Abstract
We screened 150 individuals from two recent seroconverter cohorts and found that six (4%) had CXCR4-using viruses. Clonal analysis of these six individuals, along with a seventh individual identified during clinical care as a recent seroconverter, revealed the presence of both X4- and dual-tropic variants in these recently infected adults. The ability of individual CXCR4-using variants to infect cells expressing CD4/CXCR4 or CD4/CCR5 varied dramatically. These data demonstrate that virus populations in some newly infected individuals can consist of either heterogeneous populations containing both CXCR4-using and CCR5-tropic viruses, or homogeneous populations containing only CXCR4-using viruses. The presence of CXCR4-using viruses at early stages of infection suggests that testing for viral tropism before using CCR5 antagonists may be important even in persons with known recent infection. The presence of CXCR4-using viruses in a subset of newly infected individuals could impact the efficacies of vaccine and microbicide strategies that target CCR5-tropic viruses.
Numerous studies support the widely accepted viewpoint that CCR5-using (R5-tropic) HIV-1 dominates during the early stages of infection.1–4 The protective effect of the CCR5 Δ32 homozygous mutation against HIV-1 transmission provides compelling support for the highly selective transmission, or outgrowth, of R5-tropic viruses.5–9 Viruses that use CXCR4 exclusively (X4-tropic) or both CXCR4 and CCR5 (dual-tropic) typically emerge during later stages of disease.10–12 The presence of CXCR4-using viruses (X4- and dual-tropic) has been associated with rapid CD4+ T cell decline and accelerated disease progression.1,13–16 Whether this decline is a cause or consequence of disease progression is not known. Documented cases of CXCR4-using viruses in individuals recently infected with HIV-1 have raised concern because of the well-established association between CXCR4-using viruses and disease progression.17–19 It is unclear why R5-tropic viruses dominate early HIV-1 infection. Some suggest that a higher density of CCR5-expressing cells at mucosal surfaces or in lymphoid tissues may select for R5-tropic variants during transmission or favor replication after transmission.20
Phenotypic characteristics of CXCR4-using viruses in newly infected individuals, and the frequency with which they occur, have not been well defined. Since the presence of CXCR4-using variants in recent infection may have implications for disease progression, antiretroviral drug treatment, development of vaccines and microbicides, and postexposure prophylaxis, we screened for CXCR4-using viruses in recent seroconverter panels and characterized the coreceptor usage and envelope (env) sequences of individual clones from recently infected subjects who harbored CXCR4-using subtype B viruses. Viruses were classified as R5-, X4-, or DM (dual/mixed)-tropic, based on the phenotypic results determined using the Trofile assay.21 Briefly, full-length env sequences were amplified by RT-PCR and cloned into an env expression vector as env libraries. A replication-defective HIV-1 genomic vector containing a luciferase reporter gene was then used to cotransfect human embryonic kidney cell cultures with patient env expression vectors. Coreceptor tropism of pseudoviruses was evaluated by infecting CXCR4- and CCR5-expressing cells in the presence and absence of CXCR4 and CCR5 inhibitors.
We tested 150 individuals from two separate cohorts of recent seroconverters. Four subjects (1, 2, 4, and 5) were identified in a cohort of 126 seroconverters,22 and two others (3 and 7) were identified in a second cohort of 24 seroconverters. Overall, 4% (95% CI 3.1–7.1%) of the 150 recent seroconverters had CXCR4-using viruses. We also studied an additional subject (6) who was a newly infected individual identified in routine clinical care.23 All seven subjects were men who are believed to have acquired HIV-1 through sexual contact with other men. Viruses were isolated from plasma collected at the time of diagnosis (years 2000 and 2003). One subject (7) was infected with a virus population that was predominantly comprised of X4-tropic variants, while the remaining six subjects were infected with DM-tropic virus populations that exhibited notably different levels of infectivity (relative light units, RLU) in CCR5+ and CXCR4+ cells (Table 1). The DM-tropic virus populations from subjects 1, 2, and 3 displayed lower levels of infectivity in CXCR4+ cells compared to CCR5+ cells. In contrast, the DM-tropic virus population from subject 6 exhibited higher levels of infectivity in CXCR4+ cells compared to CCR5+ cells. The DM-tropic virus populations from subjects 4 and 5 infected both CCR5+ and CXCR4+ cells with similar efficiencies.
Table 1.
CXCR4-Using Viruses Were Identified in Seven Subjects with Recent HIV-1 Infection
| |
|
|
|
|
RLU of Env poolsc |
|
Number (%) of clones with R5, X4, and dual tropism (screening assay) |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Subject | Days since last HIV-neg testa | Viral load (copies/ml) | CD4+ T cells (cells/mm3) | Tropism env poolsb | CCR5+ cells | CXCR4+ cells | Number of clones analyzed | R5 | X4 | Dual |
| 1 | 187 | N/Ad | N/A | DM | 298,606 | 176 | 57 | 13 (23) | 0 | 44 (77) |
| 2 | 138 | N/A | N/A | DM | 127,062 | 210 | 26 | 0 | 0 | 26 (100) |
| 3 | 54 | 500,000 | 510 | DM | 507,257 | 315 | 80 | 70 (88) | 1 (1) | 9 (11) |
| 4 | 159 | N/A | N/A | DM | 803,100 | 812,573 | 30 | 0 | 0 | 30 (100) |
| 5 | 181 | N/A | N/A | DM | 292,83 | 39,126 | 24 | 0 | 0 | 24 (100) |
| 6 | 14–21 | 362,000 | 399 | DM | 755 | 109,913 | 24 | 0 | 0 | 24 (100) |
| 7 | 132 | 33,721 | 660 | X4 | 98 | 75,006 | 40 | 0 | 36 (90) | 4 (10) |
Days between last negative HIV diagnostic test result and HIV-positive sample collection. Subject 6 was diagnosed based on positive HIV RNA and negative HIV antibody test results in the setting of symptomatic HIV infection.
Coreceptor tropism determined using the Trofile assay (Monogram Biosciences).
RLU, relative light units of luciferase output in the Trofile assay.
N/A, not available.
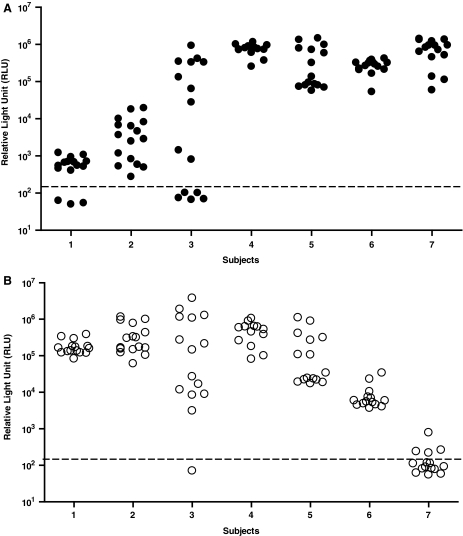
To understand the components of the CXCR4-using virus populations in these recently infected individuals, we performed env clonal analyses on viruses obtained from each of the subjects. We began by screening numerous clones from each virus population for their abilities to infect CCR5- and CXCR4-target cells to estimate the relative proportion of R5-, dual-, and X4-tropic clones (Table 1). The virus populations from subjects 2, 4, 5, and 6 were comprised exclusively of dual-tropic variants, whereas the virus populations from the remaining three subjects were comprised of mixtures of R5- and dual-tropic variants (1), X4- and dual-tropic variants (7), or R5-, X4-, and dual-tropic variants (3). To confirm the coreceptor tropism of these variants, we analyzed a subset of representative clones (103 clones total, 13–16 clones per sample) derived from each of the seven subjects using the Trofile assay. Infectivity levels (RLU) of the clones in CXCR4+ and CCR5+ cells are shown in Fig. 1. Both X4- and dual-tropic clones were identified in these seven newly infected subjects, and dual-tropic clones exhibited different abilities to infect CXCR4+ and CCR5+ cells, with infectivity ranging from 102 to 106 RLU.
FIG. 1.
Coreceptor Usage and V3 Sequences of env Clones from Seven Subjects Infected with CXCR4-Using Viruses
We previously reported that dual-tropic env clones of subtype D viruses varied in their ability to use CXCR4 and CCR5, and created two new designations, dual-X and dual-R, to describe dual-tropism based on coreceptor use and V3 amino acid sequence.24 Dual-X refers to dual-tropic clones that infect CXCR4+ cells efficiently and have V3 sequences that are distinct from R5-tropic clones in the same virus population; dual-R refers to dual-tropic clones that infect CXCR4+ cells poorly and have V3 sequences that are similar, or identical, to R5-tropic clones from the same virus population. To determine whether dual-tropic, subtype B variants from recent infections have similar characteristics, we sequenced the gp160 env of the 103 clones described above using conventional dideoxy-chain termination chemistry, and investigated the relationship among tropism, infectivity, and V3 amino acid sequence for each of the clones (Table 2). All clones from subject 2 were dual-R-tropic and contained closely related V3 sequences. All clones from subjects 4, 5, and 6 were dual-X-tropic with nearly identical V3 sequences within each population. The virus population of subject 1 included a mixture of dual-R- and R5-tropic clones sharing nearly identical V3 sequences. The virus population of subject 7 included a major subpopulation of X4-tropic and a minor subpopulation of dual-X-tropic clones with low levels of infectivity in CCR5-expressing cells; all but one of the clones shared identical V3 sequences. Subject 3 had the most complex virus population, containing R5-, dual-R-, dual-X-, and X4-tropic clones. The R5- and dual-R-tropic clones had identical V3 sequences, and the dual-X- and X4-tropic clones had similar V3 sequences (Table 2). These data are highly consistent with our previous observations in subtype D virus populations24 and extend the dual-R and dual-X subclassifications to subtype B viruses, as well as to virus populations in recently infected adults.
Table 2.
Coreceptor Usage and V3 Sequences of env Clones from Seven Subjects Infected with CXCR4-Using Viruses
| |
|
|
Infectivity measured by luciferase activityb |
Predictionsc |
V3 sequence analysisd |
||||
|---|---|---|---|---|---|---|---|---|---|
| Subject | Number of clonesa | Tropism (Trofile) | R5 RLU (median) | X4 RLU (median) | 11KR/25KR | PSSM | Net charge | PNGS | V3 amino acid sequences |
| 1 | 11 | Dual-R | 174,105 | 769 | R5 | R5 | 4 | 1 | CTRPNNNTRKsIHMGPGKAFYATGeIIGDIRKAHC |
| 2 | R5 | 120,891 | 59 | R5 | R5 | 4 | 1 | . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . | |
| 1 | R5 | 83,223 | 50 | R5 | R5 | 4 | 1 | . . . . . . . . . . . . . . . . . . . . . . . . . V . . . . . . . . . | |
| 1 | Dual-R | 164,877 | 461 | R5 | R5 | 4 | 1 | . . . . . . . . . R . . . . . . . . . . . . . . . . . . . . . . . . . | |
| 2 | 10 | Dual-R | 374,986 | 4,593 | R5 | R5 | 4 | 1 | CTRPNNNTRKgIHMGPGRVFYTTGeIIGDIRKAHC |
| 1 | Dual-R | 305,047 | 18,029 | R5 | R5 | 5 | 1 | . . . . . . . . . . . R . . . . . . . . . . . . . . . . . . . . . . . | |
| 1 | Dual-R | 104,486 | 4,637 | X4 | R5 | 6 | 1 | . . . . . . . . . . . . . . . . . . . . . . . . K . . . . . . . . . . | |
| 1 | Dual-R | 123,164 | 2,484 | R5 | R5 | 4 | 1 | . I . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . | |
| 1 | Dual-R | 164,741 | 3,654 | R5 | R5 | 4 | 1 | . . . . . . . . . . . . . V . . . . . . . . . . . . . . . . . . . . . | |
| 2 | Dual-R | 240,359 | 677 | R5 | R5 | 4 | 1 | . . . . . . . . . . . . . . . . . . . . . A . . . V . . . . . . . . . | |
| 3 | 5 | R5 | 1,082,926 | 74 | R5 | R5 | 4 | 1 | CTRPGNNTRRsITMGPGRAFYTTGeIIGDIRKAHC |
| 3 | Dual-R | 1,278,284 | 1,426 | R5 | R5 | 4 | 1 | . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . | |
| 5 | Dual-X | 8,862 | 349,224 | R5 | R5 | 7 | 1 | . . . . . . . . . . . . H . . -H . R . . . . KN . . . . . . . . . . | |
| 1 | Dual-X | 11,893 | 27,700 | R5 | R5 | 6 | 1 | . . G . . . . . . . . . H . . -H . R . . . . KN . . . . . . . . . . | |
| 1 | X4 | 71 | 328,864 | R5 | X4 | 7 | 1 | . . . . . . S . . . G . LV . -T . R . . . . RN . . . . . . . . . . | |
| 4 | 13 | Dual-X | 529,308 | 759,363 | R5 | R5 | 5 | 1 | CTRPNNNTRKgIVIGPGRSFYAARsIIGDIRQAHC |
| 5 | 14 | Dual-X | 29,245 | 208,732 | X4 | X4 | 4 | 1 | CTRPNNNTIKgIRIGPGRAIYATEkIVGDIRQAHC |
| 1 | Dual-X | 106,948 | 135,564 | X4 | X4 | 4 | 1 | . . . . . . . . . . . . . . . . . . . V . . . . . . . . . . . . . . . | |
| 6 | 13 | Dual-X | 5,930 | 313,126 | X4 | X4 | 6 | 0 | CTRPNNNIRRrIHIGPGRAFYATdITGSIRRAYC |
| 1 | Dual-X | 5.294 | 207,153 | X4 | X4 | 5 | 0 | . . . . . . . . . . . . . . . . . . G . . . . . . . . . . . . . . . . | |
| 7 | 10 | X4 | 86 | 770,172 | X4 | X4 | 5 | 0 | CTRPNEYRTRrIHIGPGRAFVTTKsITGDIRQAYC |
| 4 | Dual-X | 252 | 1,149,581 | X4 | X4 | 5 | 0 | . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . | |
| 1 | X4 | 82 | 454,024 | X4 | X4 | 5 | 0 | . . . . . D . . . . . . . . . . . . . . . . . . . . . . . . . . . . . | |
The number of clones with identical V3 sequences.
RLU, relative light units of luciferase output in the Trofile assay.
11KR-25KR, positively charged lysine and/or arginine residue at position 11 and/or 25 in the V3 loop. PSSM, position-specific scoring matrices.26
PNGS, potential N-linked glycosylation site. PNGS site located at amino acids 6–8 in V3 are underlined; amino acid positions 11 and 25 are highlighted in bold.
Next, we evaluated the accuracy of tropism predictions generated using V3 sequence-based algorithms by analyzing the V3 sequences characterized in this study. Both the 11RK/25RK rule25 and position-specific scoring matrices (PSSM)26 correctly assigned coreceptor tropism in some, but not all cases (Table 2). For example, all X4- and dual-X-tropic clones from subjects 5, 6, and 7 were correctly predicted as CXCR4-using by the 11KR/25KR rule and PSSM. Conversely, all of the dual-X-tropic clones from subjects 3 and 4 were incorrectly predicted as R5-tropic. Furthermore, all of the dual-R clones analyzed were predicted to be R5-tropic by the 11RK/25RK rule and PSSM, except for one dual-R clone from subject 2 that had a positively charged lysine (K) residue at position 25. In prior studies, a higher net charge and fewer potential N-linked glycosylation sites (PNGS) in V3 have been associated with CXCR4 use.25,27 In this study, X4- and dual-X-tropic clones generally had higher net charges (+4 to +7, median: +5) than R5- and dual-R-tropic clones (+4 to +6, median: +4). Five of the 11 V3 sequences found in X4- and dual-X-tropic clones lacked PNGS, while the remaining six had a single PNGS at amino acids 6–8. All V3 sequences in the R5- and dual-R-tropic clones also had only one PNGS at this position. Overall, the associations between V3 genotype and coreceptor tropism were observed for some, but not all of the clones analyzed (Table 2).
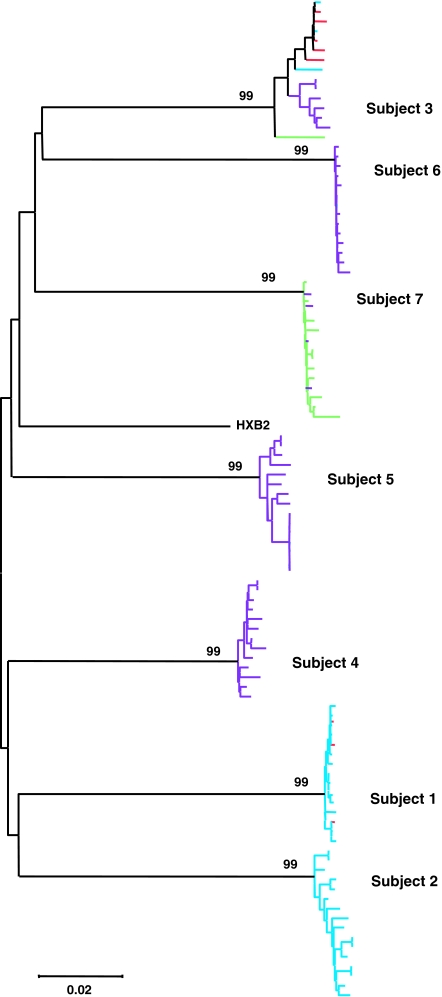
Phylogenetic analyses of gp160 nucleotide sequences of all 103 env clones from the seven subjects were performed using neighbor-joining methods (MEGA V3.0). These analyses revealed that each of the subjects was infected with phylogenetically distinct viruses (Fig. 2). All clones analyzed were subtype B (data not shown). The median within-patient env nucleotide diversity ranged from 0.19% to 1.33%, consistent with recent reports of early HIV infection.28,29 Similar to the V3 data, full-length env sequences from subject 3 exhibited the most heterogeneity among all subjects; dual-R- and R5-tropic clones clustered and were distinct from the dual-X- and X4-tropic clones in phylogenetic trees (Fig. 2). Our data indicate that individuals infected with CXCR4-using viruses can contain phylogenetically homogeneous or heterogeneous virus populations.
FIG. 2.
Phylogenetic analysis of full-length env sequences (gp160 nucleotide sequences) from seven subjects recently infected with CXCR4-using HIV-1. Replicate bootstrap resampling (1000×) of the data revealed >99% support for all seven subject nodes in neighbor-joining trees. Individual env clones are color coded for coreceptor tropism (red, R5; green, X4; blue, dual-R; purple, dual-X).
The presence of X4-tropic variants in individuals with recent seroconversion implies that either CXCR4-expressing cells exist at sites of transmission or X4-tropic viruses are carried to remote lymphoid tissues, where such target cells are available. However, our observations do not rule out the possibility that some transmitted R5-tropic variants are rapidly adapted to efficient CXCR4 use after infection. Clinical follow-up was available for subjects 6 and 7, with dual-X- and predominantly X4-tropic virus populations, respectively. Both individuals experienced rapid CD4+ T cell declines within 1 year of HIV-1 diagnosis (from 399 to 73 cells/mm3 for subject 6; 660 to 192 cells/mm3 for subject 7). Thus, these two cases support the linkage between CXCR4 use during primary or early-stage infection and accelerated disease progression. The efficiency of CXCR4 use of individual variants and the proportion of CXCR4-using variants in virus populations may impact the pathogenesis and clinical course of HIV-1 infection in recently infected individuals. Unfortunately, no follow-up information was available for the other five subjects in this study. Based on observations in sexual transmission reported here and vertical transmission reported elsewhere,30 we speculate that in addition to evolution from R5-tropic viruses, the emergence of CXCR4-using viruses during later stages of HIV-1 infection could, in some cases, result from the outgrowth of transmitted CXCR4-using variants.
The presence of CXCR4-using variants may have important implications for treatment regimens that include CCR5 inhibitors. In clinical evaluations of the CCR5 antagonists, maraviroc (Pfizer) and vicriviroc (Schering-Plough), highly treatment-experienced patients harboring only R5-tropic virus showed significant reductions in viral load,31,32 whereas patients with CXCR4-using virus did not.33–36 The utility of CCR5 antagonists in treatment-naive or early treatment settings is currently under investigation.37 Recent surveys have reported CXCR4-using virus was detected in approximately 20% of antiretroviral drug-naive HIV-1-infected patients.15,38 Here our results suggest that although R5-tropic viruses predominate in early HIV-1 infection, CXCR4-using viruses are not rare. Thus, testing for viral tropism to reduce the risk of treatment failure could be important prior to initiating CCR5 inhibitor therapy, even for patients in early stages of infection.
Acknowledgments
This study was supported in part by (1) National Institutes of Health (NIH)–U.S. National Institute of Allergy and Infectious Diseases (NIAID) Small Business Innovation Research (SBIR) Grant R44-AI048990, (2) NIH-NIAID Program Grant P01 A1071713, (3) the HIV Network for Prevention Trials (HIVNET) sponsored by the Department of Health and Human Services (DHHS) through contract N01-AI-35173 with Family Health International, contract N01-AI-45200 with the Fred Hutchinson Cancer Research Center, and contract N0I-AI-35173-417 with Johns Hopkins University, and (4) the HIV Prevention Trials Network (HPTN), sponsored by the NIAID, National Institutes of Child Health and Human Development (NICH/HD), National Institute on Drug Abuse (NIDA), National Institute of Mental Health (NIMH), and Office of AIDS Research, NIH, DHHS (U01-AI-46745, U01-AI-48054, and U01-AI-068613).
We thank the Monogram Biosciences Clinical Reference Laboratory for performing coreceptor tropism assays, Sunny Choe for critical review, and Cynthia Sedik for administrative assistance. We also thank Drs. Coates, Koblin, Chesney, and Donnell and the EXPLORE study team for providing samples and data for this study.
Disclosure Statement
Wei Huang, Jonathan Toma, Eric Stawiski, Signe Fransen, Terri Wrin, Neil Parkin, Jeanette Whitcomb, Ean Coakley, and Christos Petropoulos are employees and shareholders of Monogram Biosciences. Susan Eshleman is a Clinical Advisor for Monogram Biosciences.
References
- 1.Schuitemaker H. Koot M. Kootstra NA, et al. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J Virol. 1992;66(3):1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu T. Mo H. Wang N, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 3.van't Wout AB. Kootstra NA. Mulder-Kampinga GA, et al. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94(5):2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keele BF. Giorgi EE. Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean M. Carrington M. Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 6.Liu R. Paxton WA. Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y. Paxton WA. Wolinsky SM, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2(11):1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 8.Paxton WA. Martin SR. Tse D, et al. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat Med. 1996;2(4):412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 9.Samson M. Libert F. Doranz BJ, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 10.Melby T. Despirito M. Demasi R. Heilek-Snyder G. Greenberg ML. Graham N. HIV-1 coreceptor use in triple-class treatment-experienced patients: Baseline prevalence, correlates, and relationship to enfuvirtide response. J Infect Dis. 2006;194(2):238–246. doi: 10.1086/504693. [DOI] [PubMed] [Google Scholar]
- 11.Hunt PW. Harrigan PR. Huang W, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis. 2006;194(7):926–930. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]
- 12.Coakley E. Benhamida J. Chappey C, et al. An evaluation of tropism profiles and other characteristics among 3988 individuals screened for the A4001026, A4001027 (MOTIVATE 1) and A4001028 (MOTIVATE 2) phase 2b/3 studies of MARAVIROC; Paper presented at the 2nd International Workshop on Targeting HIV Entry; Boston, MA. Oct 20–21, 2006. [Google Scholar]
- 13.Koot M. Keet IP. Vos AH, et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118(9):681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 14.Richman DD. Bozzette SA. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169(5):968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 15.Brumme ZL. Goodrich J. Mayer HB, et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis. 2005;192(3):466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 16.Daar ES. Kesler KL. Petropoulos CJ, et al. Baseline HIV type 1 coreceptor tropism predicts disease progression. Clin Infect Dis. 2007;45(5):643–649. doi: 10.1086/520650. [DOI] [PubMed] [Google Scholar]
- 17.Markowitz M. Mohri H. Mehandru S, et al. Infection with multidrug resistant, dual-tropic HIV-1 and rapid progression to AIDS: A case report. Lancet. 2005;365(9464):1031–1038. doi: 10.1016/S0140-6736(05)71139-9. [DOI] [PubMed] [Google Scholar]
- 18.Yu XF. Wang Z. Vlahov D. Markham RB. Farzadegan H. Margolick JB. Infection with dual-tropic human immunodeficiency virus type 1 variants associated with rapid total T cell decline and disease progression in injection drug users. J Infect Dis. 1998;178(2):388–396. doi: 10.1086/515646. [DOI] [PubMed] [Google Scholar]
- 19.Masquelier B. Capdepont S. Neau D, et al. Virological characterization of an infection with a dual-tropic, multidrug-resistant HIV-1 and further evolution on antiretroviral therapy. AIDS. 2007;21(1):103–106. doi: 10.1097/QAD.0b013e3280117053. [DOI] [PubMed] [Google Scholar]
- 20.Moore JP. Kitchen SG. Pugach P. Zack JA. The CCR5 and CXCR4 coreceptors––central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20(1):111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 21.Whitcomb JM. Huang W. Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51(2):566–575. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eshleman SH. Husnik M. Hudelson S, et al. Antiretroviral drug resistance, HIV-1 tropism, and HIV-1 subtype among men who have sex with men with recent HIV-1 infection. AIDS. 2007;21(9):1165–1174. doi: 10.1097/QAD.0b013e32810fd72e. [DOI] [PubMed] [Google Scholar]
- 23.Gandhi RT. Wurcel A. Rosenberg ES, et al. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin Infect Dis. 2003;37(12):1693–1698. doi: 10.1086/379773. [DOI] [PubMed] [Google Scholar]
- 24.Huang W. Eshleman SH. Toma J, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: High prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol. 2007;81(15):7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fouchier RA. Groenink M. Kootstra NA, et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66(5):3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen MA. Li FS. van 't Wout AB, et al. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77(24):13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malenbaum SE. Yang D. Cavacini L. Posner M. Robinson J. Cheng-Mayer C. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J Virol. 2000;74(23):11008–11016. doi: 10.1128/jvi.74.23.11008-11016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salazar-Gonzalez JF. Bailes E. Pham KT, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82(8):3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottlieb GS. Heath L. Nickle DC, et al. HIV-1 variation before seroconversion in men who have sex with men: Analysis of acute/early HIV infection in the multicenter AIDS cohort study. J Infect Dis. 2008;197(7):1011–1015. doi: 10.1086/529206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Church JD. Huang W. Mwatha A, et al. HIV-1 tropism and survival in vertically infected Ugandan infants. J Infect Dis. 2008;197(10):1382–1388. doi: 10.1086/587492. [DOI] [PubMed] [Google Scholar]
- 31.Gulick RM. Lalezari J. Goodrich J, et al. Maraviroc for previously treated patients with R5 HIV-1 infection. N Engl J Med. 2008;359(14):1429–1441. doi: 10.1056/NEJMoa0803152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gulick RM. Su Z. Flexner C, et al. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-Infected, treatment-experienced patients: AIDS clinical trials group 5211. J Infect Dis. 2007;196(2):304–312. doi: 10.1086/518797. [DOI] [PubMed] [Google Scholar]
- 33.Mayer H. van der Ryst E. Saag M, et al. Safety and efficacy of MARAVIROC, a novel CCR5 antagonist, when used in combination with optimized background therapy for the treatment of antiretroviral-experienced subjects infected with dual/mixed-tropic HIV-1: 24-Week results of a phase 2b exploratory trial; Paper presented at the XVI International AIDS Conference; Toronto. Aug 13–18, 2006. [Google Scholar]
- 34.Westby M. Lewis M. Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80(10):4909–4920. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis M. Simpson P. Fransen S, et al. CXCR4-using virus detected in patients receiving maraviroc in the phase III studies MOTIVATE 1 and 2 originates from a pre-existing minority of CXCR4-using virus. Antivir Ther. 2007;12:S65. [Google Scholar]
- 36.Su Z. Reeves JD. Krambrink A, et al. Response to vicriviroc in HIV-infected treatment-experienced subjects using an enhanced version of the Trofile HIV coreceptor tropism assay: Reanalysis of ACTG 5211 results. Antivir Ther. 2008;13(Suppl 3):A98. [Google Scholar]
- 37.Saag M. Heera J. Goodrich J, et al. Reanalysis of the MERIT study with the Enhanced TrofileTM Assay; Paper presented at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapeutics/IDSA 46th Annual Meeting; Washington, DC. Oct 25–28, 2008. [Google Scholar]
- 38.Moyle GJ. Wildfire A. Mandalia S, et al. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J Infect Dis. 2005;191(6):866–872. doi: 10.1086/428096. [DOI] [PubMed] [Google Scholar]