Abstract
Influenza vaccine strains have been traditionally developed by annual reassortment between vaccine donor strain and the epidemic virulent strains. The classical method requires screening and genotyping of the vaccine strain among various reassortant viruses, which are usually laborious and time-consuming. Here we developed an efficient reverse genetic system to generate the 6:2 reassortant vaccine virus from cDNAs derived from the influenza RNAs. Thus, cDNAs of the two RNAs coding for surface antigens, haemagglutinin and neuraminidase from the epidemic virus and the 6 internal genes from the donor strain were transfected into cells and the infectious viruses of 6:2 defined RNA ratio were rescued. X-31 virus (a high-growth virus in embryonated eggs) and its cold-adapted strain X-31 ca were judiciously chosen as donor strains for the generation of inactivated vaccine and live-attenuated vaccine, respectively. The growth properties of these recombinant viruses in embryonated chicken eggs and MDCK cell were indistinguishable as compared to those generated by classical reassortment process. Based on the reverse genetic system, we generated 6 + 2 reassortant avian influenza vaccine strains corresponding to the A/Chicken/Korea/MS96 (H9N2) and A/Indonesia/5/2005 (H5N1). The results would serve as technical platform for the generation of both injectable inactivated vaccine and the nasal spray live attenuated vaccine for the prevention of influenza epidemics and pandemics.
Keywords: influenza vaccines, influenzavirus A, influenzavirus B, orthomyxoviridae
Introduction
Influenza viruses are members of orthomyxoviridae family and composed of 8 segmented negative-sense, single-stranded RNAs. Type A and B influenza viruses are the most important as human pathogens responsible for annual outbreaks of acute respiratory disease. A total of 16 haemagglutinin subtype and 9 neuraminidase subtype were identified in influenza A type (Webby and Webster, 2001; Nicholson et al., 2003). Due to segmented nature of RNA genes, the virus can exchange their RNA segments genes with other subtypes of influenza viruses causing major changes of antigenicity. In addition, the replication of the RNA genes is error-prone, allowing frequent nucleotide substitutions at genetic level (Webby and Webster, 2001). Frequent antigenic drift and shift provides the diversity of influenza viruses that causes the emergence and re-emergence of epidemic and pandemic viruses. Beside well-known human threats posed by H5N1 avian influenza, the potential for newly emerging influenza virus has been elevated with recent outbreak of Swine influenza viruses into the human (Novel Swine-Origin Influenza A Virus Investigation Team, 2009). The development of novel antiviral therapy and efficient vaccine is urgently needed.
Two different types of influenza vaccine are currently licensed and used for the prevention of seasonal influenza infection. Inactive influenza vaccines that are delivered by an intramuscular injection have been popularly used world-wide for the prevention of annual influenza epidemics. Alternatively, live vaccines are given by an intranasal route and are known to stimulates broader immunological response than the inactive vaccines providing cross-protective immune responses (Wareing and Tannock, 2001; Cox et al., 2004).
An immediate task to make inactive or live influenza vaccine is to generate reassortant vaccine strain that matches the current infection. The classical method for the generation of reassortant vaccine virus is co-infections of circulating strains with high-yielding seed strain in embryonated eggs and selecting a reassortant of particular 6:2 ratio that inherits the haemagglutinin and neuraminidase gene from the circulating strain and the other six internal genes from the high-yielding seed strain. This method relies on antibody selections, plaque isolation of many reassortant viruses followed by genotyping of individual plaque isolates for the identification of reassortant virus of desired RNA constellation (Bardiya and Bae, 2005). The method is very laborious and time-consuming and may not meet the tight schedule for the production of vaccine in a timely fashion. In contrasts, the reverse genetic approach allows direct generation of the 6:2 recombinant vaccine strains in a pre-determined RNA constellation in few weeks. This process is highly recommended for an efficient generation of vaccine strains in manufacturing both the inactive and live influenza vaccines. A/PR/8/34 has been approved by WHO as high-growth reassortant vaccine strain for the inactivation vaccines (Kilbourne and Murphy, 1960) and the reverse genetic system has been developed for A/PR/8/34 system (Hoffmann et al., 2002). Here we extended and adopted the cDNA based reverse genetic system to generate both injectable and nasal spray type live attenuated influenza vaccine (LAIV). For this purpose, we chose the X-31 virus, a reassortant of A/PR/8/34 and A/HK/6/68, as vaccine donor strain. The X-31 virus has been used for many years in influenza vaccine production because of its growth property in embryonated chicken eggs. Previously, a cold-adapted virus was generated from multiple passage of the X-31 at low temperature to yield X-31ca virus as donor strain for LAIV (Lee et al., 2006). The X-31ca strain showed the cold-adapted (ca), temperature-sensitive (ts)and attenuated (att) phenotypes as ideal attributes of live attenuated vaccine. Moreover, the vaccine strain strongly elicits both innate and adaptive immune response providing extremely early cross-protection from various influenza subtypes (Seo et al., 2007). Here, we established reverse genetic systems for both the X-31 and X-31ca virus. The utility of the system was verified by generating epidemic and pandemic influenza vaccine viruses on a routine basis. The method would serve as technical platform for the generation of both inactivated and live attenuated vaccine for the prevention of influenza epidemics and pandemics.
Results
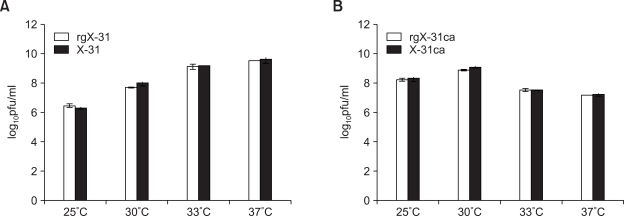
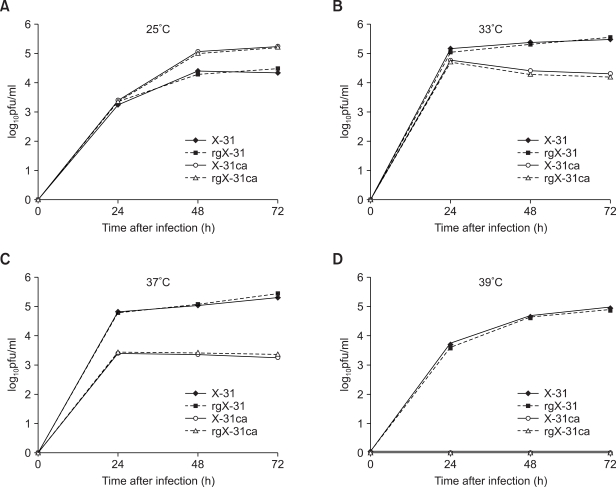
The eight plasmids set of X-31 and X-31ca were transfected into 293T cells, and after 3 days of incubation, the supernatants were collected for plaque assay on MDCK cells. Each virus was plaque-purified to ensure the genetic homogeneity. The recombinant viruses of X-31 and X-31ca were named as rgX-31 and rgX-31ca, respectively. We then inoculated eight embryonated chicken eggs with 100 pfu/100ul of X-31, X-31ca, rgX-31 and rgX-31ca viruses, respectively. Following incubation for three days at different temperatures, 25, 30, 33 and 37℃, the allantonic fluids were harvested, and the virus titer was estimated by plaque assay on MDCK cells. As shown in Figures 1A and 1B, the growth of X-31 and X-31ca virus in eggs was compared with that of rgX-31 and rgX-31ca. At all different temperatures tested, 25, 30, 33 and 37℃, respectively, the growth pattern was indistinguishable suggesting that rgX-31 and rgX-31ca were biologically identical to X-31 and X-31ca virus, respectively. Both X-31 and rgX-31 grow more efficiently at 33℃ and 37℃ than lower temperature of 30℃ and 25℃. In contract, both X-31ca and rgX-31ca grow better at low temperature (25℃, 30℃) than higher temperatures, and 30℃ was found to be the best temperature for the growth of the X-31ca and rgX-31ca. The results confirm the ca and ts phenotype of the cold-adapted virus (Lee et al., 2006). We then performed the growth of each virus on MDCK cells. The MDCK cells were infected with 0.01 m.o.i. of X-31, X-31ca, rgX-31 and rgX-31ca viruses, respectively, and the growth was compared at various temperatures (Figure 2). The X-31ca and rgX-31ca viruses replicate efficiently at 25℃ (Figure 2A), but are restricted at 37℃ (Figure 2C) and no growth was observed at 39℃ (Figure 2D). Basically, growth restriction was observed for X-31ca and rgX-31ca in a temperature-dependent fashion. In contrasts, both X-31 and rgX-31 viruses replicate efficiently at 33℃ and 37℃ (Figures 2B and 2C) and 37℃ was found to be the optimal temperature for both X-31 and rgX-31 viruses (Figure 2C). Overall, the plasmid-derived transfectant viruses (rgX-31, rgX-31ca) and their biological counterparts (X-31, X-31ca, respectively) exhibited comparable growth kinetics.
Figure 1.
Growth profiles of the X-31 and rgX-31 (A) and X-31ca and rgX-31ca (B) in embryonated chicken eggs. Viruses were grown in embryonated chicken eggs. Eggs were infected with 0.1 PFU/ml of the X-31, rgX-31, X-31ca and rgX-31ca viruses, respectively, and incubated at 25, 30, 33 and 37℃ for 3 days. Virus titers were determined by plaque assay on MDCK cells.
Figure 2.
Growth profiles of the X-31, rX-31, X-31ca and rX-31ca viruses in MDCK cells. MDCK cells were infected with 0.01 m.o.i. of the X-31, rgX-31, X-31ca and rgX-31ca viruses and incubated at 25℃ (A), 33℃ (B), 37℃ (C) and 39℃ (D) for 3 days. Supernatants were taken every 24 h for determination of viral titers by plaque assay.
Recent human infection by H5N1 and H9N2 subtypes (Peiris et al., 1999; Webby and Webster 2001; Lee and Saif, 2009) calls for the development of human vaccine strains against avian viruses. In part of testing the utility of the present reverse genetic system and its practical application for vaccine production, we generated rgX-31ca-ΔH5N1 and rgX-31ca-H9N2 reassortant viruses that have 6 internal genes of X-31ca and the two haemagglutinin and neuraminidase surface antigens derived from A/Chicken/Korea/MS96/96 and A/Indonesia/5/2005. Highly pathogenic H5N1 viruses carry multi-basic amino acids at the cleavage site of haemagglutinin gene (Cinatl et al., 2007), and the deletion of the amino acids usually leads to attenuation of virulence (Suguitan et al., 2006). For this purpose, we removed the multi-basic amino acids of haemagglutinin gene at the cleavage site of A/Indonesia/5/2005 (PQRESRRKKRG→PQREKRG). The corresponding PHW2000-haemagglutinin and PHW2000-neuraminidase genes of avian origin were mixed with the 6 internal plasmids of the X-31ca virus and tranfected into 293T cells. As shown at Figure 3, the genotyping of the plaque purified viruses by the multiplex RT-PCR method (Ha et al., 2006) showed that the genetic composition of the transfectants, rgX-31ca-ΔH5N1 and rgX-31ca-H9N2, contains six internal segments derived from X-31ca virus and the two segments encoding surface antigens, haemagglutinin and neuraminidase, derived from the wild type avian viruses.
Figure 3.
Genotyping of transfectant viruses by multiplex RT-PCR. PCR products of X-31ca reassortants, rgXca-H9N2 (between X-31ca and A/Chicken/Kores/MS96/96) and rgXca-H5N1 (between X-31ca and A/Indonesia/2005) were fractionated by 1% agarose gel and stained with ethidium bromide. Lane 1 and 4: specific for X-31caPB2 (1,002 bp): NP (853 bp): PB1 (447 bp); Lane2 and 5: specific for X-31ca-PA (730 bp): NS (521 bp): M (326 bp); Lane3: specific for haemagglutinin (981 bp) and neuraminidase (552 bp) of H9N2; Lane6: specific for haemagglutinin (510 bp) and neuraminidase (1,286 bp) of H5N1.
Discussion
Here we developed an efficient reverse genetic system by which the 6:2 reassortant influenza vaccine strains could be generated from cDNAs derived from the influenza RNAs. For this purpose, 6 internal genes were derived from the high-yielding X-31 virus strain and the two genes for haemagglutinin and neuraminidase were derived from human infecting virulent influenza viruses. Two different versions of X-31 virus, one for wild type and the other for cold-adapted virus, designed for inactivated vaccine by injection and for LAIV as nasal spray, respectively.
Especially for vaccine production, serious consideration should be given to the fact that not all influenza viruses grow well in fertilized eggs. Poor growth and relatively low yield of wild type virulent viruses in the embryonated eggs would hamper the production of seasonal influenza vaccine in a timely manner. To circumvent the problems, vaccine viruses have been traditionally generated by annual reassortment with high-growth mater strain. This process requires the generation of reassortant viruses of essentially random genetic composition followed by screening and selection of virus of desired RNA composition, which is labor intensive and time-consuming. Recently, various groups have developed reverse genetic systems by which recombinant viruses could be rescued from transfection of cDNAs derived from influenza RNA segments. Neumann et al. (1999) and Palese et al. (1997) developed 12 plasmid systems where 8 plasmids were designed to express influenza virion RNAs (vRNAs) and the four plasmids expressing the polymerase proteins (PB1, PB2, PA and NP) required for the formation of influenza RNP complexes that support replication and transcription of vRNAs. Subsequently, Hoffman et al. (2000) developed 8 bi-directional plasmids that was designed to express negative-stranded influenza vRNAs in one direction and positive-stranded mRNAs from opposite direction. The system was found useful for the generation of the 6:2 reassortant viruses for the vaccine purpose. For instance, Suguitan et al. groups (Suguitan et al., 2006) inserted modified haemagglutinin and neuraminidase genes from wild type H5N1 into ca A/Ann Arbor/6/60 virus by reverse genetics as a means to generation of reassortant vaccine strains (Bardiya and Bae, 2005).
In our work here, we adopted and further extended the eight bi-directional plasmids system for the generation of influenza vaccine strain based on the X-31 virus that affords high yield in embryonated eggs. Based on the present reverse genetic system, we generated two independent 6:2 reassortant viruses, rgX-31ca-H9N2 and rgX-31ca-ΔH5N1, and the growth properties of the viruses were examined. Both reassortant viruses grew well in the embryonic chicken eggs and they also exhibited the ca (cold-adapted) growth property at 30℃ low temperature (unpublished data). Therefore, regardless of the origin of haemagglutinin and neuraminidase genes, the reassortant viruses still maintained the high growth property in eggs and ca growth property in MDCK cells. The X-31 virus was previously described as one of the high-yielding reassortant viruses containing the six internal genes of A/PR/8/34 and the two RNAs (haemagglutinin and neuraminidase) of A/Aichi/2/68 (Kilbourne, 1969). Since this virus has been passaged many times in embryonated eggs, the virus has accumulated multiple nucleotide changes as compared to the parental A/PR/8/34 strain (Lee et al., 2001). These accumulated mutations in internal genes of X-31 are due to the repeated passage in eggs, has an effect to enhance the growth property in embryonated chicken eggs (Lee et al., 2001), and further contributed to partial attenuation (data not shown). Therefore, the X-31 backbone based reverse genetic system is expected to provide a safer process for vaccine production. Furthermore, further attenuation by cold-adaptation of the X-31 virus would be suitable for using the X-31ca reverse genetics for the generation of live attenuated vaccine that could be produced in high yield. Since attenuation phenotype is usually associated with the loss of viruses' growth, it is sometimes difficult to yield vaccine strains maintaining desirable level attenuation as well as high growth property. And yet, the X-31 virus maintained good growth as well as desired level of attenuation. In brief, X-31 and X-31ca reverse genetic system would serve as technical platforms for the generation of both injectable inactivated vaccine and the nasal spray live attenuated vaccine for the prevention of influenza epidemics and pandemics. Besides its application for a seasonal influenza vaccine, the platform could be further extended to vaccines against H1N1 pandemic virus as well as H5N1 and H9N2 avian influenza viruses.
Methods
Cells and viruses
Madin-Darby Canine kidney (MDCK) cells and 293T cells were obtained from American Type Culture Collection (ATCC) and cultured in MEM (GIBCO) supplemented with 10% FBS (GIBCO). The X-31 virus was from the University of Oxford, UK. The X-31ca virus was generated after 92 passages at lower temperature in embryonated chicken eggs (Lee et al., 2006).
Plasmids for reverse genetics
Viral RNAs from X-31 and X-31ca virus were reverse transcribed and amplified by PCR and the eight cDNAs of viral gene segments of X-31 and X-31ca were inserted between Pol I and Pol II promoter of PHW2000 vector, which was provided by Dr. Webster (Hoffmann et al., 2002). The nucleotide sequence of all plasmids was verified by multiple sequencing of the gene from opposite directions. The cDNAs of the haemagglutinin and neuraminidase gene of A/Indonesia/5/2005 were made by total chemical synthesis (GenScript Corporation). The multi-basic amino acids at the cleavage site of haemagglutinin1 and haemagglutinin2 were deleted for minimizing potential virulence of the vaccine virus. The viral RNAs were prepared from A/Chicken/Korea/MS96/96 (H9N2) using the RNA preparation kits (Qiagen) and the haemagglutinin and neuraminidase genes were amplified with specific primers and cloned into PHW2000 vectors. In the same fashion, the cDNAs of the neuraminidase and neuraminidase gene of A/Indonesia/5/2005 were cloned into PHW2000 vetor using BsmBI enzyme site.
Transfection of 293T cells
Transfection of 293T cells was carried out as previously described (Hoffmann et al., 2002). 293T cells were plated and grown on 6-well plates. The eight plasmids of mixed and transfected into 293T cell with lipofectamin reagents (Invitrogen). After 3 days of incubation, the supernatant were collected and the generation of viruses was confirmed by plaque assay on MDCK cells. Generation of reassortant viruses were also performed with 6 internal genes of X-31ca and 2 surface antigens of A/Chicken/Korea/MS96/96 and A/Indonesia/5/2005.
Genotyping of transfectant viruses by multiplex PCR
Genotyping of influenza vaccine viruses by multiplex RT-PCR was performed as previously described (Ha et al., 2006). Briefly, 0.5 µl of cDNA of reassortant viruses was added to the PCR reaction mixture containing 10 mM Tris-HCl (pH8.3), 50 mM KCl, 1.5 mM MgCl2, 1.5 µl of 10 mM each dNTP, 1 µl of 10 µM each primer set, and 1 unit i-Taq polymerase (iNtRON Biotechnology). The mixture was initially denaturized at 95℃ for 5 min and then followed by 25 cycles of 95℃ for 30 s, 55℃ for 45 s, and 72℃ for 1 min. The final incubation was at 72℃ for 7 min.
Acknowledgements
This work was supported by a grant of the Korea Healthcare Technology R&D Project (grant number: A085105) and by National Research Foundation of Korea Grant (grant number: 2009-0092970) funded by the Korean Government.
Abbreviations
- att
attenuation
- ca
cold-adapted
- rg
reverse genetic
- ts
temperature-sensitive
References
- 1.Bardiya N, Bae JH. Influenza vaccines: recent advances in production technologies. Applied Microbiology and Biotechnology. 2005;67:299–305. doi: 10.1007/s00253-004-1874-1. [DOI] [PubMed] [Google Scholar]
- 2.Cinatl J, Michaelis M, Doerr H. The threat of avian influenza A (H5N1). Part I: epidemiologic concerns and virulence determinants. Medical Microbiology and Immunology. 2007;196:181–190. doi: 10.1007/s00430-007-0042-5. [DOI] [PubMed] [Google Scholar]
- 3.Cox RJ, Brokstad KA, Ogra P. Influenza Virus: Immunity and Vaccination Strategies. Comparison of theImmune Response to Inactivated and Live, Attenuated Influenza Vaccines. Scand J Immunol. 2004;59:1–15. doi: 10.1111/j.0300-9475.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- 4.Ha SH, Kim HA, Kim YH, Kim JS, Lee KH, Park SY, Park WJ, Seong BL. A multiplex RT-PCR method for screening of reassortant live influenza vaccine virus strains. J Virol Methods. 2006;134:154–163. doi: 10.1016/j.jviromet.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine. 2002;20:3165–3170. doi: 10.1016/s0264-410x(02)00268-2. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann E, Neumann G, Hobom G, Webster RG, Kawaoka Y. "Ambisense" Approach for the Generation of Influenza A Virus: vRNA and mRNA Synthesis from One Template. Virology. 2000;267:310–317. doi: 10.1006/viro.1999.0140. [DOI] [PubMed] [Google Scholar]
- 7.Kilbourne ED. Future influenza vaccines and the use of genetic recombinants. Bull World Health Organ. 1969;41:643–645. [PMC free article] [PubMed] [Google Scholar]
- 8.Kilbourne ED, Murphy JS. Genetic studies of influenza viruses. I. Viral morphology and growth capacity as exchangeable genetic traits. Rapid in ovo adaptation of early passage Asian strain isolates by combination with PR8. J Exp Med. 1960;111:387–406. doi: 10.1084/jem.111.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CW, Saif YM. Avian influenza virus. Comparative Immunology. Microbiology and Infectious Diseases. 2009;32:301–310. doi: 10.1016/j.cimid.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Lee KH, Seo SU, Song JM, Lee CM, Kim HA, Seong BL. Characterization of live influenza vaccine donor strain derived from cold-adaptation of X-31 virus. Vaccine. 2006;24:1966–1974. doi: 10.1016/j.vaccine.2005.10.051. [DOI] [PubMed] [Google Scholar]
- 11.Lee KH, Youn JW, Kim HJ, Seong BL. Identification and characterization of mutations in the high growth vaccine strain of influenza virus. Arch Virol. 2001;146:369–377. doi: 10.1007/s007050170181. [DOI] [PubMed] [Google Scholar]
- 12.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson KG, Wood JM, Zambon M. Influenza. Lancet. 2003;362:1733–1745. doi: 10.1016/S0140-6736(03)14854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novel Swine-Origin Influenza A Virus Investigation Team. Emergence of a Novel Swine-Origin Influenza A (H1N1) Virus in Humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 15.Palese P, Zavala F, Muster T, Nussenzweig RS, Garcia-Sastre A. Development of novel influenza virus vaccines and vectors. J Infect Dis. 1997;176(Suppl 1):S45–S49. doi: 10.1086/514175. [DOI] [PubMed] [Google Scholar]
- 16.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PLS, Lai RWM, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 17.Seo SU, Lee KH, Byun YH, Kweon MN, Seong BL. Immediate and broad-spectrum protection against heterologous and heterotypic lethalchallenge in mice by live influenza vaccine. Vaccine. 2007;25:8067–8076. doi: 10.1016/j.vaccine.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Suguitan AL, Jr, McAuliffe J, Mills KL, Jin H, Duke G, Lu B, Luke CJ, Murphy B, Swayne DE, Kemble G, Subbarao K. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 2006;3:e360. doi: 10.1371/journal.pmed.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wareing MD, Tannock GA. Live attenuated vaccines against influenza; an historical review. Vaccine. 2001;19:3320–3330. doi: 10.1016/s0264-410x(01)00045-7. [DOI] [PubMed] [Google Scholar]
- 20.Webby RJ, Webster RG. Emergence of influenza A viruses. Philos Trans R Soc Lond B Biol Sci. 2001;356:1817–1828. doi: 10.1098/rstb.2001.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]