Abstract
The HCV core protein is implicated in diverse aspects of HCV-induced pathogenesis. There is a paucity of information on core in acute hepatitis C infection. We analyzed core gene sequences and protein functions from 13 patients acutely infected with HCV genotype 1. While core isolates differed slightly between patients, core quasispecies were relatively homogeneous within a patient. In 2 of 4 patients studied temporally, core quasispecies did not change over time. Comparison with more than 2700 published core isolates indicated that amino acid changes from a prototype reference strain found in acute core isolates were present in chronically infected persons at low frequency (6.4%, range 0-32%). Core isolates associated with lipid droplets (LDs) to similar degrees in Huh7 cells. Core diffusion in cells was not affected by non-conservative changes F130L and G161S in the lipid targeting domain of core. Core isolates inhibited ISRE- and NF-κB-dependent transcription, and TNF-α- induced nuclear translocation of NF-κB and were also secreted from Huh7 cells. The data suggest that upon transmission, core quasispecies undergo genetic homogenization associated with amino acid changes that are rarely found in chronic infection, and that despite genetic variation, acute core isolates retain similar functions in vitro.
Keywords: hepatitis C, HCV, core, acute infection
INTRODUCTION
Acute HCV infection is frequently subclinical and not associated with symptoms, making recognition of infection in the early stages problematic. Unfortuantely, acute HCV infection frequently progresses to chronic infection. A number of mechanisms can be forwarded to explain these results including immune escape from humoral and cellular responses by rapidly evolving HCV quasispecies, exhaustion of viral specific T cells, and induction of Treg cells[1]. An unanswered question is whether the ability to clear acute infection occurs earlier and depends on the interaction of HCV with innate antiviral and cellular responses mediated by the infected hepatocyte.
HCV core is a structural protein, binding viral RNA and forming the nucleocapsid [2]. The core protein represents the first protein in the polyprotein, followed by two glycoproteins, E1 and E2. The immature form of HCV core contains 191 amino acids, and has been separated into three general domains [3]. Domain I (aa 1 – 117) is highly basic and hydrophilic, and thought to be responsible for binding RNA and mediating capsid assembly. Domain II (aa118-171) is hydrophobic and mediates interactions with lipids and membrane proteins. Domain III (aa 172 - 191) is at the extreme C-terminus of the immature core protein, and corresponds to the signal sequence (SS) for E1.
After core is translated, the nascent polyprotein is targeted to the ER translocation channel by the E1 SS. A host enzyme located in the ER, signal peptidase, cleaves just proximal to the E1 SS, releasing the immature form of core from the polypeptide [4, 5]. A different endoplasmic reticulum (ER) enzyme, signal peptide peptidase (SPP), then cleaves just before the E1 SS liberating the mature form of HCV core at the cytoplasmic face of the ER [3, 6]. After cleavage, core can undergo a number of possible fates, including assembly into virions[7], targeting to other organelles, and interaction with host proteins resulting in modulation of various cellular processes[8].
Core is a major player in HCV assembly at lipid droplets[9, 10], and also perturbs many signaling pathways [2]. Moreover, core quasispecies differ between early and advanced liver disease and liver tumor and non-tumor tissue [11-16], suggesting that core quasispecies display varied functions, which can be observed when isolates are expressed in cultured cells [2, 17]. In the current report, we hypothesized that core quasispecies isolated from patients with acute infection would have distinct phenotypes when expressed in cultured hepatocytes.
MATERIALS AND METHODS
Patients
Thirteen patients were analyzed in the current study. Of these 13 patients, we performed temporal analysis on 2 or 3 timepoints in 4 patients. Patients with acute HCV were recruited from sites including academic medical centers (Seattle, Portland), blood banks (Seattle) and an IDU research site (Seattle). This study was approved by the Institutional Review Boards for all participating institutions. Informed consent was obtained from all participants. Two methods were used to identify patients with acute HCV: (1) a positive HCV antibody or HCV RNA PCR assay in a participant with a documented negative anti-HCV test within the past year; (2) a positive anti-HCV assay in a participant with clinical hepatitis, detectable serum HCV RNA, serum ALT > 10 times the upper limit of normal, and negative tests for hepatitis B surface antigen and hepatitis A IgM antibody, as described recently [18]. Diagnoses of symptomatic acute hepatitis C were made after patients sought medical attention for symptoms including nausea, anorexia, abdominal pain, malaise, fever, and jaundice. Serum samples analyzed in this study represented the first HCV RNA positive timepoint where the HCV core gene could be amplified.
Core Amplification and Cloning
RNA was extracted from patient serum as described [19]. Core was amplified from viral RNA by RT-PCR using various primers (available on request), and using primers sets previously described [20]. Core was initially cloned into pENTR/D-TOPO plasmid (Gateway system, Invitrogen, Carlsbad, CA). Core genes were then recombined into pcDNA3.1/nV5 DEST vector according to manufacturer’s specifications. All clones were verified by sequencing. Sequences were submitted to Genbank under accession numbers FJ911699 to FJ911736.
Core genes were also subcloned from pDEST as non-tagged core proteins. For this, we used PCR followed by restriction enzyme digestion to move core genes into pcDNA3.1. Domain 2 of core genes 6-8-2, 4-7-1, and 3-9-5 was also cloned as a EGFP fusion protein using primers tagged with BglII and KpnI and ligated into similarly digested pEGFP-C1 (Clontech).
Phylogenetic Analysis
Core genes were analyzed by Neighbor Joining method and standard bootstrap analysis (with 1000 replicates) with reference gentoype 1a and 1b strains (accession numbers M62321 and AF333324.1, respectively) using MacVector 10.2 software. Core sequences were also compared to a FASTA alignment of 2000+ sequences downloaded from the European HCV database (EuHCVdb®) at http://euhcvdb.ibcp.fr/euHCVdb/.
Core Expression and Functional Analyses
Endotoxin free core plasmids were purified and transfected with control plasmids into Huh7 cells as described [21-23]. Twenty-four to 48 hours later, cells were subjected to additional manipulations or treatments and protein lysates, western blots, and luciferase reporter gene assays were performed as described [24].
For immunofluorescent detection of core, Huh7 cells were plated on chamber slides, transfected with core plasmids, and fixed in 4% paraformaldehyde at 48 hours after transfection. Fixed cells were blocked and incubated 2 hours with primary antibodies followed by Alexa Fluor 488 or 568 labeled secondary antibodies for 1 hour. Lipid droplets were detected by staining cells with BODIPY 493/503 dye (D3922; Invitrogen) during the final wash. Lipid droplets were also detected by staining for ADRP, a lipid droplet associated protein, as described [25]. Images were recorded using a laser scanning confocal microscope (Bio-Rad LS2000).
For detection of core secretion, Huh7 cells were transfected with core genes or control plasmids. As a positive control for core secretion, cells were infected with JFH-1 at an m.o.i. of 0.01. Culture supernatants were harvested and core protein was detected by ELISA (Ortho-HCV antigen, Ortho-Clinical Diagnostics).
Fluorescence Recovery After Photobleaching (FRAP) Analysis
Huh7 cells were seeded onto glass bottom microwell dishes and transfected with plasmids expressing the GFP-tagged proteins. FRAP analysis and image recording were conducted 12-14 hours after transfection with a LSM510 META inverted confocal microscope (Zeiss).
Prior to photobleaching, cell media was replaced with prewarmed (to 37°C) DMEM lacking phenol red and supplemented with 2% fetal calf serum. For photobleaching, selected regions of cells were bleached at 100% laser power (488nm laser line) for approximatey 1 second. Images were taken at 1 second intervals using 2% laser power. The intensity of fluorescence for each cell was expressed as a percentage of the pre-bleach level and plotted against time. FRAP data were generated from 6-8 cells for each GFP-tagged protein and the average values for fluorescence intensity at each time point were calculated.
RESULTS
Patients with Acute HCV Infection
The characteristics of the 13 patients are shown in Table 1. Six patients were male, seven patients were female, and patients had viral loads ranging from 45,000-6.5 million copies/ml. All patients were infected with HCV genotype 1, 10 patients with HCV-1a and 3 with HCV-1b. Of the 13 patients, all but 1 progressed to chronic HCV infection.
TABLE 1. Patient clinical and virological characteristics.
Samples highlighted in pink represent isolates from different timepoints from the same patient. Genetic changes in core represent changes compared to prototype genotype 1a core sequences (Genbank accession number M62321). Conservative changes are defined as mutations that maintain amino acid charge and polarity, while non-conservative changes are defined as mutations that result in a change in amino acid charge and polarity.
| Denver ID |
Sex | Sample month |
Genotype | Titer (10E3/ml) |
Outcome | Seattle ID |
Clone ID |
Conservative Changes |
Non-conservative Changes |
|---|---|---|---|---|---|---|---|---|---|
| TN 106 | F | m0 | 1a | 5186 | Resolver | 1 | 1-3-2 | R101H, A147V | - |
| HS 124 | F | m0 | 1b | 52 | Chronic | 3 | 3-9-5 | V187I | T75A, T110N |
| HS111 | M | m0 | 1a | 53 | Chronic | 4 | 4-7-1 | F130L | - |
| HS 111 | M | m2 | 1a | 3054 | Chronic | 21 | 21-4-3 | T110S, F130L | - |
| HS 108 | F | m0 | 1a | 66 | Chronic | 5 | 5-4-1 | R43K | C91R |
| TN 103 | M | m0 | 1a | 6520 | Chronic | 8 | 8-1-1 | A189V, A191L | S190R, |
| HS 102 | M | m0 | 1b | 620 | Chronic | 12 | 12-2-1 | C91M, T110S, V187I | R70Q, T75A |
| PD 121 | F | m2 | 1a | 2451 | Chronic | 14 | 14-3-1 | T110S | A191P |
| TN 104 | M | m0 | 1b | 608 | Chronic | 15 | 15-3-2 | A147V, V187I | D68A |
| TN 104 | M | m6 | 1b | 806 | Chronic | 29 | 29-10-1 | A147V, V187I | D68A |
| HS 115 | F | m0 | 1a | 114 | Chronic | 16 | 16-5-1 | - | - |
| TN108 | M | m6 | 1a | na | Chronic | 19 | 19-4-1 | R101H, A147V | - |
| PD 106 | M | m0 | 1a | 884 | Chronic | 6 | 6-8-2 | - | P49T, G161S |
| PD 106 | M | m2 | 1a | 1009 | Chronic | 30 | 30-4-4 | - | G45S, P49T, G161S |
| PD 106 | M | m12 | 1a | 1223 | Chronic | 24 | 24-10-1 | - | P49T, G161S |
| PD108 | F | m9 | 1a | 119 | Chronic | 41 | 41-2-3 | - | T75A |
| TN101 | F | m2 | 1a | Chronic | 28 | 28-6-1 | WT | - | |
| TN 101 | F | m4 | 1a | 1248 | Chronic | 27 | 27-9-5 | WT | - |
Core Isolates Are Conserved in Acute Infection
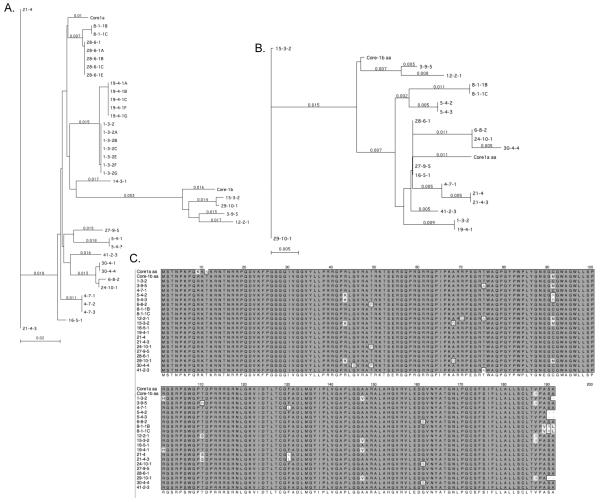
Phylogenetic analysis of core nucleotide sequences aligned relative to HCV-1 is shown in Figure 1A. Two patients were infected with identical sequences (Figure 1; clone 16-5-1 from patient HS115 and clones 27-9-5 and 28-6-1 from patient TN101). In four patients (HS111, TN104, PD106, and TN101), we analyzed clones from 2 or 3 time points during a 12 month period. Clones 27-9-5 and 28-6-1 from patient TN101 isolated 2 months apart occupied different branches of the nucleotide tree (Figure 1A), but had identical amino acid sequences (Figure 1B,C). Clones 4-7-1 and 21-4-3 from patient HS111 isolated 2 months apart, were also genetically distinct at the nucleotide level, but at the protein level, were highly related, except for the acquisition of a T110S amino acid change at 2 months. Clones 15-3-2 and 29-10-1 from patient TN104 were highly related at both the nucleotide and amino acid levels (Figure 1A,B). Clones 6-8-2, 30-4-4, and 24-10-1 were isolated from patient PD106 at months 0, 2, and 12 of acute infection. 6-8-2 was more closely related to 24-10-1, and both clones were slightly distinct from 30-4-4, because of the acquisition of the G45S change in clone 30-4-4 at month 2. Moreover, sequencing of 3-8 clones per sample in select patients revealed that HCV core nucleotide and amino acid sequences were homogeneous during acute infection (Figure 1 and data not shown). Thus, there was little genetic variation in the HCV core gene during acute infection within a patient, and in 2 of 4 patients studied, core quasispecies did not change over time.
Figure 1. Phylogenetic analysis of HCV core quasispecies.
Core genes were analyzed by Neighbor Joining method and standard bootstrap analysis (with 1000 replicates) with reference gentoype 1a and 1b strains (accession numbers M62321 and AF333324.1, respectively) using MacVector 10.2 software. A, nucleotide alignments. B, amino acid alignments. C, Alignment of HCV core proteins isolated from acute infection. Clustal alignments were performed with MacVector.
Unique Spectrum of Genetic Variation During Acute Infection
Table 1 and Figure 1C summarizes the genetic variation in the core protein. Relative to HCV-1a, we found 19 mutations among all clones, with 10/19 representing non-conservative amino acid changes, defined as changes in amino acid polarity. Of the ten non-conservative mutations, seven mutations mapped to the domain 1 in the amino terminus of the protein (G45S, R49T, D68A, R70Q, T75A, C91R, T110N).
An A147V amino acid change in the D2 domain of core in JFH1 affects the binding strength of D2 to lipid droplets significantly increasing virus production [26]. Interestingly, patients 1, 15, 19, and 29 contained this substitution, but they did not appear to have higher viral loads compared with other patients. We also observed several changes in Domain 3 of core in the signal peptide (V187I, A189V, A191L, S190R, A191P).
The vast majority of published core sequences are derived from chronically infected patients. Table 2 compares the genetic variation in core identified in the current study with 2757 published core sequences. Amino acid changes are relative to the prototype genotype 1a strain, accession number M62321. Changes C91R and S190R were not found in any published sequences. Changes G45S, G161S, and A191P were found in published core sequences, but at low frequency (0.07 to 0.98%). Of the 11 amino acid changes found in acutely infected patients, 7 were found in chronically infected patients less than 5% of the time. Amino acid changes T75A, T110N, F130L and P49T detected in our acutely infected cohort were found more frequently in isolates from chronically infected patients. The data suggest that the spectrum of genetic variation in core quasispecies during acute infection is different from chronic infection.
TABLE 2. Frequency of genetic changes in acute core isolates relative to chronic isolates.
Calculations are based on comparison to 2757 core sequences from the European HCV Database. Amino acid change in core reflect changes compared to prototype genotype 1a core sequences (Genbank Accession number M62321).
| Amino Acid Change in Acute HCV |
Number of Amino Acid Changes in Chronic HCV Isolates |
Frequency of Amino Acid Change in Chronic HCV |
|---|---|---|
| P49T | 184 | 6.67 |
| G45S | 2 | 0.07 |
| D68A | 96 | 3.48 |
| R70Q | 122 | 4.43 |
| T75A | 886 | 32.14 |
| C91R | 0 | 0.00 |
| T110N | 404 | 14.65 |
| F130L | 226 | 8.20 |
| G161S | 27 | 0.98 |
| S190R | 0 | 0.00 |
| A191P | 2 | 0.07 |
| AVG | 6.43 |
Acute HCV Core Isolates Associate With Lipid Droplets
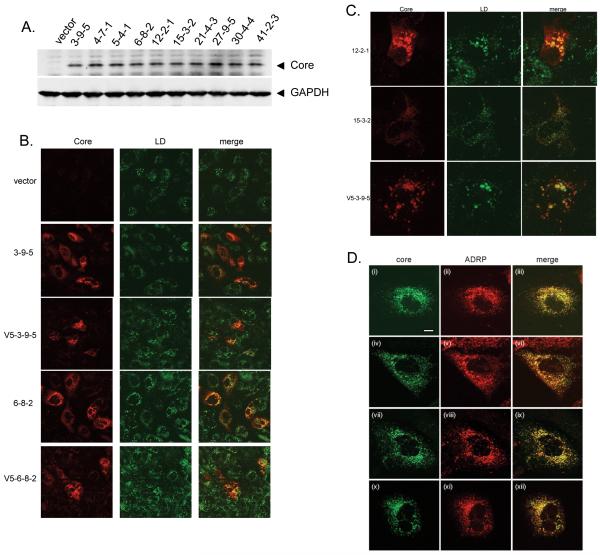
Nontagged and V5-Tagged Core isolates were readily expressed in Huh7 cells (Figure 2A and data not shown). Since core has been shown to associate with lipid droplets [27], the site of HCV assembly [10], we examined the ability of core isolates to associate with the organelles. Figure 2B demonstrates that the core isolates associated with lipid droplets, which were stained with BODIPY 493/503 dye, as evidenced by orange staining consistent with colocalization. Figure 2C provides higher magnification images of the colocalization of core with the lipid droplets. In fact, the concentric dye staining pattern was consistent with core surrounding lipid droplets [10]. Moreover, core isolates consistently co-localized with the lipid droplet-associated protein, ADRP (Figure 2D).
Figure 2. Expression and lipid targeting of HCV core isolates.
A, Huh7 cells were transfected with 1.0μg of each core plasmid and cytoplasmic protein lysates were harvested 24 hours later. Core protein was detected by western blotting. B, Confocal microscopy of core associated with lipid droplets. Core plasmids were transected into Huh7 cells and 48 hours later, cells were fixed and core protein detected with a core monoclonal antibody, followed by AlexaFluor568 conjugated anti-mouse antibody. Lipid droplet specific BODIPY 493/503 dye was added during the final wash. C, higher magnification images show core colocalization with lipid droplets. D, Colocalization of core with ADRP at the surface of lipid droplets. Huh-7 cells were transfected with plasmids for 2 days and then fixed with methanol at −20°C. Cells were probed with core (R308) and anti-ADRP antibodies. Images were recorded by confocal microscopy and are shown for core expressed by the following constructs: panels i-iii, 3-9-5; panels iv-vi, 4-7-1; panels vii-ix, 6-8-2; panels x-xii, 12-2-1. The scale bar in panel i represents 10μm.
Core Diffusion and Secretion In Huh7 Cells
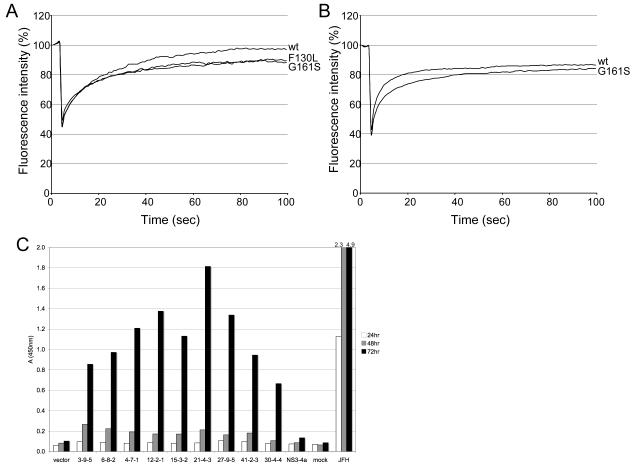
In a recent report [26], members of our team established that domain 2 (D2) of core is mobile on lipid droplets. The extent of mobility can be determined by measuring the recovery of fluorescence following photobleaching of selected regions in live cells that express D2 domains linked to GFP [26]. Sequences encoding the genotype 1 D2 domains of 3-9-5 (wt), 6-8-2 (G161S) and 4-7-1 (F130L) were cloned downstream of GFP. The constructs were transfected into Huh-7 cells and FRAP analysis was conducted in live cells. For comparative purposes, we also performed FRAP analysis with GFP chimeras containing the wt D2 domain of genotype 2a JFH1 and a G161S mutant that was equivalent to 6-8-2.
All of the GFP fusion proteins for the wt and mutant forms of D2 were directed to lipid droplets (data not shown). As observed for other HCV strains [26], the D2 domain of 3-9-5 was mobile and fluorescence recovery to about 90% of pre-bleach levels was achieved by 40 seconds after bleaching (Figure 3A). Both G161S and F130L mutants gave similar patterns as the wt D2 sequence although there was a slight reduction (5%) in the extent of fluorescence recovery (Figure 3A). Similarly, fluorescence recovery for the D2 domain of JFH1 reached 85% by 40 seconds and mutation of glycine to serine at position 161 lowered fluorescence recovery by about 5% (Figure 3B). The data are consistent with our observation that introduction of 161S has no significant effect on the amount of infectious virus produced by JFH1 in cell culture (McLauchlan et al. unpublished data).
Figure 3. Core diffusion and secretion in Huh7 cells.
A, B, FRAP analysis of GFP chimeras linked to wild type (wt) and mutant D2 domains. Huh-7 cells were transfected with plasmids for 12-14 hours prior to photobleaching. FRAP analysis was conducted in live cells at 37°C. A, fluorescence recovery curves for wt (black line), G161S (red line) and F130L (blue line) genotype 1 D2 domains. B, fluorescence recovery curves for wt (black line) and G161S (red line) D2 domains for JFH1 (genotype 2a). Data were obtained from 6-8 cells for each construct. C, Core isolates are efficiently secreted from liver cell cultures. Core plasmids were transfected into Huh7 cells. As controls, cells were separately transfected with vector (pcDNA3.1), pcDNA3.1-NS3-4a, or infected with JFH-1 at an m.o.i. of 0.01. Supernatants were harvested at the indicated times and core detected by ELISA.
Since core has been shown to be secreted from cells [28, 29], we examined the core isolates for secretion into culture supernatants following transient transfection into Huh7 cells. Core was measured by ELISA. Figure 3C shows that all core isolates were efficiently secreted from cells by 72 hours post-transfection. However, core protein secretion from the individual clones was significantly lower than when cells were infected with JFH-1, where core secretion was detected as early as 24 hours and increased over time. The data are consistent with the involvement of other HCV proteins, such as NS5A, in lipid targeting and virus assembly [10, 30].
Core Isolates Disrupt Antiviral and Inflammatory Signaling
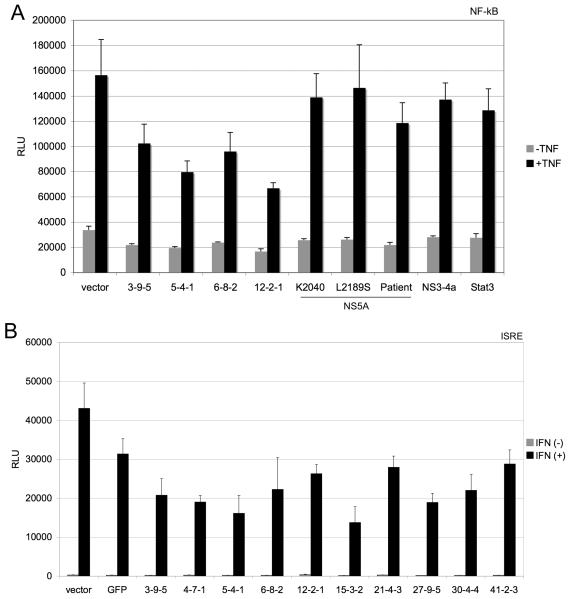
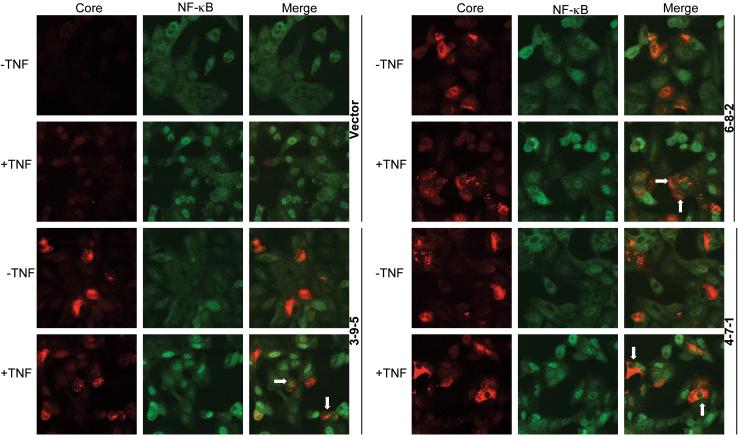
The 10 core genes that had amino acid differences were examined for modulation of NF-κB and ISRE signaling pathways since they represent key lines of inflammatory and antiviral defenses. The 10 core genes blocked TNF-α induced activation of the NF-κB transcription, as assessed by measuring transcription from the pRDII region of the IFN-B promoter (data not shown). To determine the specificity of the NF-κB blockade, the 4 core genes showing the greatest suppression of NF-κB transcription (3-9-5, 5-4-1, 6-8-2, and 12-2-1) were retested for inhibition of NF-κB transcription along side 3 different NS5A constructs, the NS3-4a gene, and the Stat-3 protein (Figure 4A). Only the core genes suppressed TNF-α induction of NF-κB transcription, suggesting that the effect is specific to the HCV core protein. Furthermore, the 10 core genes also blocked IFN-α induced signaling to the ISRE (Figure 4B). Thus, HCV core protein isolates from patients with acute hepatitis C appear to block innate antiviral and inflammatory signal transduction. When the core genes were expressed as V5-tagged proteins, similar inhibition of ISRE, NF-κB, and CXCL-8 (an NF-κB responsive gene) signaling were observed (data not shown). We further examined TNF-α induced nuclear translocation of NF-κB by immunofluorescence. Expression of core isolates 3-9-5, 6-8-2, and 4-7-1 was associated with reduced nuclear translocation of NF-κB following TNF-α treatment (Figure 5). Similar results were observed for the other core isolates, and we did not observe any significant difference in the behavior of isolates with different mutations (data not shown). Thus, the core isolates, despite small degrees of genetic variation, exhibited similar effects on innate antiviral and inflammatory signaling pathways.
Figure 4. Core perturbs innate inflammatory and antiviral signal transduction.
One-hundred nanograms of core genes were co-transfected with luciferase reporter genes under control of NF-κB (panel A), or ISRE (panel B) promoters. Twenty-four hours post-transfection, cells were treated with 10ng/ml TNF-α or 100u/ml of IFN-α (Roferon) for 4 hours before luciferase activity was measured by Britelite assay. Error bars represent standard deviations of quadruplicate transfections.
Figure 5. Core expression perturbs TNF-α induced nuclear translocation of NF-κB.
Huh7 cells were transfected with core genes and 24 hours later, were treated with 10ng/ml TNF-α for 20 minutes. Cells were fixed in 4%paraformaldehyde and core and the p65 subunit of NF-κB detected with mouse and rabbit antibodies, respectively, followed by AlexaFlour568-anti-mouse and AlexaFluor488-anti-rabbit antibodies. Images were captured on a Biorad confocal microscope. Arrows indicate reduced nuclear staining of NF-κB p65.
DISCUSSION
In the current study, we found that core genes were relatively conserved among different patients with acute HCV infection, and that all clones analyzed from a single patient at a single timepoint were identical at the nucleotide and amino acid level. This homogeneity could reflect a genetic bottleneck [31] or selection of the most-fit viruses during transmission. Analysis of a higher number of clones from infected individuals may reveal minor quasispecies for the core coding region that have not been identified in our study. Indeed, a previous study that analyzed a larger number of clones revealed genetic variability in the HCV genome during acute infection [32].
Our results indicate that despite the presence of relatively rare mutations in the core protein during acute infection, essential functions of core, such as immune-perturbing activities and targeting to lipid droplets, were generally the same for all isolates. Whether immune perturbing functions of core are critical for establishing chronic infection is currently unknown as all but one of our patients progressed to chronic infection. Nonetheless, mutations at amino acid 91 of core have been associated with liver cancer and virological non-responsiveness to IFN therapy[33, 34].
We found that expression of isolates of core from acute infection inhibited TNF-α induced nuclear translocation of NF-κB and NF-κB dependent transcription. This finding is in agreement with a previous study[35], but at odds with other studies which found core activates NF-κB [36-45]. Similarly, core has been reported to have opposing effects on the TNF-α system[46]. The differences may relate to different cell line and expression systems or the different core sequences used.
The extent of mobility of the lipid targeting domain 2 (D2) of the HCV core protein is likely governed by the strength of binding of D2 to the surface of lipid droplets [26]. From our FRAP analysis, substitutions at positions 130 (leucine replacing phenylalanine) and 161 (serine replacing glycine), both of which represent relatively conservative changes, did not dramatically affect mobility. These residues in D2 are located in amphipathic helices (position 130 in Helix I and position 161 in Helix II). However, mutations that affect the amphipathic nature of the helices, and hence their interaction with lipid on the surface of lipid droplets, have a more pronounced influence on mobility, reduce virus production, and can considerably decrease the stability of core [47] (McLauchlan et al. unpublished). Indeed, alteration of phenylalanine at position 130 to glutamic acid induces proteasomal degradation of core and abrogates production of infectious virus [9]. Therefore, the nature of the interaction of D2 with lipid droplets plays an important role in the process of assembly and release of virions. The possible exception is the A147V mutation found in 4 patients, which has been shown to enhance virus release in the JFH-1 system [26]. Moreover, we showed that all core proteins were efficiently secreted from Huh7 cells, but the kinetics were significantly delayed in comparison to low m.o.i. infection with JFH-1. It is possible that the kinetics of core secretion change in the presence of other HCV proteins or in the context of virus replication. Indeed, NS5A is known to target to lipid droplets, where it interacts with core to facilitate virus assembly [10, 30].
With regard to the effect of the amino acid changes in core on virus replication and assembly, as discussed above, the G161S change in Domain 2 does not affect virus production in JFH1. With regard to the F130L substitution, while changing this residue to a glutamate abolishes virus production, a phenylalanine to leucine change would be predicted to be conservative and therefore unlikely to significantly affect virus production. It certainly had very little impact in the FRAP analysis and the ability of the domain to target droplets. As far as the changes in the signal peptide are concerned (Domain 3), we predict that the V187I change would have little impact. Previous studies with JFH1 indicate that changing the adjacent residue from threonine to leucine (T186L refererred to as mut3 in [7] had barely any effect on virus production. Moreover, Murray et al [48]made 3 alanine scanning mutations in the signal peptide (mutants 181-184, 185-188 and 189-191). All 3 mutants were viable and had little impact on virus production using the J6/JFH1 chimera. Thus, a single valine to isoleucine mutation at position 187 is unlikely to have any effect. Indeed, the Murray paper illustrates how difficult it can be to introduce mutations into the signal peptide and influence assembly because of the reported degeneracy in the cleavage site for SPP [7]. For the changes in Domain 1, it is also difficult to predict any outcome on virus production. While the Murray paper [48] introduced a set of 4 consecutive alanines into sequences across core and found that Domain 1 was sensitive to such alterations, these engineered sequences are not like a natural sequences. Indeed, in the case of the core sequences isolated from acutely infected patients, the changes were maximally at 4 amino acids scattered throughout domain 1. Some of the changes are rather conservative (arginine to lysine at positions 9 and 43; glycine to serine at position 45; arginine to histidine at position 101; threonine to serine at position 110) and therefore unlikely to be deleterious to virus production. Furthermore, an aspartate at position 68 (an alanine in most sequences) and alanine at position 75 (a threonine in most sequences) are found in both JFH1 and HC-J6 sequences and the core proteins from these strains produce virus in tissue culture. Moreover, the variable residue at position 91 (cysteine, methionine and arginine) is a leucine in JFH1 and HC-J6. Thus, most of the changes are very unlikely to have any significant on virus production.
In summary, core quasispecies tended to be conserved during acute HCV infection, both within a patient and over time. Despite demonstrable amino acid changes, core proteins had similar immune perturbing and lipid targeting functions. Acute core isolates had unique amino acid changes compared to core isolates from chronic infection.
Acknowledgments
This research was supported by NIH grants U19 AI066328 to HRR, SJP, and YSH, R01 DK062187 to SJP, and RO1 DK066754 to YSH. JM was supported by the Medical Research Council (UK).
We thank Francois Penin for assistance with the European HCV database
Footnotes
No authors have any conflicts of interest to disclose.
References
- 1.Bowen DG, Walker CM. Mutational escape from CD8+ T cell immunity: HCV evolution, from chimpanzees to man. J Exp Med. 2005;201:1709–14. doi: 10.1084/jem.20050808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polyak SJ, Kline K, Shoji I, Miyamura T, Lingappa J. Assemble and Interact: Pleiotropic Functions of the HCV Core Protein. In: Tan SL, editor. Hepatitis C Viruses: Genomes and Molecular Biology. Horizon Scientific Press; 2006. pp. 89–120. [PubMed] [Google Scholar]
- 3.McLauchlan J. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. Journal Of Viral Hepatitis. 2000;7:2–14. doi: 10.1046/j.1365-2893.2000.00201.x. [DOI] [PubMed] [Google Scholar]
- 4.Santolini E, Migliaccio G, La MN. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631–41. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991;88:5547–51. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. Embo J. 2002;21:3980–8. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Targett-Adams P, Hope G, Boulant S, McLauchlan J. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J Biol Chem. 2008;283:16850–9. doi: 10.1074/jbc.M802273200. [DOI] [PubMed] [Google Scholar]
- 8.Dubuisson J. Hepatitis C virus proteins. World J Gastroenterol. 2007;13:2406–15. doi: 10.3748/wjg.v13.i17.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulant S, Targett-Adams P, McLauchlan J. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol. 2007;88:2204–13. doi: 10.1099/vir.0.82898-0. [DOI] [PubMed] [Google Scholar]
- 10.Miyanari Y, Atsuzawa K, Usuda N, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–97. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 11.Qin H, Shire NJ, Keenan ED, et al. HCV quasispecies evolution: association with progression to end-stage liver disease in hemophiliacs infected with HCV or HCV/HIV. Blood. 2005;105:533–41. doi: 10.1182/blood-2004-04-1452. [DOI] [PubMed] [Google Scholar]
- 12.Alam SS, Nakamura T, Naganuma A, et al. Hepatitis C virus quasispecies in cancerous and noncancerous hepatic lesions: the core protein-encoding region. Acta Med Okayama. 2002;56:141–7. doi: 10.18926/AMO/31716. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K, Iwata K, Matsumoto M, et al. Hepatitis C virus (HCV) genotype 1b sequences from fifteen patients with hepatocellular carcinoma: the ‘progression score’ revisited. Hepatol Res. 2001;20:161–171. doi: 10.1016/s1386-6346(00)00141-8. [DOI] [PubMed] [Google Scholar]
- 14.Nagayama K, Kurosaki M, Enomoto N, et al. Time-related changes in full-length hepatitis C virus sequences and hepatitis activity. Virology. 1999;263:244–253. doi: 10.1006/viro.1999.9924. [DOI] [PubMed] [Google Scholar]
- 15.Nagayama K, Kurosaki M, Enomoto N, Miyasaka Y, Marumo F, Sato C. Characteristics of hepatitis C viral genome associated with disease progression. Hepatology. 2000;31:745–50. doi: 10.1002/hep.510310327. [DOI] [PubMed] [Google Scholar]
- 16.Ogata S, Nagano-Fujii M, Ku Y, Yoon S, Hotta H. Comparative sequence analysis of the core protein and its frameshift product, the F protein, of hepatitis C virus subtype 1b strains obtained from patients with and without hepatocellular carcinoma. J Clin Microbiol. 2002;40:3625–30. doi: 10.1128/JCM.40.10.3625-3630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan XB, Mei L, Feng X, et al. Hepatitis C virus core proteins derived from different quasispecies of genotype 1b inhibit the growth of Chang liver cells. World J Gastroenterol. 2008;14:2877–81. doi: 10.3748/wjg.14.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang CC, Krantz E, Klarquist J, et al. Acute hepatitis C in a contemporary US cohort: modes of acquisition and factors influencing viral clearance. J Infect Dis. 2007;196:1474–82. doi: 10.1086/522608. [DOI] [PubMed] [Google Scholar]
- 19.Polyak SJ, Sullivan DG, Austin MA, et al. Comparison of amplification enzymes for Hepatitis C Virus quasispecies analysis. Virol J. 2005;2:41. doi: 10.1186/1743-422X-2-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao E, Tavis JE. A general method for nested RT-PCR amplification and sequencing the complete HCV genotype 1 open reading frame. Virol J. 2005;2:88. doi: 10.1186/1743-422X-2-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo BC, McPoland P, Wagoner JP, Kane OJ, Lohmann V, Polyak SJ. Relationships between hepatitis C virus replication and CXCL-8 production in vitro. J Virol. 2006;80:7885–93. doi: 10.1128/JVI.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller K, McArdle S, Gale MJ, Jr., et al. Effects of the hepatitis C virus core protein on innate cellular defense pathways. J Interferon Cytokine Res. 2004;24:391–402. doi: 10.1089/1079990041535647. [DOI] [PubMed] [Google Scholar]
- 23.Plumlee CR, Lazaro CA, Fausto N, Polyak SJ. Effect of ethanol on innate antiviral pathways and HCV replication in human liver cells. Virol J. 2005;2:89. doi: 10.1186/1743-422X-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polyak SJ, Morishima C, Shuhart MC, Wang CC, Liu Y, Lee DY. Inhibition of T-cell inflammatory cytokines, hepatocyte NF-kappaB signaling, and HCV infection by standardized silymarin. Gastroenterology. 2007;132:1925–36. doi: 10.1053/j.gastro.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 25.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic. 2008;9:1268–82. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 26.Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem. 2007;282:37158–69. doi: 10.1074/jbc.M707329200. [DOI] [PubMed] [Google Scholar]
- 27.Barba G, Harper F, Harada T, et al. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proceedings Of The National Academy Of Sciences Of The. 1997;175:740–744. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabile A, Perlemuter G, Bono F, et al. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999 Oct;30:1064–1076. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 29.Maillard P, Krawczynski K, Nitkiewicz J, et al. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J Virol. 2001;75:8240–50. doi: 10.1128/JVI.75.17.8240-8250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, Lai MM. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 2002;292:198–210. doi: 10.1006/viro.2001.1225. [DOI] [PubMed] [Google Scholar]
- 31.Laskus T, Wilkinson J, Gallegos-Orozco JF, et al. Analysis of hepatitis C virus quasispecies transmission and evolution in patients infected through blood transfusion. Gastroenterology. 2004;127:764–76. doi: 10.1053/j.gastro.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Netski DM, Mao Q, et al. Accurate representation of the hepatitis C virus quasispecies in 5.2-kilobase amplicons. J Clin Microbiol. 2004;42:4223–9. doi: 10.1128/JCM.42.9.4223-4229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akuta N, Suzuki F, Kawamura Y, et al. Amino acid substitutions in the hepatitis C virus core region are the important predictor of hepatocarcinogenesis. Hepatology. 2007;46:1357–64. doi: 10.1002/hep.21836. [DOI] [PubMed] [Google Scholar]
- 34.Akuta N, Suzuki F, Sezaki H, et al. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and non-virological response to interferon-ribavirin combination therapy. Intervirology. 2005;48:372–80. doi: 10.1159/000086064. [DOI] [PubMed] [Google Scholar]
- 35.Joo M, Hahn YS, Kwon M, Sadikot RT, Blackwell TS, Christman JW. Hepatitis C virus core protein suppresses NF-kappaB activation and cyclooxygenase-2 expression by direct interaction with IkappaB kinase beta. J Virol. 2005;79:7648–57. doi: 10.1128/JVI.79.12.7648-7657.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung YM, Park KJ, Choi SY, Hwang SB, Lee SY. Hepatitis C virus core protein potentiates TNF-alpha-induced NF-kappaB activation through TRAF2-IKKbeta-dependent pathway. Biochem Biophys Res Commun. 2001;284:15–9. doi: 10.1006/bbrc.2001.4936. [DOI] [PubMed] [Google Scholar]
- 37.de Lucas S, Bartolome J, Amaro MJ, Carreno V. Hepatitis C virus core protein transactivates the inducible nitric oxide synthase promoter via NF-kappaB activation. Antiviral Res. 2003;60:117–24. doi: 10.1016/j.antiviral.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Kanda T, Yokosuka O, Nagao K, Saisho H. State of hepatitis C viral replication enhances activation of NF-kB- and AP-1-signaling induced by hepatitis B virus X. Cancer Lett. 2006;234:143–8. doi: 10.1016/j.canlet.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 39.Mann EA, Stanford S, Sherman KE. Prevalence of mutations in hepatitis C virus core protein associated with alteration of NF-kappaB activation. Virus Res. 2006;121:51–7. doi: 10.1016/j.virusres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Ray RB, Steele R, Basu A, et al. Distinct functional role of Hepatitis C virus core protein on NF-kappaB regulation is linked to genomic variation. Virus Res. 2002;87:21–9. doi: 10.1016/s0168-1702(02)00046-1. [DOI] [PubMed] [Google Scholar]
- 41.Sato Y, Kato J, Takimoto R, et al. Hepatitis C virus core protein promotes proliferation of human hepatoma cells through enhancement of transforming growth factor alpha expression via activation of nuclear factor-kappaB. Gut. 2006;55:1801–8. doi: 10.1136/gut.2005.070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida H, Kato N, Shiratori Y, et al. Hepatitis C virus core protein activates nuclear factor kappa B-dependent signaling through tumor necrosis factor receptor-associated factor. J Biol Chem. 2001;276:16399–405. doi: 10.1074/jbc.M006671200. [DOI] [PubMed] [Google Scholar]
- 43.You LR, Chen CM, Lee YH. Hepatitis C virus core protein enhances NF-kappaB signal pathway triggering by lymphotoxin-beta receptor ligand and tumor necrosis factor alpha. J Virol. 1999;73:1672–81. doi: 10.1128/jvi.73.2.1672-1681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu N, Khoshnan A, Schneider R, et al. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691–7. doi: 10.1128/jvi.72.5.3691-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu N, Ware CF, Lai MM. Hepatitis C virus core protein enhances FADD-mediated apoptosis and suppresses TRADD signaling of tumor necrosis factor receptor. Virology. 2001;283:178–87. doi: 10.1006/viro.2001.0896. [DOI] [PubMed] [Google Scholar]
- 46.Polyak SJ. Hepatitis C virus-cell interactions and their role in pathogenesis. Clin Liver Dis. 2003;7:67–88. doi: 10.1016/s1089-3261(02)00075-2. [DOI] [PubMed] [Google Scholar]
- 47.Boulant S, Montserret R, Hope RG, et al. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J Biol Chem. 2006;281:22236–47. doi: 10.1074/jbc.M601031200. [DOI] [PubMed] [Google Scholar]
- 48.Murray CL, Jones CT, Tassello J, Rice CM. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J Virol. 2007;81:10220–31. doi: 10.1128/JVI.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]