Hillen H, Blohm D, Moller A, Jacobsen H, Kirchner H. Antiviral effects of recombinant tumour necrosis factor in vitro. Nature. 1986;323:816–819. doi: 10.1038/323816a0. [DOI] [PubMed] [Google Scholar]
- 25.Benedict CA, Banks TA, Senderowicz L, Ko M, Britt WJ, Angulo A, Ghazal P, Ware CF. Lymphotoxins and cytomegalovirus cooperatively induce interferon-beta, establishing host-virus detente. Immunity. 2001;15:617–626. doi: 10.1016/s1074-7613(01)00222-9. [DOI] [PubMed] [Google Scholar]
- 26.Elkon KB, Liu CC, Gall JG, Trevejo J, Marino MW, Abrahamsen KA, Song X, Zhou JL, Old LJ, Crystal RG, et al. Tumor necrosis factor alpha plays a central role in immune-mediated clearance of adenoviral vectors. Proc Natl Acad Sci USA. 1997;94:9814–9819. doi: 10.1073/pnas.94.18.9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweeney SE, Mo L, Firestein GS. Antiviral gene expression in rheumatoid arthritis: role of IKKepsilon and interferon regulatory factor 3. Arthritis Rheum. 2007;56:743–752. doi: 10.1002/art.22421. [DOI] [PubMed] [Google Scholar]
- 28.Tassiulas I, Hu X, Ho H, Kashyap Y, Paik P, Hu Y, Lowell CA, Ivashkiv LB. Amplification of IFN-alpha-induced STAT1 activation and inflammatory function by Syk and ITAM-containing adaptors. Nat Immunol. 2004;5:1181–1189. doi: 10.1038/ni1126. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-alpha/beta revisited. Nat Rev Mol Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 30.Dhodapkar KM, Banerjee D, Connolly J, Kukreja A, Matayeva E, Veri MC, Ravetch JV, Steinman RM, Dhodapkar MV. Selective blockade of the inhibitory Fcgamma receptor (FcgammaRIIB) in human dendritic cells and monocytes induces a type I interferon response program. J Exp Med. 2007;204:1359–1369. doi: 10.1084/jem.20062545. [DOI] [PMC free article] [PubMed] [Google Scholar]