Abstract
Background:
Previously described molecular biology techniques used to detect periprosthetic infections have been complicated by false-positive results. We have reported the development of a messenger RNA (mRNA)-based procedure to reduce these false-positive results. The limitations of this procedure are the lack of a universal target and reduced sensitivity due to a low concentration of bacterial mRNAs in test samples. The objective of the present study was to determine whether reverse transcription-quantitative polymerase chain reaction (RT-qPCR) using universal primers can be used to detect the more abundant bacterial ribosomal RNA (rRNA) as an indicator of periprosthetic infection.
Methods:
Serial dilutions of simulated synovial fluid infections were analyzed with rRNA RT-qPCR to determine the detection limit of this assay. Escherichia coli cultures treated with gentamicin were analyzed with RT-qPCR over a twenty-day time course to determine the degradation of rRNA as compared with the decrease in the viable cell count as determined by means of cell plating. As a proof of concept, group-specific polymerase chain reaction primers were developed for Streptococcus species and were tested against fifteen orthopaedically relevant organisms to show the potential for speciation with this assay. Sixty-four patients with a symptomatic effusion at the site of a total knee arthroplasty were enrolled, and complete patient information was documented in a prospective manner. Synovial fluid analysis with rRNA RT-qPCR was performed in a blind fashion.
Results:
The rRNA RT-qPCR assay was able to detect as few as 590 colony forming units/mL of Staphylococcus aureus and 2900 colony forming units/mL of Escherichia coli. The rRNA RT-qPCR signal closely followed cell death, pointing to its potential use as a viability marker. Three group-specific primer sets correctly identified their intended targets without amplifying closely related species. Clinically, the test correctly identified all six patients with a confirmed infection and all fifty patients who clearly did not have an infection. Eight patients had some laboratory or clinical signs of infection, but their status could not be confirmed. Infection was indicated by rRNA RT-qPCR in three of these patients who had elevated synovial fluid white blood-cell counts but negative results on culture. For statistical purposes, all patients who were categorized as indeterminate were considered to have an infection for the purpose of analysis, for a prevalence of 22% in this cohort.
Conclusions:
With respect to current diagnostic tests, rRNA-based RT-qPCR demonstrated 100% specificity and positive predictive value with a sensitivity equivalent to that of intraoperative culture. The RT-qPCR signal followed bacterial culture trends but exhibited detectable level for seven days after sterilization, allowing for the detection of infection after the antibiotic administration. These findings indicate that rRNA RT-qPCR is a sensitive and reliable test that retains the universal detection and speciation of DNA-based methods while functioning as a viability indicator.
Level of Evidence:
Diagnostic Level I. See Instructions to Authors for a complete description of levels of evidence.
Periprosthetic joint infection is one of the most common and catastrophic complications of total joint arthroplasty. Infection is the documented cause of total hip arthroplasty revisions in 7% of cases1 and in as many as 18% of total knee failures2,3. Some studies involving the use of molecular techniques and bacterial staining have demonstrated that as many as 73% of total hip arthroplasty failures were caused by indolent infections even when only 22% of these infections were diagnosed with current clinical methods4. Early identification of acute infections allows implant retention if irrigation and débridement is performed in a timely fashion5. In the case of a chronic infection, a two-stage reconstruction involving removal of the implant, placement of an antibiotic spacer, and later reimplantation offers the best chance for long-term implant survival6,7.
Unfortunately, one of the most difficult aspects of treating periprosthetic joint infections is obtaining an early and accurate diagnosis when the patient presents with pain at the site of the arthroplasty. Although culture of a joint aspirate or intraoperative tissue has been considered to be the closest to a true gold standard, the rate of false-positive results has recently been called into question8. A considerable number of false-negative results also have been noted, especially if the patient has received antibiotics prior to joint aspiration9. Because of the time delay and inconsistency of culture results, many surgeons use a protocol based on a combination of clinical suspicion and microbiological, radiographic, and serological findings10. Radiographs, while useful, are neither specific nor sensitive for infection as a cause of failure11. Biochemical markers such as the C-reactive protein level and the erythrocyte sedimentation rate are ubiquitous markers of inflammation that have high sensitivity for infection but questionable specificity, especially in patients with inflammatory arthropathies or other causes of inflammation, as the preoperative values are often above the published diagnostic criteria12.
In the last decade, many studies have investigated the use of traditional polymerase chain reaction, a technology that enzymatically amplifies deoxyribonucleic acid (DNA) by means of sequence-specific oligodeoxynucleotide primers, to detect bacterial DNA in order to identify joint infections13-15. The results have shown high sensitivity but have been limited by the number of potential false-positive results16. With the persistence of bacterial DNA after cell death, any bacterial contamination, even in an antibiotic-cleared infection, could cause a positive result in an otherwise sterile sample. In our recent study, we explored the utility of messenger ribonucleic acid (mRNA) as a viability indicator with use of reverse transcription-quantitative polymerase chain reaction (RT-qPCR)17. In that study of simulated joint infections, there were no false-positive results and all detectable signals quickly approached the sterile baseline. However, the sensitivity of this method is limited by the naturally low number of mRNA transcripts in the bacteria present in a clinical sample and the high rate of degradation of mRNA after cell death18. In the current study, we explored the utility of detecting ribosomal RNA (rRNA) for the diagnosis of periprosthetic joint infection. We hypothesized that rRNA-based detection would have higher sensitivity compared with mRNA as ribosomes are abundant intracellular organelles and rRNAs make up >95% of the total RNA content. Conversely, mRNAs constitute <1% of total RNA content. rRNA is also a more robust RNA species and will survive the extraction process at a considerably higher concentration than will mRNA19. Interestingly, the biodegradation kinetic profile of rRNAs has been shown to be similar to that of mRNAs, suggesting that rRNAs may serve well as a cell viability marker20.
Since rRNAs are highly conserved among bacterial species, rRNA-based detection targeting the conserved sequences in the rRNA should provide universal coverage for all orthopaedically relevant bacterial species. On the other hand, the mRNA-based methodology is by its very nature more species-specific. It should be noted that rRNA genes also contain variable regions, thus allowing for species identification of the infecting organism21.
Materials and Methods
RNA Isolation
Total RNA was isolated from samples of simulated septic arthritis for the purposes of determining the diagnostic sensitivity, rRNA degradation patterns, and speciation characteristics of the assay. The same protocol was used to evaluate unknown clinical samples. Combined enzymatic and mechanical bacterial lysis was performed to ensure release of all intracellular RNA species in the samples. One milliliter of each sample was pelleted by means of centrifugation at 14,400 times gravity for ten minutes. The cell pellet was then incubated for thirty minutes at 37°C in 100 μL of lysis solution containing 20 mg/mL of both lysozyme and proteinase K (Sigma-Aldrich, St. Louis, Missouri) in RNase-free water. Next, zirconia beads (RiboPure Bacteria Kit; Applied Biosystems/Ambion, Austin, Texas) were added and the samples were vigorously agitated by means of vortexing for ten minutes. The RNeasy Mini Kit (QIAGEN, Valencia, California) was used for column purification of total RNA. Poly(A) RNA (20 ng/5 μL) was used as a carrier species and was added to the specimen before the introduction of zirconia beads and again before using the RNeasy column to improve RNA yield with dilute samples22. DNA contamination was eliminated by means of on-column DNase digestion prior to elution of total RNA from the column with 120 μL of RNase-free water.
Sensitivity of rRNA Detection
To demonstrate the sensitivity of this protocol, sterile synovial fluid obtained aseptically from the knee of a patient with osteoarthritis was inoculated with known quantities of both Staphylococcus aureus and Escherichia coli (ATCC 29213 and ATCC 53508, respectively; American Type Culture Collection, Manassas, Virginia), to create a factor of ten dilution series from 109 to 101 colony-forming units (CFU)/mL for each organism. Total RNA extraction was performed as described above on each sample in the dilution series for subsequent RT-qPCR analysis with universal rRNA primers as described below. The same total RNA sample was also analyzed with mRNA-based RT-qPCR with use of previously described primers17. The bacterial concentration was plotted against the polymerase chain reaction signal to develop quantifiable standard curves. Sterile synovial fluid was used as a negative control, and all samples were also analyzed without the addition of reverse transcriptase in the reaction mixture to ensure the complete removal of DNA during RNA isolation.
Demonstration of rRNA Degradation as a Function of Bacterial Viability
Cultures of Escherichia coli (ATCC 53508; American Type Culture Collection) were grown to a final bacterial concentration of 1.4 × 109 CFU/mL, at which time gentamicin (1 mg/mL) was added to each sample. Total RNA extraction was performed at time points over a twenty-day period. The bacterial concentration (CFU/mL) was determined by plating at each of these time points. The cells were pelleted by means of centrifugation and were resuspended in clean growth broth three times to ensure that there was no remaining antibiotic, which would have interfered with the plating results. On confirmation of cell death by means of plating, a 100-μL aliquot of the antibiotic-free sample was inoculated in 5 mL of culture broth and was further incubated to confirm the absence of viable cells.
Speciation and Primer Design
Conserved 16S rRNA primers were used as a universal screen for bacterial infection. qPCR primers, forward 5′-ATTAGATACCCTGGTAGTCCACGCC-3′ and reverse 5′-CGTCATCCCCACCTTCCTCC-3′, amplified a 387-base-pair segment (based on the Escherichia coli genome database, http://www.uni-giessen.de/cgi-bin/cgiwrap/gx1052/ecfasta.pl). Hypervariable regions of the 16S rRNA sequence were also aligned and analyzed with AlleleID (PREMIER Biosoft International, Palo Alto, California). This analysis generated group-specific primers for group-A Streptococcus (forward 5′-AATACCGCATAAGAGAGACTAACG-3′ and reverse 5′-CTCGCTAGAGTGCCCAACTTA-3′), group-B Streptococcus (forward 5′-CTTTCTCTTCGGAGCAGAA-3′ and reverse 5′-CTCGCTAGAGTGCCCAACTTA-3′), and alpha-hemolytic Streptococcus (forward 5′-GTGAGAGTGGAAAGTTCACACTGT-3′ and reverse 5′-AGCCTTTAACTTCAGACTTATCTAAC-3′). These primers were tested on a panel of fifteen different bacteria (Table I). 1.0 × 107 CFU of each species were extracted for RNA according to the protocol described above. A sterile sample was also processed as a negative control. All of the samples and primers were analyzed by means of RT-PCR with use of the protocol described below.
TABLE I.
Organisms Used to Assess Speciation*
| Organism | ATCC No. |
| Staphylococcus aureus | 29213 |
| Methicillin-resistant Staphylococcus aureus | BAA-41 |
| Staphylococcus epidermidis | 14990 |
| Staphylococcus saprophyticus | 15305 |
| Listeria monocytogenes | 19115 |
| Enterococcus faecalis | 19433 |
| Streptococcus pneumoniae | 6301 |
| Streptococcus pyogenes | 14289 |
| Streptococcus agalactiae | 12386 |
| Streptococcus oralis | 9811 |
| Escherichia coli | 53508 |
| Citrobacter freundii | 8090 |
| Proteus mirabilis | 4630 |
| Pseudomonas aeruginosa | 9721 |
| Acinetobacter baumannii | 19606 |
Clinically relevant bacterial species, identified according to the genus and species as well as the American Type Culture Collection (ATCC) number, were used to assess the utility of species-specific primer sets.
RT-qPCR
A 5-μL aliquot of total bacterial RNA was analyzed with use of the iScript one-step RT-PCR Kit with SYBR Green on an iCycler Thermal Cycler (Bio-Rad, Hercules, California). The cycling conditions were 50°C for ten minutes and 95°C for five minutes, followed by forty-five cycles of 95°C for ten seconds and 62°C for thirty seconds. For all samples, the cycle number at which the fluorescence values became logarithmic (Ct) was determined. The ΔCt value was calculated for each sample as the difference between the sample Ct and the sterile synovial fluid control Ct.
Clinical Sample Analysis
With informed consent and institutional review board approval, synovial fluid samples were collected from sixty-four patients who had pain at the site of a total knee arthroplasty and from eight patients with knee effusions but without previous knee surgery. This group of patients represents the consecutive experience of four surgeons (including one of the authors [W.G.H.]) over two years at a single institution. All patients with a symptomatic effusion after total knee arthroplasty who underwent aspiration for which a minimum of 1 mL of synovial fluid remained after culture and cell counts were approached for participation in the study. The samples were given an identification number and were analyzed in the laboratory without any information pertaining to the infection status of the patient. Information was collected from the patients at the time of the initial office visit, including the history, serological findings (C-reactive protein level, erythrocyte sedimentation rate, and white blood-cell count), and the results of synovial fluid analysis (culture and white blood-cell count with differential). If surgery was performed, surgical details regarding the intraoperative appearance, culture results, Giemsa staining of cryosections, Gram staining, surgical treatment, and clinical impression were also recorded. This information was recorded and was sent separately in sealed envelopes to remove bias from sample analysis. Total RNA extraction was performed as described above with use of 1 mL of the synovial fluid from each subject.
After samples were analyzed with RT-qPCR, the clinical information was reviewed and the patients were divided into three categories: positive, negative, and indeterminate. Patients who met the criteria described by Spangehl et al.23 and who had a positive clinical impression as determined by the attending surgeon were categorized as positive. Those who met none of the criteria of Spangehl et al.23 and who had a negative clinical impression were categorized as negative. Those who had a positive clinical impression and/or partially met criteria of Spangehl et al. were categorized as indeterminate.
Statistical Methods
For the purposes of statistical analysis, the samples in the indeterminate group were considered to be infected. According to these criteria, the prevalence of infection in the cohort was 22%. The sensitivity, specificity, positive predictive value, and negative predictive value were calculated for the rRNA RT-qPCR assay as well as for the C-reactive protein level, erythrocyte sedimentation rate, intraoperative cell culture result, and joint fluid cell count and differential. The C-reactive protein level was considered to be positive if the value was >10 mg/L, and the erythrocyte sedimentation rate was considered to be positive if the value was >30 mm/hr23. The cell count was considered to be positive if the synovial fluid total white blood-cell count was >2500/mm3 with >60% polymorphonuclear cells24. A 95% confidence interval was also determined for each test with use of the “constant chi-square boundaries” calculation25.
Source of Funding
This research was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Intramural Research Program (NIH ZO1 AR41131).
Results
Sensitivity of rRNA Detection
From the Staphylococcus aureus and the Escherichia coli dilution series, the detection limits were found to be 590 and 2900 CFU/mL, respectively, with use of rRNA RT-qPCR. For both species, there was a linear relationship between logarithmic bacterial concentration and ΔCt. The same total RNA samples were also analyzed by means of quantitative mRNA-based RT-qPCR17. The latter method yielded a detection limit of 60,000 CFU/mL for Escherichia coli and 110,000 CFU/mL for Staphylococcus aureus. For both the mRNA and rRNA assays, a two-cycle difference from the sterile baseline was used to assign a detectable result.
rRNA Degradation
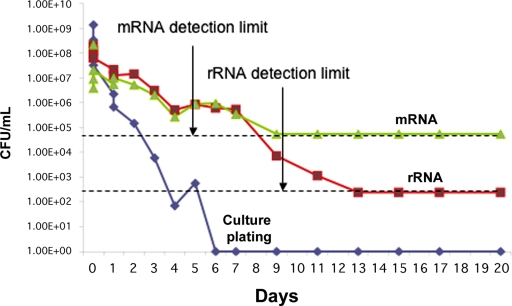
Cell death occurred over a period of six days in the presence of gentamicin. At Day 6 and beyond, inoculation of 100 μL of the culture, after removal of antibiotic, in fresh culture medium failed to yield any growth despite optimal bacterial growth conditions. By Day 13, the rRNA ΔCt was <2, indicating that within one week after successful antibiotic treatment of an infection, rRNA RT-qPCR accurately identified a clean sample. In parallel, the total RNA isolate was also analyzed by means of mRNA-based RT-qPCR with use of the appropriate mRNA primers17. By Day 9, the mRNA ΔCt was <2, indicating bacterial death. Both the mRNA and rRNA-derived polymerase chain reaction signals closely matched bacterial viability measured throughout antibiotic treatment (Fig. 1). While the mRNA signal reached baseline four days earlier (Fig. 1), the sensitivity of mRNA-based detection is poor relative to rRNA modalities with the same specimen.
Fig. 1.
Degradation of mRNA and rRNA with cell death. Cell cultures of Escherichia coli (1.4 × 109 CFU/mL) were treated at Day 0 with 1 mg/mL gentamicin. Over a twenty-day period, the concentration of cells (CFU/mL) was calculated by means of a traditional agar plating technique (blue line). The cells were pelleted by means of centrifugation and were resuspended in clean growth media three times to ensure that there was no remaining antibiotic (which would have interfered with the plating results). In addition, at each time point, total RNA extraction was performed and cell concentration was predicted with use of mRNA RT-qPCR (green line) and rRNA RT-qPCR (red line). The absence of viable bacteria is predicted by RT-qPCR when the calculated cell concentration reaches the detection threshold indicated by the dashed lines (6.0 × 104 for mRNA and 2.9 × 103 for rRNA). (For the rRNA data, although a theoretical limit of 2.9 x 102 CFU/mL is indicated based on theoretical extrapolation, the experimental data could only reliably detect 2.9 x 103 CFU/mL, while the difference between 2.9 x 102 and zero could not be accurately ascertained.)
Speciation
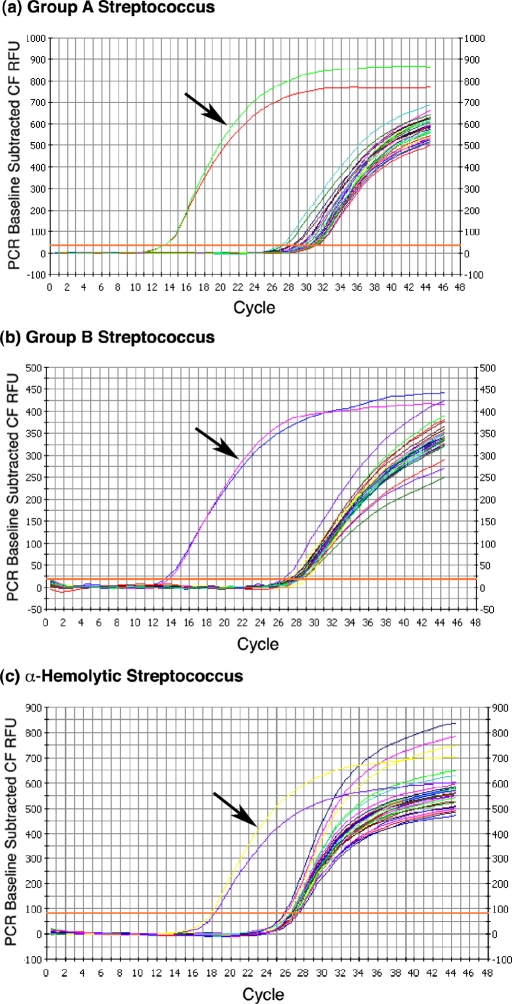
For each of the three group-specific primers, the targeted organism was the only species of the fifteen that was correctly identified on the basis of low ΔCt values. The ΔCt value for the group-A and group-B Streptococcus primers was 14, and the value for the alpha-hemolytic Streptococcus primers was 8 (Fig. 2).
Fig. 2.
Species-specific rRNA-based RT-qPCR. rRNA RT-qPCR analysis of RNA samples was performed on total RNA obtained from fifteen clinically relevant bacterial pathogens with use of group-specific primers. All organisms were grown to a concentration of 1 × 107 CFU/mL prior to total RNA extraction. The three graphs show the polymerase chain reaction outcome with use of primers designed for (a) group-A Streptococcus, (b) group-B Streptococcus, and (c) α-hemolytic Streptococcus. Initiation of the logarithmic rise in the analysis curve at a lower cycle indicates identification of the labeled species. Arrows indicate resultant Ct curves from polymerase chain reactions with use of RNA from the intended target organisms. All reactions were run with duplicate samples.
Clinical Sample Analysis
Of the sixty-four unknown samples, six were from patients with a confirmed infection at the site of a total knee arthroplasty. All patients with a corroborated infection were correctly identified on the basis of the 16S rRNA RT-qPCR test with use of universal primers. All fifty patients who were negative for infection were also correctly identified. Of the eight patients in the indeterminate group, three were characterized as positive and five were characterized as negative by means of RT-qPCR. Table II presents the complete clinical, serological, and histopathological data for the eight patients in the indeterminate group. In general, excellent concordance is seen between the RT-qPCR data and these data.
TABLE II.
Pathological and Clinical Data on Patients with Indeterminate Infection Status*
| Aspirate |
|||||||||||||
| Case | Sex, Age (yr) | Pertinent History | White Blood-Cell Count (cells/mm3) | Erythrocyte Sedimentation Rate (mm/hr) | C-Reactive Protein (mg/L) | White Blood-Cell Count (cells/mm3) | Polymorphonuclear Cell Concentration (% of white blood cells) | Frozen Section | Gram Stain | Culture | Procedure | rRNA RT-PCR Assay | Details |
| 1 | M, 84 | Diabetes mellitus, hypertension | 8.2 | 45 | 2.5 | 19,550 | 78 | 4 to 5 white blood cells/high-power field | Rare white blood cells, 0 organisms | − | Revision total | + | Treatment with 5 weeks of intravenous antibiotics |
| 2 | F, 77 | Hypertension, chronic obstructive pulmonary disease | 6.1 | 20 | 2.1 | 146,000 | 90 | Rare white blood cells, 0 organisms | − | Two stage revision | + | ||
| 3 | M, 68 | Diabetes mellitus, gout | 8.3 | 48 | 3 | 27,625 | 85 | − | + | Patient lost to follow-up | |||
| 4 | M, 85 | Diabetes mellitus, hypertension, coronary artery disease, revision total knee arthroplasty, receiving chronic antibiotic suppression with Duricef for Staphylococcus lugdunensis | 7.1 | 75 | 2.3 | 12,674 | 84 | 0 white blood cells, 0 organisms | Staphylococcus haemolyticus | Débridement with polyethylene liner exchange | − | Chronic antibiotics changed to doxycycline | |
| 5 | F, 61 | Breast cancer, bronchitis, rheumatoid arthritis | 4.7 | 50 | 6.1 | 5000 | 93 | >20 white blood cells /high-power field | Enterobacter | Two-stage revision | − | ||
| 6 | M, 55 | Three débridements with liner exchange at three years for Staphylococcus species | 8 | 7 | 0.2 | 55 | 8 | No acute inflammation | 0 white blood cells, 0 organisms | Methicillin-resistant Staphylococcus aureus + diphtheroids (delayed in broth) | Revision total | − | Treated with 6 weeks of Zyvox after culture results |
| 7 | M, 69 | Lupus anticoagulant +, pulmonary embolism, atrial fibrillation, hypertension | 4.7 | 22 | 0.2 | 2100 | 45 | 1/2 broth with methicillin-resistant Staphylococcus aureus | Revision tibia only | − | 6 weeks parenteral antibiotics after culture results | ||
| 8 | M, 74 | Cement spacer in place, receiving intravenous antibiotics for methicillin-resistant Staphylococcus aureus | 7.1 | 25 | 2.2 | 3937 | 44 | Chronic synovitis | − | Exchange of antibiotic spacer | − | ||
Eight patients had an indeterminate infection status on the basis of the microbiological, radiographic, and serological data. Three patients (Cases 1, 2, and 3) had negative synovial fluid cultures, but an infection was suspected on the basis of increased white blood-cell count in the synovial fluid aspirate. Two patients (Cases 4 and 5) had positive cultures with indeterminate results on blood and synovial fluid analysis. Two patients (Cases 6 and 7) had positive cultures with otherwise negative workups. One patient (Case 8) had a history of infection that was being treated with an antibiotic spacer. At the time of aspiration, the clinical suspicion was ongoing infection, although the objective data were uncertain. With all data available, five patients (Cases 1 through 5) were considered by the researchers to have an infection, whereas three patients (Cases 6, 7, and 8) were not.
Of these eight indeterminate samples, the first five were likely infected. Although those five patients did not meet the criteria of Spangehl et al.23 for periprosthetic infection at the site of total hip arthroplasty, they all were considered to be positive for infection according to the criteria of Mason et al.24. Two patients (Cases 1 and 2) were managed with revision and intravenous antibiotics as though they had an infection at the site of the arthroplasty. One patient (Case 3) was scheduled for the same procedure but was lost to follow-up. All three patients were identified as having an infection on the basis of the RT-qPCR test. Another patient (Case 4) had been receiving long-term suppressive antibiotic treatment with Duricef (cefadroxil) but this treatment was failing; the patient was taken to the operating room for débridement and liner exchange and the suppressive antibiotic was changed to doxycycline. A fifth patient (Case 5) was diagnosed with an Enterobacter infection and was managed with a two-stage revision. Two patients (Cases 4 and 5) were considered to have a false-negative result on the RT-qPCR test.
The final three patients who were categorized as indeterminate did not meet the criteria of Spangehl et al.23 or Mason et al.24 for the diagnosis of periprosthetic infection. Two patients (Cases 6 and 7) were noted to have positive late growth on culture after undergoing a revision arthroplasty for the treatment of presumed aseptic loosening. Both patients were managed prophylactically with antibiotics for six weeks. These two patients were considered to have a true-negative result. The third patient (Case 8) had previously received an antibiotic-impregnated spacer for the treatment of a methicillin-resistant Staphylococcus aureus periprosthetic infection. At the time of surgery, the spacer was exchanged as the tissue showed signs of chronic synovitis. In an attempt to be as conservative as possible in the analysis of our data, we have included as positive all specimens in the confirmed positive group and the indeterminate group. The calculated sensitivity, specificity, positive predictive value, and negative predictive value of the rRNA RT-qPCR assay were 71%, 100%, 100%, and 93%, respectively. The overall accuracy was 94%. For comparison, the respective values for C-reactive protein level, erythrocyte sedimentation rate, tissue culture, and joint aspiration are shown in Table III. The cell count and differential of joint fluid showed high accuracy. Sensitivity according to the criteria of Mason et al. was 71%, with a 98% specificity and an overall accuracy of 92%. Overall, serological tests had a worse outcome than previously described. The C-reactive protein level had a sensitivity of only 21%, although the specificity was 94%. The erythrocyte sedimentation rate had a higher sensitivity of 71% but was only 82% specific.
TABLE III.
Comparison of Molecular and Laboratory Tests Used to Diagnose Periprosthetic Infections*
| Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value | Accuracy | |
| rRNA RT-PCR | 0.71 (0.55 to 0.71) | 1.00 (0.96 to 1.00) | 1.00 (0.77 to 1.00) | 0.93 (0.88 to 0.93) | 0.94 (0.87 to 0.94) |
| C-reactive protein | 0.21 (0.08 to 0.34) | 0.94 (0.90 to 0.98) | 0.50 (0.20 to 0.80) | 0.81 (0.78 to 0.84) | 0.78 (0.72 to 0.84) |
| Erythrocyte sedimentation rate | 0.71 (0.49 to 0.87) | 0.82 (0.76 to 0.86) | 0.53 (0.36 to 0.64) | 0.91 (0.84 to 0.96) | 0.80 (0.70 to 0.87) |
| Intraoperative culture | 0.71 (0.54 to 0.77) | 0.96 (0.87 to 0.99) | 0.91 (0.68 to 0.98) | 0.86 (0.78 to 0.89) | 0.88 (0.75 to 0.92) |
| Synovial fluid cell count with differential | 0.71 (0.53 to 0.77) | 0.98 (0.93 to 1.00) | 0.91 (0.68 to 0.98) | 0.93 (0.88 to 0.94) | 0.92 (0.84 to 0.95) |
On the basis of the analysis of the sixty-four clinical samples, the sensitivity, specificity, positive predictive value, negative predictive value, and accuracy of the detection of infection with use of rRNA RT-qPCR were calculated. In addition, the same values were calculated with the use of C-reactive protein, erythrocyte sedimentation rate, intraoperative culture, and synovial fluid cell count with differential as diagnostic tools. For the purposes of analysis, all clearly positive and indeterminate samples were analyzed as being infected, giving a prevalence of 22%. Values are given as the mean, with the range in parentheses.
Discussion
The diagnosis of periprosthetic joint infections remains a challenging clinical problem. There is no true gold standard, and clinicians must depend on an array of tests that are limited in terms of sensitivity and accuracy. There has been an increasing focus on the use of existing modalities to improve diagnostic accuracy by either redefining the positive values for these tests26 or using them in combination27. The application of polymerase chain reaction-based molecular biology technology has been an exciting development as a potential method for the detection of orthopaedic infections; however, at the present time, it remains a research endeavor only and has not been widely adopted for clinical practice. The progress in translating this technology into practice has been limited by a number of factors, including the time required for the test, technical equipment requirements, and cost, but it has been most severely limited by the number of potential false-positive results associated with polymerase chain reaction-based methods. As the major cause of the false-positive results is related to the intrinsically high sensitivity of the DNA-based assay, DNA that is present in dead bacteria or in the recombinantly-prepared reagents would also be amplified and detected. Thus, in principle, an RNA-based polymerase chain reaction method would be preferable as RNAs are more labile and would be associated primarily with viable cells. Our recent study involving mRNA-based RT-qPCR illustrates the feasibility of this approach17, albeit with an expected reduced sensitivity in comparison with the DNA-based methods.
The results reported here show that quantitative RT-qPCR detection of bacterial 16S rRNA offers several unique advantages. As rRNA has highly conserved regions, practically all of the orthopaedically relevant bacterial pathogens can be rapidly identified with the universal primers used in the present study. Similar highly conserved regions in mRNAs are infrequent, making universal bacterial detection more difficult with mRNAs, which are also found at a considerably lower concentration in the cell in comparison with rRNAs. This is clearly supported by our results, which demonstrated a considerable difference in sensitivity between the two methods. The higher sensitivity of bacterial detection with use of rRNA RT-qPCR suggests its potential to transition from clinical research to clinical utility. Other studies have shown that the hypervariable regions of the 16S rRNA gene are useful in DNA-based polymerase chain reaction protocols for group and species-level bacterial identification21. In the present study, as an additional proof of concept, we have shown that, when using the rRNA-based RT-qPCR method, the hypervariable regions of 16S rRNA can indeed be used to discriminate between Streptococcus groups. In the future, once species-specific rRNA primers are designed for all bacterial families, rapid and sensitive speciation should be possible. These factors, combined with the lower sensitivity of mRNA detection and the higher false-positive rate of DNA detection, indicate that in adopting RNA-based RT-qPCR as a modality for bacterial detection, the rRNA template may be preferred over mRNA.
From the rRNA degradation data, it is clear that reduction in rRNA abundance follows trends in antibiotic-induced bacterial cell death. Interestingly, there is a delay of approximately one week during which rRNA detection persists although the samples remain culture-negative. This characteristic most likely represents what has previously been described as “viable but non-culturable” or “septic but unculturable” bacterial infections28. Thus, a negative rRNA RT-qPCR assay may be used as reassurance to the surgeon that the infection has been adequately treated and the patient is no longer in the “viable but non-culturable” period. This lag could also be useful clinically for assessing infections that have been partially treated with antibiotics, as the results of culture are notoriously poor for these patients9. In principle, the results of rRNA RT-qPCR analysis of serial aspirations for a patient who has undergone débridement with the placement of an antibiotic spacer could also be used to determine the appropriate timing of reimplantation. With further clinical validation, the ability of the RT-qPCR test to detect only viable bacteria makes this an attractive option in the future.
The clinical portion of the present study provides exciting data suggesting the feasibility of applying the rRNA RT-qPCR test in the clinical setting. The molecular diagnostic test correctly identified all obvious infections and those that were considered to be definitive aseptic failures. Of the eight patients who were characterized as indeterminate, five were thought to have a clinical infection. Two other patients had positive culture results and were managed with intravenous antibiotics after a revision arthroplasty for the treatment of presumed aseptic loosening. The last patient was already being managed for a known infection with an antibiotic-impregnated cement spacer. At the time of surgery, the decision was made to replace the spacer and not to reimplant a prosthesis because of the appearance of the tissues. No laboratory abnormalities were reported in the synovial fluid from this patient.
Three patients with negative culture results had positive RT-qPCR results. All three of these patients had elevated white blood-cell counts. Two of the three patients had an elevated erythrocyte sedimentation rate. Of the five patients with negative RT-qPCR results, two had no signs of infection other than the late growth of bacteria in broth medium in the laboratory. Both had undergone standard revisions for the treatment of presumed aseptic loosening. On the basis of the culture results, six weeks of antibiotics were added to the treatment. The basis of such spurious false-positive results was recently reviewed8, and we believe that these should be considered to be true-negative RT-qPCR results. The other three patients with negative RT-qPCR results could be considered to have false-negative results. As mentioned previously, one patient was scheduled for a reimplantation after the placement of an antibiotic spacer for the treatment of infection. The tissues surrounding the spacer did not appear to be healthy, and therefore the decision was made to replace the antibiotic spacer. The second patient had had a previous two-stage revision for the treatment of infection and was having a failure of long-term antibiotic suppression. These individuals received a polyethylene liner exchange with a change in the antibiotic suppression regimen. The final patient was treated as though he had an infection and was managed with a two-stage revision by the surgeon and had positive cultures as well as an elevated erythrocyte sedimentation rate, a positive result on Gram staining, and a positive white blood-cell count and differential on the joint aspirate. Thus, in spite of these few false-negative results, the rRNA-based RT-qPCR test outperformed existing modalities.
In summary, while molecular diagnostic tests face many challenges in clinical adoption, rRNA-based RT-qPCR offers excellent diagnostic sensitivity within a few hours after a sample is obtained. In this small series, there were no false-positive results as has been the case with other nucleic acid amplification methods, and the overall sensitivity and accuracy were improved in comparison with culture and serological testing. Retaining the cell viability-sensitive degradation patterns of RNA while keeping the ability to speciate infections that has been the hallmark of DNA-based methods makes rRNA a uniquely ideal target for RT-qPCR detection of periprosthetic infections.
Footnotes
Disclosure: The authors did not receive any outside funding or grants in support of their research for or preparation of this work. One or more of the authors, or a member of his or her immediate family, received, in any one year, payments or other benefits or a commitment or agreement to provide such benefits from commercial entities in excess of $10,000 (DePuy, a Johnson and Johnson company) and less than $10,000 (Lifenet). This research was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Intramural Research Program (NIH ZO1 AR41131).
References
- 1.Clohisy JC, Calvert G, Tull F, McDonald D, Maloney WJ. Reasons for revision hip surgery: a retrospective review. Clin Orthop Relat Res. 2004;429:188-92 [DOI] [PubMed] [Google Scholar]
- 2.Mulhall KJ, Ghomrawi HM, Scully S, Callaghan JJ, Saleh KJ. Current etiologies and modes of failure in total knee arthroplasty revision. Clin Orthop Relat Res. 2006;446:45-50 [DOI] [PubMed] [Google Scholar]
- 3.Barrack RL, McClure JT, Burak CF, Clohisy JC, Parvizi J, Sharkey P. Revision total knee arthroplasty: the patient's perspective. Clin Orthop Relat Res. 2007;464:146-50 [PubMed] [Google Scholar]
- 4.Tunney MM, Patrick S, Curran MD, Ramage G, Hanna D, Nixon JR, Gorman SP, Davis RI, Anderson N. Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16S rRNA gene. J Clin Microbiol. 1999;37:3281-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva M, Tharani R, Schmalzried TP. Results of direct exchange or debridement of the infected total knee arthroplasty. Clin Orthop Relat Res. 2002;404:125-31 [DOI] [PubMed] [Google Scholar]
- 6.Booth RE, Jr, Lotke PA. The results of spacer block technique in revision of infected total knee arthroplasty. Clin Orthop Relat Res. 1989;248:57-60 [PubMed] [Google Scholar]
- 7.Insall JN, Thompson FM, Brause BD. Two-stage reimplantation for the salvage of infected total knee arthroplasty. J Bone Joint Surg Am. 1983;65:1087-98 [PubMed] [Google Scholar]
- 8.Barrack RL, Aggarwal A, Burnett RS, Clohisy JC, Ghanem E, Sharkey P, Parvizi J. The fate of the unexpected positive intraoperative cultures after revision total knee arthroplasty. J Arthroplasty. 2007;22:94-9 [DOI] [PubMed] [Google Scholar]
- 9.Barrack RL, Jennings RW, Wolfe MW, Bertot AJ. The value of preoperative aspiration before total knee revision. Clin Orthop Relat Res. 1997;345:8-16 [PubMed] [Google Scholar]
- 10.Bauer TW, Parvizi J, Kobayashi N, Krebs V. Diagnosis of periprosthetic infection. J Bone Joint Surg Am. 2006;88:869-82 [DOI] [PubMed] [Google Scholar]
- 11.Tigges S, Stiles RG, Roberson JR. Appearance of septic hip prostheses on plain radiographs. AJR Am J Roentgenol. 1994;163:377-80 [DOI] [PubMed] [Google Scholar]
- 12.Laiho K, Mäenpää H, Kautiainen H, Kauppi M, Kaarela K, Lehto M, Belt E. Rise in serum C reactive protein after hip and knee arthroplasties in patients with rheumatoid arthritis. Ann Rheum Dis. 2001;60:275-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenollar F, Roux V, Stein A, Drancourt M, Raoult D. Analysis of 525 samples to determine the usefulness of PCR amplification and sequencing of the 16S rRNA gene for diagnosis of bone and joint infections. J Clin Microbiol. 2006;44:1018-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moojen DJ, Spijkers SN, Schot CS, Nijhof MW, Vogely HC, Fleer A, Verbout AJ, Castelein RM, Dhert WJ, Schouls LM. Identification of orthopaedic infections using broad-range polymerase chain reaction and reverse line blot hybridization. J Bone Joint Surg Am. 2007;89:1298-305 [DOI] [PubMed] [Google Scholar]
- 15.Tarkin IS, Henry TJ, Fey PI, Iwen PC, Hinrichs SH, Garvin KL. PCR rapidly detects methicillin-resistant staphylococci periprosthetic infection. Clin Orthop Relat Res. 2003;414:89-94 [DOI] [PubMed] [Google Scholar]
- 16.Mariani BD, Martin DS, Levine MJ, Booth RE, Jr, Tuan RS. Polymerase chain reaction detection of bacterial infection in total knee arthroplasty. Clin Orthop Relat Res. 1996;331:11-22 [DOI] [PubMed] [Google Scholar]
- 17.Birmingham P, Helm JM, Manner PA, Tuan RS. Simulated joint infection assessment by rapid detection of live bacteria with real-time reverse transcription polymerase chain reaction. J Bone Joint Surg Am. 2008;90:602-8 Erratum in: J Bone Joint Surg Am. 2008;90:1337 [DOI] [PubMed] [Google Scholar]
- 18.Sheridan GE, Masters CI, Shallcross JA, MacKey BM. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Vliet GM, Schepers P, Schukkink RA, van Gemen B, Klatser PR. Assessment of mycobacterial viability by RNA amplification. Antimicrob Agents Chemother. 1994;38:1959-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aellen S, Que YA, Guignard B, Haenni M, Moreillon P. Detection of live and antibiotic-killed bacteria by quantitative real-time PCR of specific fragments of rRNA. Antimicrob Agents Chemother. 2006;50:1913-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69:330-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Russell DW. The condensed protocols from molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. p 7.69 [Google Scholar]
- 23.Spangehl MJ, Masri BA, O'Connell JX, Duncan CP. Prospective analysis of preoperative and intraoperative investigations for the diagnosis of infection at sites of two hundred and two revision total hip arthroplasties. J Bone Joint Surg Am. 1999;81:672-83 [DOI] [PubMed] [Google Scholar]
- 24.Mason JB, Fehring TK, Odum SM, Griffin WL, Nussman DS. The value of white blood cell counts before revision total knee arthroplasty. J Arthroplasty. 2003;18:1038-43 [DOI] [PubMed] [Google Scholar]
- 25.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes in C. The art of scientific computing. 2nd ed.Cambridge, UK: Cambridge University Press; 1992. p 693-4 [Google Scholar]
- 26.Greidanus NV, Masri BA, Garbuz DS, Wilson SD, McAlinden MG, Xu M, Duncan CP. Use of erythrocyte sedimentation rate and C-reactive protein level to diagnose infection before revision total knee arthroplasty. A prospective evaluation. J Bone Joint Surg Am. 2007;89:1409-16 [DOI] [PubMed] [Google Scholar]
- 27.Bernard L, Lübbeke A, Stern R, Bru JP, Feron JM, Peyramond D, Denormandie P, Arvieux C, Chirouze C, Perronne C, Hoffmeyer P; Groupe d'Etude sur l'Ostéite. Value of preoperative investigations in diagnosing prosthetic joint infection: retrospective cohort study and literature review. Scand J Infect Dis. 2004;36:410-6 [DOI] [PubMed] [Google Scholar]
- 28.González-Escalona N, Fey A, Höfle MG, Espejo RT, A Guzmán C. Quantitative reverse transcription polymerase chain reaction analysis of Vibrio cholerae cells entering the viable but non-culturable state and starvation in response to cold shock. Environ Microbiol. 2006;8:658-66 [DOI] [PubMed] [Google Scholar]