Abstract
All type I interferons (IFNs) bind to a common cell-surface receptor consisting of two subunits. IFNs initiate intracellular signal transduction cascades by simultaneous interaction with the extracellular domains of its receptor subunits IFNAR1 and IFNAR2. In this study we mapped the surface of IFNα2 interacting with the extracellular domain of IFNAR1 (IFNAR1-EC) by following changes in or the disappearance of the [1H,15N]-TROSY-HSQC cross peaks of IFNα2 caused by the binding of the extracellular domain of IFNAR1 (IFNAR1-EC) to the binary complex of IFNα2 with IFNAR2-EC. The NMR study on the 89 kDa complex was conducted at pH 8 and 308 K using an 800 MHz spectrometer. IFNAR1 binding affected a total of 47 out of 165 IFNα2 residues contained in two large patches on the face of the protein opposing the binding site for IFNAR2 and in a third patch located on the face containing the IFNAR2 binding site. The first two patches form the IFNAR1 binding site and one of these matches the IFNAR1 binding site previously identified by site-directed mutagenesis. The third patch partially matches the IFNα2 binding site for IFNAR2-EC indicating allosteric communication between the binding sites for the two receptor subunits.
Type I Interferons (IFNs) are a family of homologous helical cytokines which constitute a major component of the innate immune response (1). This family consists of 13 IFNα isotypes as well as single forms of IFNβ, IFNε, IFNκ, and IFNω (1). IFNs initiate a strong antiviral and antiproliferative activity and provide the first line of defense against viral infection. All human type I IFNs share a common cell surface receptor consisting of two subunits, IFNAR1 and IFNAR2 (2, 3). The extracellular domain of the IFNAR2 subunit (IFNAR2-EC) binds IFNs with high affinity (KD=10 nM for IFNα2 (4)) in the absence of the extracellular domain of IFNAR1 (IFNAR1-EC). The affinity of the human IFNAR1-EC subunit to IFNs is much lower with a KD for IFNα2 of 5 μM (5). IFN-induced association of the receptor subunits results in reciprocal trans-phoshorylation of the IFNAR1-associated Tyk2 protein and the IFNAR2-associated Jak1 protein. These processes initiate an intracellular signal transduction cascade leading to strong antiviral and antiproliferative responses (6–11).
Several mechanisms, involving pre-association of the receptor chains and ligand-induced changes, were postulated based on other cytokine receptor systems (6–11). Studies of the receptor subunits tethered to solid-supported lipid bilayers confirmed a two step binding mechanism in which IFN binds first to IFNAR2-EC and then recruits IFNAR1-EC to form the ternary complex (12, 13). High-resolution three-dimensional structures of the ternary complex formed by IFNAR1-EC, IFNAR2-EC and any one of the type I IFNs, as well as of the structure of the individual proteins and that of a IFNAR2-EC/IFN heterodimer should enhance our understanding of how the various IFNs differ in their mode of binding to IFNAR1-EC and IFNAR2-EC, and help understand the initial steps in interferon signaling. At present, atomic resolution structures of the ternary complex, IFNAR1-EC/IFN and IFNAR2-EC/IFN heterodimers are not known. However, the structures of three IFN molecules, IFNα2, IFNβ and IFNτ were solved by X-ray crystallography and NMR (14–18) revealing a bundle of five anti-parallel helices. The structure of IFNAR2-EC was solved using multi-dimensional NMR techniques. This structure consists of two perpendicular fibronectin domains connected by a rigid hinge region (19, 20).
In the absence of high resolution three dimensional structures of the binary and ternary complexes formed by IFNs and the receptor subunits, several laboratories have attempted to map the binding interfaces on IFNAR2-EC, IFNα/β and IFNAR1-EC. Mutational analysis of IFNAR2-EC identified four receptor residues in the β3–β4 loop as “hot-spots” for IFNα2 binding, with some contributions from residues in the β5–β6 loop and the hinge region (4). In a parallel study mutations of IFNAR2-EC residue E77 in the β5–β6 loop, and of three residues in the hinge region were found to abolish response to IFNα2 with no effect on the response to IFNβ (21). Double mutant cycle experiments revealed interactions between five IFNAR2-EC residues and corresponding IFNα2 residues (22).
Changes in IFNAR2-EC 1H and 15N amide chemical shifts upon IFNα2 binding were also used to map the IFNα2 binding site on IFNAR2-EC (34). This binding site was found to be formed by a strip of hydrophobic residues flanked by two strips of polar residues. Overall the binding site for IFNα2 consists of the β3–β4 loop, the beginning of the β5–β6 loop, the end of the β6 strand and the hinge region of IFNAR2-EC. NMR mapped the binding site for IFNα2 on IFNAR2-EC to a larger area than that identified previously by mutational studies and included most residues previously implicated in IFNα2 binding.
Even before the three dimensional structure of IFNα2 was available, the AB-loop and the D-helix of the cytokine were identified as important for IFN activity and for its interactions with IFNAR2-EC. Knowledge of the structure of IFNα2 (16, 17) provided the basis for additional mutational studies of the binding site for IFNAR2-EC on IFNα2. Several IFNα2 residues were identified as the hot-spot residues for the cytokine binding site to IFNAR2-EC (4). Residues in the AB-loop, the D-helix, the DE-loop and the E-helix were also found to be involved in IFNAR2-EC binding (4, 22, 23). NMR studies revealed the entire continuous surface of IFNα2 involved in IFNAR2-EC binding (24). Overall, the NMR analysis mapped the IFNα2 binding site for IFNAR2-EC to the AB-loop, and the E-helix of the cytokine. The NMR mapping is in a good agreement with the earlier mutational results (22, 23) and highlights additional residues as being part of the IFNα2 binding site for IFNAR2-EC.
The binding site for IFNAR1-EC on IFNα2 has been investigated by site-directed mutagenesis. This site was mapped to the B and C-helices on IFNα2 opposite to the IFNα2 binding site for IFNAR2-EC (25). This is similar to what has been shown for hGH, EPO and IL4 where the cytokine binds to two receptor subunits through two distinct binding surfaces on opposing faces of the cytokines (26). Mutations of residues F64, N65, T69 in the B-helix, and L80, Y85 and Y89 in the C-helix were found to reduce binding to IFNAR1-EC or reduce the biological activity of IFNα2 by more then two-fold (25, 27, 28). R120 was previously identified to be involved in IFNAR1 binding. Mutation of R120 to alanine decreased antiviral activity to 1–3% and antiproliferative activity to about 0.05% and the charge-reversal mutation R120E lead to a total loss of activity (29). In contrast, mutations of H57, E58 and Q61 on the B-helix to alanine were found to increase binding to IFNAR1-EC (25) and specifically enhanced its anti-tumor activity (30, 31). The IFNα2-mutants H57Y, E58N and Q61S were found to be optimal for IFNAR1-EC binding and were similar in their activity and binding properties to IFNβ (31).
IFNAR1-EC consists of four fibronectin domains (D1–D4) of which the three N-terminal domains were implicated in IFN binding (5). Site directed mutagenesis studies have shown that residues in D2 and in D3 participate in forming the binding site for IFNα2 (32). The segment 62FSSLKLNVY70 in D1 was identified as being involved in IFN binding and in the induction of biological activity (33).
Different models for the IFNα2 complex with IFNAR2-EC have been calculated (22, 24). The model incorporating the most recent experimental data used the NMR mapping of the binding sites of IFNAR2-EC and IFNα2, NOEs between D35 of IFNα2 and K48 of IFNAR2-EC and previously published double mutant cycle constraints (22) to dock IFNα2 into the IFNAR2-EC binding site using the three-dimensional NMR structures of the two molecules (24). In all these models the N-terminal domain and the hinge region of IFNAR2-EC interact with a complementary surface on IFNα2.
A low-resolution structure of the IFNAR2-EC/IFNα2/IFNAR1-EC ternary complex was obtained recently using electron microscopy (34). The NMR-based model of the IFNAR2-EC/IFNα2 complex (24) was compatible with this EM structure without any modifications. Homology modeling was used to calculate a model for IFNAR1-EC that was fitted into the EM structure as well (34). The EM-based model for the ternary complex suggested that the three N-terminal fibronectin domains of IFNAR1-EC, D1, D2 and D3, interact with IFN while the fourth extra-cellular domain of IFNAR1-EC (C-terminal) interacts with the second (C-terminal) domain of IFNAR2-EC.
In the present study, we use NMR spectroscopy to determine the entire binding site for IFNAR1-EC on IFNα2 and probe changes in IFNα2 upon binding of IFNAR1-EC to the binary complex between IFNα2 and IFNAR2-EC. The mapping of the binding site for IFNAR1-EC on IFNα2 reveals two adjacent binding surfaces for IFNAR1-EC one of which is in excellent agreement with mutational data and the other which is a new region that was not previously revealed in mutational analyses. In addition to changes in chemical shift that are associated with the IFNα2 binding surface for IFNAR1-EC, significant changes were observed also in the IFNα2 face containing the binding site for IFNAR2-EC. These changes indicate that IFNAR1-EC binding causes allosteric changes in the binding site for IFNAR2-EC. This communication between the two binding sites found on opposing surfaces of IFNα2 could be important for the mutual orientation of the two receptor subunits for the initiation of the intra-cellular signal transduction cascade.
EXPERIMENTAL PROCEDURES
Uniformly 15N,2H-labeled IFNα2, unlabeled IFNAR2-EC (U-IFNAR2-EC) and unlabeled IFNAR1-EC (U-IFNAR1-EC) were expressed and purified as described previously (24, 35). IFNAR1-EC in its truncated form that contains D1, D2 and D3 was prepared as previously described (5).
IFNAR2-EC and IFNα2 at a 1.1:1 molar ratio and at approximately 0.5 μM concentration were incubated for 1–2 h in 25 mM deuterated Tris pH 8 solution containing 0.02% NaN3. For the formation of the ternary complex, IFNAR1-EC was added to the 0.5 μM solution of the IFNAR2-EC/IFNα2 complex in a 1:1 molar ratio with respect to IFNα2 and incubated for another 2 h. The binary and ternary complexes were then concentrated using Vivaspin concentrators (10 kDa MWCO, GE Healthcare). The final concentration of the binary IFNα2/IFNAR2-EC complex was 0.3 mM in 25 mM deuterated Tris 95% H2O/5% D2O pH 8 solution containing 0.02% NaN3. The final concentration of the ternary complex was 0.2 mM in 25 mM deuterated Tris pH 8 in 95% H2O/5% D2O solution containing 0.02% NaN3 and 150 mM NaCl. The binary and the ternary complex solutions differed in the salt concentration since the binary complex was most stable without the addition of NaCl and the ternary complex containing IFNAR1-EC was most stable in the presence of 150 mM NaCl. Control HSQC measurements to evaluate the effect of NaCl on the chemical shifts of IFNα2 were performed on samples containing 50 μM 2H,15N-IFNα2/U-IFNAR2-EC in 25 mM deuterated Tris at pH 8, 95% H2O/5% D2O, 0.02% NaN3 as well as 0 mM NaCl and 150 mM NaCl.
All NMR measurements were recorded at 308 K on a Bruker DRX 800 MHz spectrometers equipped with a x,y,z-gradient triple resonance probe. Data were processed and analyzed using NMRPipe (36) and NMRView (37).
The 2D [1H,15N]-TROSY-HSQC spectra of 2H,15N-IFNα2/U-IFNAR2-EC and U-IFNAR1-EC/2H,15N-IFNα2/U-IFNAR2-EC were acquired using 256 (160) t1 increments with a sweep width of 1,622 (1,622) Hz and 1024 (1024) t2 points with a sweep with of 10,417 (10,417) Hz. The number of scans in each experiment was 16 (16) and the relaxation delay was 1.4 sec (2.0 sec) (numbers in parenthesis are those used for the ternary complex). Water flip-back and Watergate were used for water suppression. For the processing of the binary complex data 256 t1 increments (zero filed to 512) and 700 t2 points (zero filed to 1400) were used. For the processing of the ternary complex data 90 t1 increments (zero filed to 180) and 700 t2 points (zero filed to 1400) were used. In both spectra Sine bell window function with π/2 offset was used. The control 2D [1H,15N]-TROSY-HSQC spectra of 2H,15N-IFNα2/U-IFNAR2-EC with and without NaCl were acquired using 96 scans. All other parameters were the same as described above. For simplicity, in what follows we will refer to [1H,15N]-TROSY-HSQC as TROSY-HSQC.
Sequence alignment was performed using ClustalW (38) and structure alignment of FNα2 and IFNβ was performed using the Combinatorial Extension method (39) provided online at http://cl.sdsc.edu/. Molecular pictures were created using Pymol (40).
RESULTS
Changes in the TROSY-HSQC spectrum of IFNα2 in IFNα2/IFNAR2-EC complex upon IFNAR1-EC binding
NMR is a very powerful technique to map the binding sites of proteins and to assess their conformational changes upon ligand binding. This can be achieved by measuring 1H,15N-HSQC spectra of the protein alone and in complex with its ligand, and subsequent analysis of the changes in chemical shifts as a result of ligand binding. Sequential assignment of the free protein is a prerequisite for this analysis in a residue specific manner. However, sequential assignment of the complex may not be necessary for determining the residues involved in ligand binding. This is especially advantageous when studying large ternary complexes such as IFNAR1-EC/IFNα2/IFNAR2-EC with a molecular weight of 102 kDa.
Deuteration of IFNα2 considerably reduced the width of the IFNα2 amide cross peaks in the HSQC of the binary and ternary complexes. Nevertheless we thought it would be beneficial to further reduce the linewidth of the resonances in the ternary complex. As it has been suggested that only the three N-terminal fibronectin domains of IFNAR1-EC, D1, D2, and D3, interact with IFNα2 (5, 34) and since our labeling strategy in this study allowed observation of only changes in the cytokine, we used a construct of IFNAR1-EC missing the fourth domain (C- domain), thus reducing the total size of the complex to 89 kDa and minimizing the change in the widths of the HSQC cross peaks due to addition of the IFNAR1-EC protein.
Mapping of the binding site for IFNAR1-EC on free IFNα2 was not possible since IFNα2 aggregates under the conditions in which the binary IFNAR1-EC/ IFNα2 is stable (pH 7–8). In any case, the binding site for IFNAR1-EC on the binary complex IFNAR2-EC/ IFNα2 is more relevant since most probably IFNα2 binds first to IFNAR2-EC, and then recruits IFNAR1-EC to form the ternary complex (12, 13).
To map the binding site for IFNAR1-EC on the IFNα2 molecule bound to IFNAR2-EC we measured the HSQC spectra of uniformly 2H,15N labeled IFNα2 in complex with U-IFNAR2-EC before and after the addition of U-IFNAR1-EC under the same pH and temperature conditions. Measurements were carried out in 95% H2O/5% D2O to enable detection of the 1H-15N amide cross peaks in the HSQC spectra. It should be noted that sequential assignment for 85% of the amide 1H and 15N nuclei was previously obtained for IFNα2 in complex with IFNAR2-EC at pH 8 and 308 K (24).
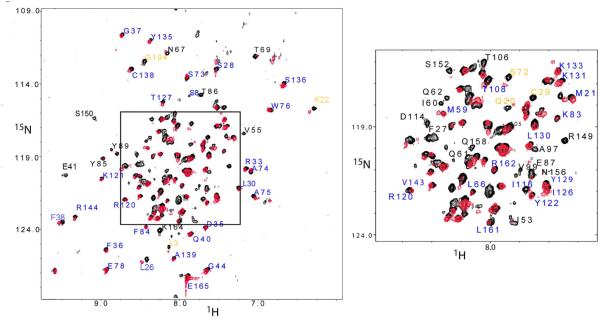
An overlay of HSQC spectra of the U-IFNAR1-EC/2H,15N-IFNα2/U-IFNAR2-EC ternary complex and the 2H,15N-IFNα2/ U-IFNAR2-EC binary complex is presented in Figure 1. The changes in chemical shift were calculated according to the formula . The majority of the unambiguously assigned amide HSQC cross peaks of 2H,15N-IFNα2 exhibited no-changes or just small changes in chemical shift (Δδ < 0.05 ppm) upon the formation of the ternary complex. This group contains 65 residues which were labeled and colored deep blue in Figures 1, 2 and 3. A second group containing 10 residues revealed significant changes in chemical shift (between 0.05 – 0.10 ppm, labeled and colored in yellow in Figures 1, 2 and 3. For a third group of residues the HSQC cross peaks either disappeared or could not be assigned in the ternary complex due to large differences in the chemical shifts of these IFNα2 residues between the binary and the ternary complexes. This group includes 37 residues which are labeled in black in Figure 1 and colored gray in Figures 2 and 3. For three of these 37 residues designated as “disappeared”, namely N65, F67 and T69 the HSQC cross-peaks were weakened by 80%, 72% and 87%, respectively while the other 34 cross peaks were not detected at all. In addition, there were two more groups of residues: The fourth group included IFNα2 residues that could be assigned in theIFNα2/IFNAR2-EC complex using 3D NMR spectra, however their HSQC cross-peaks showed overlap with cross peaks of other residues. One of the overlapping cross peaks either experienced significant change in chemical shift or disappeared (these residues are colored green in Figures 2 and 3). The fifth group included IFNα2 residues whose HSQC cross peaks could not be assigned in the IFNα2/IFNAR2-EC binary complex (colored cyan in Figures 2 and 3). Residues from groups 1–5 were mapped on the structure of IFNα2 (Figure 3). The sequence and secondary structure of IFNα2 is presented in Figure 4.
FIGURE 1.
Overlay of the [15N,1H]-TROSY-HSQC spectra of IFNα2/IFNAR2-EC binary complex (black) and IFNAR1-EC/IFNα2/IFNAR2-EC ternary complex (red) (A) and expansion of the central (boxed) region of this spectrum (B). Residues that did not change their chemical shift significantly upon IFNAR1-EC binding are labeled in blue, residues that underwent significant changes in chemical shift upon IFNAR1-EC binding are labeled in yellow (0.05<Δδ<0.10 ppm); residues that could not be assigned due to large changes in chemical shift or disappearance upon IFNAR1-EC binding are labeled in black. Spectra were measured at 308 K and pH 8.
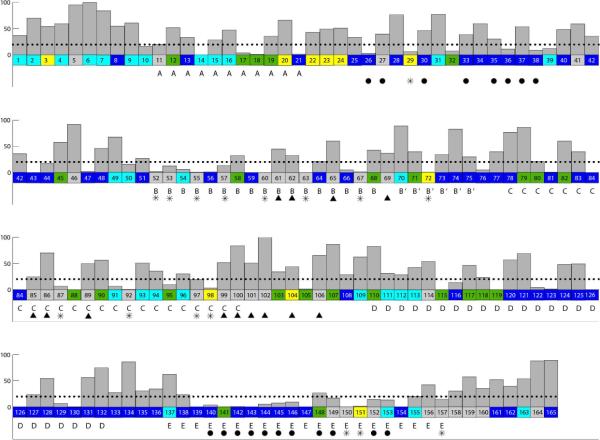
FIGURE 2.
A summary of the changes in IFNα2 HSQC cross peaks upon IFNAR1-EC binding to the binary IFNα2/IFNAR2-EC complex. Residues located in helices are marked with the corresponding helix label (A, B, C, D and E). Residues that did not change their chemical shift significantly upon IFNAR1-EC binding are colored in deep blue, residues that underwent significant chemical shift changes upon IFNAR1-EC binding are colored in yellow (0.05<Δδ<0.10 ppm); residues that could not be assigned due to large chemical shift changes or disappearance upon IFNAR1-EC binding are colored in grey, overlapping cross peaks which include one cross peak that disappeared or underwent a large chemical shift change but could not be assigned due to overlap are colored green and residues whose HSQC cross peaks could not be assigned in the IFNα2/IFNAR2-EC binary complex are colored in cyan. The exposure of IFNα2 residues (backbone and sidechain included) in the binary complex are given by grey bars with a scale from 0–100%. A dotted line represents 20% exposure and below this value of exposure residues are defined as buried. Filled black triangles indicate residues that are not buried and are implicated in IFNAR1-EC binding by the present study. Buried residues that underwent significant changes in chemical shift or disappeared upon IFNAR1-EC binding and were not found to be involved in IFNAR2-EC binding are marked with asterisks. Filled black circles indicate residues found by NMR and site directed mutagenesis to interact with IFNAR2-EC in the binary complex (4, 22, 24).
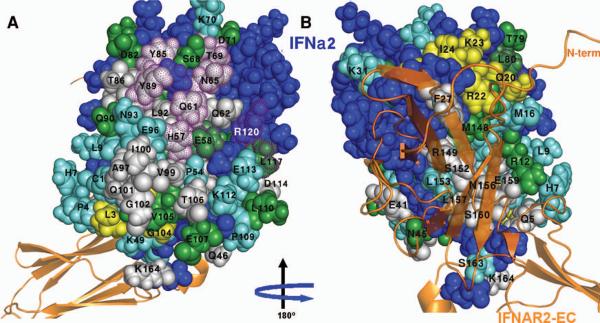
FIGURE 3.
Mapping of the residues that exhibited significant chemical shift changes or disappeared upon IFNAR1-EC binding on the surface of IFNα2. A) The face of IFNα2 opposing the binding site for IFNAR2-EC and that contain the two patches involved in IFNAR1-EC binding according to the present study. B) The face of IFNα2 containing the surface previously found to be involved in IFNAR2-EC binding. IFNAR2-EC bound to IFNα2 is presented by an orange ribbon diagram according to the previously calculated model of the binary complex (24). Residues previously identified by site directed mutagenesis as being involved in IFNAR1-EC binding (25) are marked with dotted purple surfaces. Other color coding is the same as in Figure 2. All molecular pictures were created using Pymol (40).
FIGURE 4.
Comparison of IFNα2 (top) and IFNβ (bottom) residues implicated in IFNAR1-EC binding. Bold characters indicate IFNα2 and IFNβ residues found in the helices. Residues in boxes are IFNα2 residues implicated in IFNAR1-EC binding in the present study. Black triangles designate residues of IFNα2 that were determined by mutagenesis to be in the binding site for IFNAR1-EC (25, 29). Filled black circles indicate residues of IFNβ that were determined by mutagenesis to be in the binding site of IFNAR1-EC (28).
The most significant change to the binary complex observed upon IFNAR1-EC binding is the disappearance of a large number of IFNα2 cross-peaks in the HSQC spectrum of the complex. This change could be due to very large changes in chemical shift in IFNα2 cross peaks upon the formation of the ternary complex or broadening beyond detection of residues in the binding interface. In the HSQC spectrum of the binary complex we counted 158 IFNα2 cross peaks while in the ternary complex 129 cross peaks were counted indicating that the vast majority of the 37 cross peaks in group 3 (grey in Figures 2 and 3) actually vanished rather than changed their chemical shift upon IFNAR1-EC binding. The weak binding of IFNAR1-EC to the binary complex and its fast off-rate (25) could contribute to the broadening of cross peaks in the IFNα2/IFNAR1-EC binding interface.
The binary and ternary complexes were measured using different NaCl concentrations (0 mM and 150 mM, respectively) due to sample stability issues. In order to check whether the salt concentration had an effect on the chemical shifts of the binary IFNα2/IFNAR2-EC complex, we measured HSQC spectra of 2H,15N-IFNα2/U-IFNAR2-EC with and without 150 mM NaCl at low protein concentration (50 μM). Even at the low concentration used, we observed signal loss due to precipitation of the complex in the sample containing 150 mM NaCl. Nonetheless, we could clearly observe, that the vast majority of cross peaks was not affected by the addition of NaCl. Only two residues, L26 and F38, showed a minor change in chemical shift. This change in chemical shifts due to the addition of salt was taken into account in our analysis of chemical shift changes between the binary and ternary complex.
Mapping the IFNAR1-EC binding site on IFNα2
Earlier mutational studies identified the possible binding surface for IFNAR1-EC on the IFN face opposing the binding site for IFNAR2-EC (25, 29, 41). IFNα2 residues that have the potential to interact with IFNAR1-EC must be exposed in the IFNα2/IFNAR2-EC complex. The exposure of the IFNα2 residues in the binary complex is shown in Figure 2 as grey bars. Figure 3 highlights residues whose chemical shifts were affected upon IFNAR1-EC binding. As shown in Figure 3A, the face of IFNα2 opposing the binding site for IFNAR2-EC comprises a large exposed surface affected by IFNAR1-EC binding. This surface can be divided into two distinct areas. The upper surface contains residues Y85, T86, Y89 and L92 in the C-helix, residues H57, Q61, Q62, and N65 in the B-helix and T69 which connects the B and the B' helices. The lower adjacent surface contains residues A97, V99, and I100 in the C-helix, Q101, G102 and T106 in the CD-loop. Residues L3 and G104 which exhibit pronounced changes in chemical shifts upon IFNAR1-EC binding (colored yellow in Figure 3) are adjacent to the second patch. On the basis of the changes in the HSQC spectrum, the exposure of many of the cytokine residues in these two surfaces, the involvement of the majority of the residues in the upper patch in IFNAR1 binding (25, 29, 41) and the proximity of the two surfaces, we suggest that both of these adjacent patches form the binding site for IFNAR1-EC.
Residues that are marked as “overlapping” are colored green in Figures 2 and 3. They are marked as such when one of two or three overlapping peaks exhibited a change in chemical shift upon IFNAR1-EC binding or when one of the peaks disappeared, but due to cross peak overlap we could not conclude which of these was affected by IFNAR1-EC binding. Statistically, these residues have a 33–50% chance of being in the IFNAR1-binding interface. Not surprisingly there are numerous such residues that are adjacent to the patches colored grey or yellow. One of these IFNα2 residues, E58, has been implicated in IFNAR1 binding by site directed mutagenesis studies (25). Other residues assigned as “overlapping” include S68, D71, D82, V105 and E107 that surround the surfaces identified in the present study.
Changes in the HSQC cross-peaks of residues in the face of IFNα2 containing the binding site for IFNAR2-EC
The face of IFNα2 on which the binding site for IFNAR2-EC is located contains 8 residues that disappeared upon IFNAR1-EC binding to the binary complex (colored grey in Figure 1–3) as well as four residues that significantly changed their chemical shift (colored yellow in Figures 1–3). These residues include Q20 in the end of the A-helix, R22, K23, I24 and F27 in the AB-loop, R149, S152, N156 and S157 in the E-helix and E159 and S160 in the flexile C-terminal tail and Q5 from the N-terminal tail. Of these residues F27, R149 and S152 were previously identified by site-directed mutagenesis and by NMR as being in the IFNα2 binding site for IFNAR2-EC (22–24). It has been suggested that the flexible C-terminal tail is also involved in IFNAR2-EC binding in some IFNs (42).
Changes in the HSQC cross peaks of buried residues
The HSQC spectrum of IFNα2 in the binary complex contains many cross peaks corresponding to buried residues (less than 20% exposure) that underwent significant changes upon binding of IFNAR1-EC (labeled by asterisks in Figure 2) and are not located in the binding sites for IFNAR1-EC and IFNAR2-EC. These residues include: C29 in the AB-loop, T52, I53, V55, I60, Q63 and F67 in the B-helix, S72 in the B'-helix, E87, L92, A97 (19.7% exposure), and C98 in the C-helix and S150, F151 and L157 in the E-helix. The changes in the HSQC cross peaks of these residues, indicate that they are located in internal regions of the protein that underwent some changes in their environment or conformation upon IFNAR1-EC binding. Changes in local structure, which result in changes in chemical shifts, are necessary in order to facilitate the proposed allosteric interaction between the IFNAR1-EC and IFNAR2-EC binding interfaces.
DISCUSSION
Mapping the binding site for IFNAR1-EC
The mapping of the binding site for IFNAR1-EC on IFNα2 bound to IFNAR2-EC presents a major challenge for NMR investigations due to the high molecular weight of the ternary complex (89 and 102 kDa for the complex with three and four IFNAR1-EC domains, respectively), the low concentration of the binary and ternary complexes that are soluble in aqueous solution (0.2 mM), the helical structure of IFNα2 that results in considerable overlap in the HSQC spectrum and the high pH at which the binary and the ternary complexes are stable (pH 8) resulting in fast exchange of exposed amide protons.
To map the binding site for IFNAR1-EC on IFNα2 we analyzed the perturbation in the 1H and 15N chemical shifts of IFNα2 bound to IFNAR2-EC upon the formation of the ternary complex IFNAR1-EC/ IFNα2/IFNAR2-EC. The HSQC spectrum of the ternary complex revealed that the cross-peaks of 34 residues vanished and three others experienced considerable reduction in intensity. Only a small number of residues (10 residues) exhibited changes in chemical shifts upon IFNAR1-EC binding and these changes were rather small (less than 0.1 ppm) indicating that they correspond to residues in the periphery of the regions affected by IFNAR1-EC binding.
Chemical shift perturbation is an indirect method for the determination of residues involved in the binding interface. Changes in chemical shift can be the result of direct binding or of other effects, such as allosteric changes, conformational changes upon binding etc. Other NMR methods like cross saturation transfer or deuterium exchange would allow the identification of residues directly involved in binding. However, these methods were not suitable for the IFNAR1-EC/IFNα2/IFNAR2-EC system, since most of the cross-peaks that were affected by IFNAR1-EC binding disappeared beyond detection in the ternary complex (37 residues).
By comparing the HSQC spectra of the binary and ternary complexes of IFNα2 we were able to map two main surfaces of IFNα2 that were affected upon the binding of IFNAR1-EC to the binary IFNα2/IFNAR2-EC complex. One surface that consists of two adjacent large patches is implicated in IFNAR1-EC binding and the other is located on the face of IFNα2 containing the binding site for IFNAR2-EC.
The upper section of the surface involved in IFNAR1-EC binding (Figure 3) consists of exposed residues at the center and C-terminal end of the B-helix as well as residues at the center of the C-helix (Figure 2). This patch overlaps very well with the IFNAR1-EC binding surface previously identified by site directed mutagenesis which suggested that residues H57, E58, Q61, F64, N65 and T69 in the B-helix and L80, Y85, Y89 located in the C-helix are involved in IFNAR1-EC binding to IFNα2 (25). The present study adds residues Q62, T86 and L92 to this site resulting in a significant increase in the protein-protein interface.
The NMR analysis could not confirm the involvement of L117 and R120 in IFNAR1-EC binding. These two residues were found to be involved in IFNAR1-EC binding by site-directed mutagenesis experiments. Mutation of residue L117, located in the D helix, resulted in a decrease in antiviral and antiproliferative activities (29). Unfortunately, the cross peak for L117 overlaps that of L88 and the cross-peak of one of these residues (It is not known which of them) exhibits changes in chemical shift upon IFNAR1-EC binding. Residue R120, located adjacent to residues E58, Q62, E113, and L117 was previously implicated in IFNAR1-EC binding (29). The R120A mutation and the charge reversal R120E mutation significantly decreased the antiviral and antiproliferative activity of IFNα2. However, no significant change in chemical shift upon IFNAR1-EC binding was observed for R120. Thus, R120 may play a structurally significant role rather than being involved in binding IFNAR1-EC directly.
The lower part of the IFNα2 binding site for IFNAR1-EC includes L3 at the N-terminal segment, A97, V99 and L100 in the C-terminal end of the C-helix, and Q101, G102, G104 and T106 in the CD-loop. This patch has not been implicated in IFNAR1-EC-binding previously. We suggest that this large surface containing 6 residues that disappeared upon IFNAR1-EC binding and two residues that significantly changed their chemical shift is involved in IFNAR1-EC binding in addition to the upper patch. Support for the existence of two IFNα2 patches involved in IFNAR1-EC binding comes from the observation that two of the four fibronectin domains of IFNAR1-EC are major contributors to IFN binding (43). This was later demonstrated also by the EM-derived model for the ternary complex (34). It could be that one of the two patches identified in this study interacts with one of IFNAR1-EC fibronectin domains while the other cytokine patch interacts with a second IFNAR1-EC fibronectin domain similar to what has been suggested by the model for the ternary complex that was based on cryo-EM (see below, (34)).
Evidence that binding of IFNs to IFNAR1-EC involves more than a single localized region of the cytokine was obtained also by studying IFNAR1-EC binding to IFNβ. A systematic mutational analysis of human IFNβ suggests involvement of residues in the B-helix (L63, I66, F67 and F70), the B'-helix (Q72), the C-helix (H93, N96, K99, T100) and the D-helix (H121, R128, L130, H131, K134) as well as in the DE-loops (K136 and E137) in the binding to IFNAR1-EC (28) (see Figure 4).
Sequence alignment (Figure 4) of IFNα2 and IFNβ (Figure 4) shows that the binding site for IFNAR1-EC on IFNα2 identified by NMR partially overlaps with the binding site of IFNAR1-EC on IFNβ determined by site-directed mutagenesis (28). Involvement of the B- and C-helices of IFNβ in IFNAR1-EC binding agrees with the present NMR study. However, the binding site on IFNβ involves also residues in the D-helix and the DE-loop which showed no changes in the HSQC spectrum of IFNα2 upon IFNAR1-EC binding.
Comparison with the EM structure of IFNAR2-EC/IFNα2/IFNAR1-EC
A model for the three-dimensional structure of the IFNAR2-EC/IFNα2/IFNAR1-EC complex was previously obtained (34) using the NMR derived model for the IFNAR2-EC/IFNα2 binary complex (24), and an homology model for IFNAR1-EC fitted into the low resolution electron density map of the ternary complex (34). Although the model of the ternary complex was not obtained on the basis of specific interactions between IFNAR1-EC and IFNα2, the fitting of the homology model of IFNAR1-EC into the EM structure is revealing in terms of identifying regions that could potentially participate in IFNAR1-EC binding. Overall the model suggests that three regions of IFNα2 are involved in IFNAR1-EC binding and these are: a) the N-terminal flexible tail of IFNα2, b) helix-B and c) helix-C (mainly residues 92–100) and the CD-loop (34). Indeed L3 and Q5 underwent changes in the HSQC spectrum upon IFNAR1-EC binding to the binary complex. However, most of the N-terminal tail residues could not be assigned. Therefore, there is no firm conclusion in the present study regarding the involvement of the N-terminus of the cytokine in the interaction with IFNAR1. Region b overlaps the Helix-B surface implicated in IFNAR1-EC binding in the present study. Region-C coincides with the large patch containing V99, I100, Q101, G102 and T106 found to be involved in IFNAR1-EC binding in the present study, thus providing further support that this patch is involved in IFNα2 binding rather than disappearing because of conformational changes.
Allostery in IFNAR1 binding to the IFNα2/IFNAR2-EC complex
Binding of IFNAR1-EC to the binary IFNα2/IFNAR2-EC complex caused significant changes in the cross-peaks of IFNα2 residues in the face of the molecule that contains the binding site for IFNAR2-EC. This patch includes three IFNα2 residues that were previously implicated in IFNAR2-EC binding by saturation-transfer experiment (F27, R149 and S152) (see Figure 2 and (24)). IFNα2 residues N156, L157, Q158, E159 and S160 form a patch of five residues the cross peaks of which disappeared upon IFNAR1-EC binding to the binary complex IFNα2/IFNAR2-EC. This patch is flanked on one side by R149 and S152 and on the other side by R162 and E165 that were previously found to be involved in IFNAR2-EC binding in the NMR-derived model for the binary complex (24).
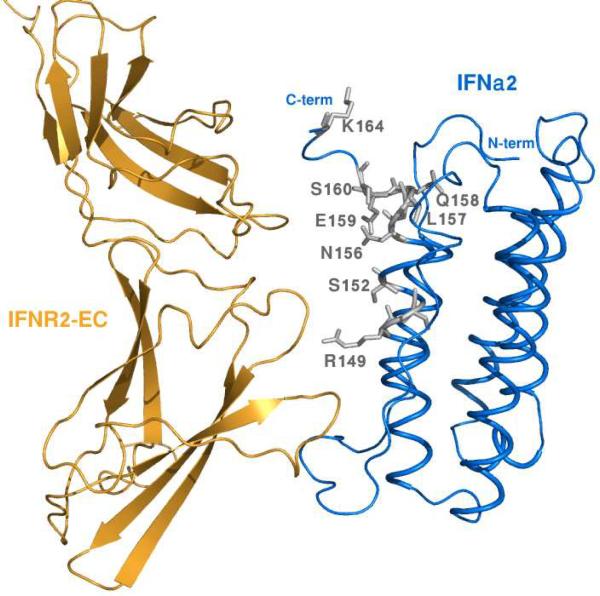
Figure 5 shows the proximity between the IFNα2 C-terminal tail and IFNAR2-EC. The closest distance between N156, E159 and S160 and IFNAR2-EC atoms is 3.0, 8.0 and 6.0 Å, respectively. It has been reported that mutations in the C-terminal tail caused up to 20-fold difference in the binding affinity of IFNs to IFNAR2-EC (41). Although this tail does not contribute significantly to IFNα2 binding to IFNAR2-EC it does contribute to IFNα8 binding to IFNAR2-EC (42). It has been suggested that the flexible C-terminal tail becomes structured upon IFNAR2-EC binding (42). As shown in Figure 5 IFNα2 residues N156, E159 and S160 could potentially interact with the β9–β10 and/or β13–β14 loops of IFNAR2-EC if IFNα2 was tilted slightly towards the C-fibronectin domain of IFNAR2-EC or if the flexible IFNα2 C-terminal tail adopted a structure that brought it closer to IFNAR2-EC. Therefore, we propose that the IFNα2 binding site for IFNAR2-EC undergoes allosteric conformational changes upon IFNAR1-EC binding to the binary IFNα2 /IFNAR2-EC complex.
FIGURE 5.
A side view of the IFNα2/IFNAR2-EC binding interface. The IFNα2 and the IFNAR2-EC are colored blue and orange respectively. The E-Helix and the C-terminal tail residues that disappeared upon IFNAR1-EC binding to the binary complex are shown in stick representation and in gray.
Ten residues out of 17 residues in the IFNα2 B-helix disappeared upon IFNAR1-EC binding to the binary complex. Of these ten residues, only three, namely Q61, Q62 and N65 are exposed and H57 is marginally exposed (17%). The other six residues within helix-B that disappeared upon IFNAR1-EC binding are buried. The disappearance of such a large number of buried residues indicates that helix B, which forms together with the E-helix the center of the five-helix bundle, undergoes significant movement upon IFNAR1-EC binding to the binary complex. Interestingly also 6 residues of the 9-residue C-terminal segment of the E-helix undergo considerable changes in chemical shift or disappear upon IFNAR1-EC binding. Of these 6 residues 4 are buried and do not interact with IFNAR2-EC. It is plausible that these changes in the central helices B and E reflect their involvement in modulating the allosteric changes in IFNα2 that are transmitted to the IFNAR2-EC binding site. This movement could involve also the C-helix.
The allosteric changes in the IFNAR2-EC binding site caused by IFNAR1-EC binding might affect the orientation of IFNAR2 with respect to IFNAR1 and thereby influence the cross-phosphorylation of the IFNAR1-associated Tyk2 protein and the IFNAR2-assciated Jak1. These phosphorylations initiate the intracellular signal transduction cascade leading to the antiviral and anti-proliferative response to interferon.
In a previous study we reported that H57 and adjacent residues (V55, M59, V60, Y89 and E96) undergo chemical shift changes upon binding of IFNAR2-EC to IFNα2 (24). These residues are located on the face of IFNα2 opposing the binding site for IFNAR2-EC. Of these residues H57, V60 and Y89 were mapped in this study as being in the binding site for IFNAR1-EC. However, in the earlier investigation we could not draw any firm conclusion regarding allostery since the free IFNα2 spectrum was measured at pH 3.5 and that of the binary IFNα2/IFNAR2-EC complex was measured at pH 8.0. A change in the protonation state of H57 could well have been the reason for the observed changes in its chemical shift between IFNα2 and the IFNα2/IFNAR2-EC complex. The present study provides unequivocal evidence for the existence of an allosteric interaction between IFNAR1-EC and IFNAR2-EC binding sites mediated by the IFNα2 cytokine since all HSQC spectra were measured under the same conditions.
Conclusions
The present study together with previous site-directed mutagenesis and EM studies suggest that the binding site for IFNAR1-EC on IFNα2 consists of two large patches located on the IFNα2 face opposing the binding site for IFNAR2-EC. Most importantly, the present NMR study provides an unequivocal indication for the communication between the IFNα2 binding sites for IFNAR1-EC and IFNAR2-EC resulting in some changes in conformation in the binding site for IFNAR2-EC caused by IFNAR1-EC binding. These conformational changes caused by allosteric effects, which might be translated to the cytoplasmic part of the receptor, likely play an important role in the initiation of the intra-cellular signal transduction cascade.
ACKNOWLEDGMENTS
We are most grateful to Ms. Rina Levy for the preparation of the IFNAR2-EC and the labeled IFNα2 and to Dr. Fred Naider for extensive discussions and comments on the manuscript.
This study was supported by the Israel Science Foundation, NIH Grant GM53329 and the Kimmelman Center. J.A. is the Dr. Joseph and Ruth Owades Professor of Chemistry.
Abbreviations
- D1, D2, D3, and D4 the first, second, third and fourth fibronectin subdomains in the extracellular region of IFNAR1, respectively
- Δδ
chemical shift difference
- EM
electron microscopy
- EPO
erythropoietin
- hGH
human growth hormone
- HSQC
heteronuclear single-quantum coherence
- IFN
interferon
- IFNAR1-EC
the extracellular domain of subunit-1 of the receptor for α- and β-Interferons
- IFNAR2-EC
the extracellular domain of subunit-2 of the receptor for α- and β-Interferons
- IL-4
interleukin-4
- Jak1
Janus kinase 1
- KD
the equilibrium dissociation constant
- NOE
Nuclear Overhauser enhancement
- TROSY
transverse relaxation optimized spectroscopy
- Tyk2
tyrosine kinase 2
- U-
preceding the molecule's name indicates unlabeled molecule.
References
- 1.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 2.Novick D, Cohen B, Rubinstein M. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell. 1994;77:391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 3.Uze G, Lutfalla G, Mogensen KE. Alpha and beta interferons and their receptor and their friends and relations. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- 4.Piehler J, Schreiber G. Mutational and structural analysis of the binding interface between type I interferons and their receptor Ifnar2. J Mol Biol. 1999;294:223–237. doi: 10.1006/jmbi.1999.3230. [DOI] [PubMed] [Google Scholar]
- 5.Lamken P, Gavutis M, Peters I, Van der Heyden J, Uze G, Piehler J. Functional cartography of the ectodomain of the type I interferon receptor subunit ifnar1. J Mol Biol. 2005;350:476–488. doi: 10.1016/j.jmb.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Ozbek S, Grotzinger J, Krebs B, Fischer M, Wollmer A, Jostock T, Mullberg J, Rose-John S. The membrane proximal cytokine receptor domain of the human interleukin-6 receptor is sufficient for ligand binding but not for gp130 association. J Biol Chem. 1998;273:21374–21379. doi: 10.1074/jbc.273.33.21374. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham BC, Ultsch M, De Vos AM, Mulkerrin MG, Clauser KR, Wells JA. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science. 1991;254:821–825. doi: 10.1126/science.1948064. [DOI] [PubMed] [Google Scholar]
- 8.Gent J, Van Den Eijnden M, Van Kerkhof P, Strous GJ. Dimerization and signal transduction of the growth hormone receptor. Molecular endocrinology (Baltimore, Md. 2003;17:967–975. doi: 10.1210/me.2002-0261. [DOI] [PubMed] [Google Scholar]
- 9.Krause CD, Pestka S. Evolution of the Class 2 cytokines and receptors, and discovery of new friends and relatives. Pharmacology & therapeutics. 2005;106:299–346. doi: 10.1016/j.pharmthera.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Bernat B, Pal G, Sun M, Kossiakoff AA. Determination of the energetics governing the regulatory step in growth hormone-induced receptor homodimerization. Proc Natl Acad Sci U S A. 2003;100:952–957. doi: 10.1073/pnas.0235023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remy I, Wilson IA, Michnick SW. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 12.Gavutis M, Lata S, Lamken P, Muller P, Piehler J. Lateral ligand-receptor interactions on membranes probed by simultaneous fluorescence-interference detection. Biophys J. 2005;88:4289–4302. doi: 10.1529/biophysj.104.055855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavutis M, Jaks E, Lamken P, Piehler J. Determination of the two-dimensional interaction rate constants of a cytokine receptor complex. Biophys J. 2006;90:3345–3355. doi: 10.1529/biophysj.105.072546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karpusas M, Nolte M, Benton CB, Meier W, Lipscomb WN, Goelz S. The crystal structure of human interferon beta at 2.2-A resolution. Proc Natl Acad Sci U S A. 1997;94:11813–11818. doi: 10.1073/pnas.94.22.11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senda T, Shimazu T, Matsuda S, Kawano G, Shimizu H, Nakamura KT, Mitsui Y. Three-dimensional crystal structure of recombinant murine interferon-beta. Embo J. 1992;11:3193–3201. doi: 10.1002/j.1460-2075.1992.tb05396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaus W, Gsell B, Labhardt AM, Wipf B, Senn H. The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution. J Mol Biol. 1997;274:661–675. doi: 10.1006/jmbi.1997.1396. [DOI] [PubMed] [Google Scholar]
- 17.Radhakrishnan R, Walter LJ, Hruza A, Reichert P, Trotta PP, Nagabhushan TL, Walter MR. Zinc mediated dimer of human interferon-alpha 2b revealed by X-ray crystallography. Structure. 1996;4:1453–1463. doi: 10.1016/s0969-2126(96)00152-9. [DOI] [PubMed] [Google Scholar]
- 18.Radhakrishnan R, Walter LJ, Subramaniam PS, Johnson HM, Walter MR. Crystal structure of ovine interferon-tau at 2.1 A resolution. J Mol Biol. 1999;286:151–162. doi: 10.1006/jmbi.1998.2480. [DOI] [PubMed] [Google Scholar]
- 19.Chill JH, Quadt SR, Levy R, Schreiber GE, Anglister J. NMR structure of the human type I interferon receptor reveals the molecular basis for ligand binding. Structure(Camb.) 2003;11:791–802. doi: 10.1016/s0969-2126(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 20.Chill JH, Quadt SR, Anglister J. NMR backbone dynamics of the human type I interferon binding subunit, a representative cytokine receptor. Biochemistry. 2004;43:10127–10137. doi: 10.1021/bi049606g. [DOI] [PubMed] [Google Scholar]
- 21.Chuntharapai A, Gibbs V, Lu J, Ow A, Marsters S, Ashkenazi A, De Vos A, Jin Kim K. Determination of residues involved in ligand binding and signal transmission in the human IFN-alpha receptor 2. J Immunol. 1999;163:766–773. [PubMed] [Google Scholar]
- 22.Roisman LC, Piehler J, Trosset J-Y, Scheraga HA, Schreiber GE. Structure of the interferon-reeptor complex determined by distance constraints from double mutant cycles and flexible docking. Proc. Natl. Acad. Sci. USA. 2001;98:13231–13236. doi: 10.1073/pnas.221290398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piehler J, Roisman LC, Schreiber G. New structural and functional aspects of the IFN - receptor interaction revealed by comprehensive mutational analysis of the binding interface. J Biol Chem. 2000;275:40425–40433. doi: 10.1074/jbc.M006854200. [DOI] [PubMed] [Google Scholar]
- 24.Quadt-Akabayov SR, Chill JH, Levy R, Kessler N, Anglister J. Determination of the human type I interferon receptor binding site on human interferon-alpha2 by cross saturation and an NMR-based model of the complex. Protein Sci. 2006;15:2656–2668. doi: 10.1110/ps.062283006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roisman LC, Jaitin DA, Baker DP, Schreiber G. Mutational analysis of the IFNAR1 binding site on IFNalpha2 reveals the architecture of a weak ligand-receptor binding-site. J Mol Biol. 2005;353:271–281. doi: 10.1016/j.jmb.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 26.Stroud RM, Wells JA. Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci STKE. 2004;2004:re7. doi: 10.1126/stke.2312004re7. [DOI] [PubMed] [Google Scholar]
- 27.Hu R, Bekisz J, Schmeisser H, McPhie P, Zoon K. Human IFN-alpha protein engineering: the amino acid residues at positions 86 and 90 are important for antiproliferative activity. J Immunol. 2001;167:1482–1489. doi: 10.4049/jimmunol.167.3.1482. [DOI] [PubMed] [Google Scholar]
- 28.Runkel L, deDios C, Karpusas M, Betzenhauser M, Muldowney C, Zafari M, Benjamin CD, Miller S, Hochman PS, Whitty A. Systematic mutational mapping of sites on human interferon-beta-1a that are important for receptor binding and functional activity. Biochemistry. 2000;39:2538–2551. doi: 10.1021/bi991631c. [DOI] [PubMed] [Google Scholar]
- 29.Pan M, Kalie E, Scaglione BJ, Raveche ES, Schreiber G, Langer JA. Mutation of the IFNAR-1 receptor binding site of human IFN-alpha2 generates type I IFN competitive antagonists. Biochemistry. 2008;47:12018–12027. doi: 10.1021/bi801588g. [DOI] [PubMed] [Google Scholar]
- 30.Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, Van der Heyden J, Uze G, Schreiber G. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol Cell Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalie E, Jaitin DA, Abramovich R, Schreiber G. An interferon alpha2 mutant optimized by phage display for IFNAR1 binding confers specifically enhanced antitumor activities. J Biol Chem. 2007;282:11602–11611. doi: 10.1074/jbc.M610115200. [DOI] [PubMed] [Google Scholar]
- 32.Cutrone EC, Langer JA. Identification of critical residues in bovine IFNAR-1 responsible for interferon binding. J Biol Chem. 2001;276:17140–17148. doi: 10.1074/jbc.M009663200. [DOI] [PubMed] [Google Scholar]
- 33.Cajean-Feroldi C, Nosal F, Nardeux PC, Gallet X, Guymarho J, Baychelier F, Sempe P, Tovey MG, Escary JL, Eid P. Identification of residues of the IFNAR1 chain of the type I human interferon receptor critical for ligand binding and biological activity. Biochemistry. 2004;43:12498–12512. doi: 10.1021/bi049111r. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Strunk JJ, Lamken P, Piehler J, Walz T. The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J Mol Biol. 2008;377:715–724. doi: 10.1016/j.jmb.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chill JH, Nivasch R, Levy R, Albeck S, Schreiber GE, Anglister J. The human interferon receptor: NMR-based modeling of the IFN-a2 binding site, and observed ligand-induced tightening. Biochemistry. 2002;41:3575–3585. doi: 10.1021/bi011778f. [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based On Unix Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Johnson BA, Blevins RA. NMRView - a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 38.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 40.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 41.Uze G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Curr Top Microbiol Immunol. 2007;316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- 42.Slutzki M, Jaitin DA, Yehezkel TB, Schreiber G. Variations in the unstructured C-terminal tail of interferons contribute to differential receptor binding and biological activity. J Mol Biol. 2006;360:1019–1030. doi: 10.1016/j.jmb.2006.05.069. [DOI] [PubMed] [Google Scholar]
- 43.Goldman LA, Cutrone EC, Dang A, Hao X, Lim JK, Langer JA. Mapping human interferon-alpha (IFN-alpha 2) binding determinants of the type I interferon receptor subunit IFNAR-1 with human/bovine IFNAR-1 chimeras. Biochemistry. 1998;37:13003–13010. doi: 10.1021/bi980073j. [DOI] [PubMed] [Google Scholar]