Abstract
The revolution in DNA sequencing technologies has now made it feasible to determine the genome sequences of many individuals; i.e., “personal genomes.” Genome sequences of cells and tissues from both normal and disease states have been determined. Using current approaches, whole human genome sequences are not typically assembled and determined de novo, but, instead, variations relative to a reference sequence are identified. We discuss the current state of personal genome sequencing, the main steps involved in determining a genome sequence (i.e., identifying single-nucleotide polymorphisms [SNPs] and structural variations [SVs], assembling new sequences, and phasing haplotypes), and the challenges and performance metrics for evaluating the accuracy of the reconstruction. Finally, we consider the possible individual and societal benefits of personal genome sequences.
Keywords: Human genome sequencing, resequencing, personal genomics, personalized medicine
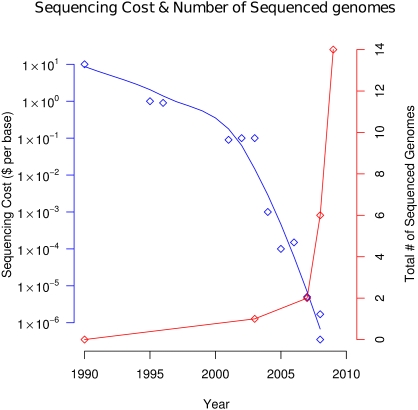
With the cost of DNA sequencing decreasing dramatically (Fig. 1), we are approaching a revolution in human biology and medicine. It is now possible for genome sequences to be determined for a large number of individuals, and the potential use of this information for discovery and medicine is enormous. Fourteen genome sequences have been reported to date (Table 1; Levy et al. 2007; Bentley et al. 2008; Ley et al. 2008; Wang et al. 2008; Wheeler et al. 2008; Ahn et al. 2009; Drmanac et al. 2009; Kim et al. 2009; Mardis et al. 2009; McKernan et al. 2009; Pushkarev et al. 2009; Pleasance et al. 2010a,b). Or have they? What exactly has been described? What is the optimal coverage of a human genome sequence to ensure an acceptable level of sequence accuracy? Given the implications of this biomedical revolution, it is worth reflecting on what constitutes a genome sequence, what one wishes to learn from that genome sequence, and what is the best approach to go about obtaining it.
Figure 1.
Cost of DNA sequencing and cumulative number of genomes sequenced as a function of time. The blue points and the fitted line show the per-base sequencing cost, and the red points show the total number of sequenced genomes.
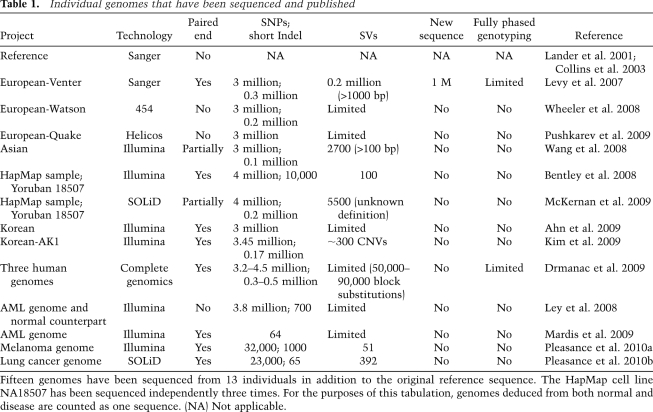
Table 1.
Individual genomes that have been sequenced and published
Fifteen genomes have been sequenced from 13 individuals in addition to the original reference sequence. The HapMap cell line NA18507 has been sequenced independently three times. For the purposes of this tabulation, genomes deduced from both normal and disease are counted as one sequence. (NA) Not applicable.
What does it currently mean to sequence an individual human genome?
Existing high-throughput DNA sequencing technologies generate relatively short reads (∼35–450 base pairs [bp]), and typically do not allow one to sequence a genome in its entirety using de novo assembly; i.e., constructing the genome sequence based solely on the sequencing reads without any other prior knowledge. Although improvements in sequencing technology and computational methods may soon permit routine use of de novo assembly (e.g., see Li et al. 2010), at the present time, sequencing a human genome typically refers to generating extensive sequence information using high-throughput DNA sequencing methods and relating this information to a reference haploid genome sequence to identify variations. The reference genome was deduced in the original international genome sequencing project from a collection of DNAs from anonymous individuals and assembled into a mosaic “haploid” genome (Lander et al. 2001). It is primarily of European origin and is estimated to be ∼99.99% accurate with ∼210 gaps (Genome Reference Consortium2009; http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?ORG=hum&MAPS=ideogr,est,loc&LINKS=ON).
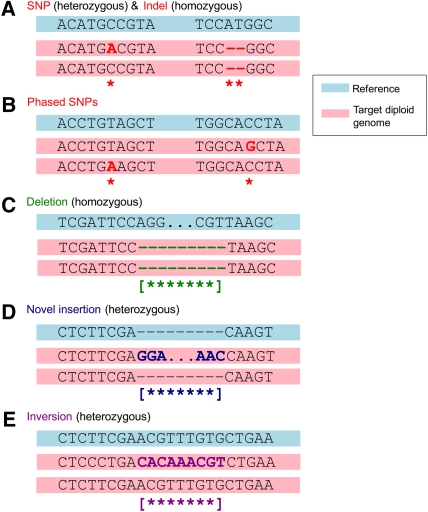
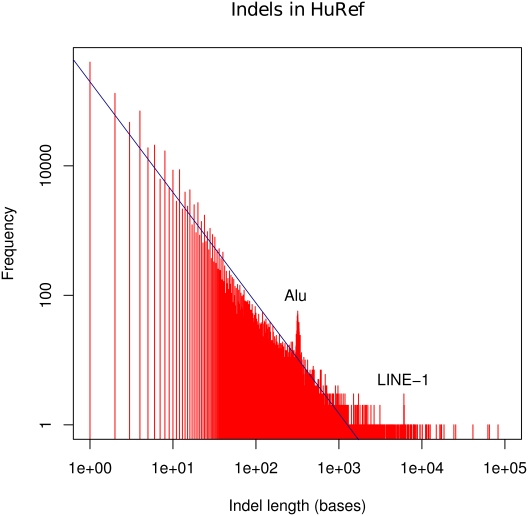
The determination of a new genome sequence relative to a reference genome is often referred to as “resequencing.” In each resequencing project, there are generally five parameters that can be evaluated (Fig. 2): (1) Single-nucleotide polymorphisms (SNPs). There are ∼3–4 million SNPs present in a given individual relative to the reference genome (Levy et al. 2007; Bentley et al. 2008; Wang et al. 2008; Wheeler et al. 2008; Ahn et al. 2009; McKernan et al. 2009; Pushkarev et al. 2009). (2) Small insertion and deletions (Indels) of size 2–1000 bp. There are ∼0.3–0.6 million Indels present in a given individual relative to the reference genome, and 320 Indels 1–2 kb in size (Fig. 3; Levy et al. 2007). (3) Large structural variations (SVs). Typically defined as deletions, insertions, and inversions >1000 bp, and can be >1 Mb in size. These SVs include transposons such as L1s, which comprise about one-third of the large SVs. There are at least 1000 SVs >2 kb in size present in a given individual relative to the reference genome (Korbel et al. 2007). (4) New sequences. These are DNA sequences that are present in an individual's genome sequence, but not in the reference genome. It is unclear how many of these exist. The identification of new sequences is confounded by the fact that the reference sequence (GRCh37) still contains ∼210 gaps, some of which are likely to contain new sequences (Genome Reference Consortium 2009; http://www.ncbi.nlm.nih.gov/mapview/maps.cgi). (5) Genotype/haplotype. Variants of a given sequence are assigned to particular chromosomes. It should be noted that the frequencies indicated in points 1–5– are subject to ascertainment bias and may not be representative.
Figure 2.
Types of variations present in a human genome sequence. The reference genome is shown at the top of each subfigure, with the individual's diploid genome shown below it. (A) A heterozygous insertion SNP and a homozygous small deletion. (B) Two phased SNPs. (C) A homozygous deletion in the target genome. (D) A heterozygous novel insertion. (E) A heterozygous inversion event.
Figure 3.
Length distribution of Indels. The figure (in log–log scale) shows the size distribution of all of the homozygous and heterozygous Indels in the HuRef genome (Levy et al. 2007) as compared with the NCBI reference.
Challenges in current personal genome sequencing
In general, SNPs and small Indels are relatively easy to identify, assuming that they can be captured in a single sequence read and correcting for sequencing errors (e.g., Li et al. 2009). However, the reference human genome sequence contains a number of DNA sequencing errors (0.01%) (Lander et al. 2001), and thus not all deviations found in a target genome sequence relative to the reference are necessarily SNPs.
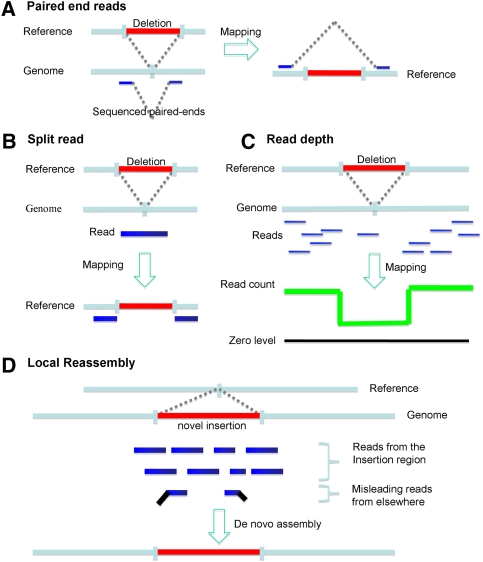
Large deletion SVs can be identified using reads long enough to have confidence that the deletion has been spanned; these reads map to adjacent, but discontinuous, regions of the genome (“split reads”) (Figs. 2, 4). The extent of altered sequences at the deletion breakpoint and the presence of repetitive sequences can make SV detection difficult. For breaks in unique DNA, 75- to 100-nt reads are likely to capture many of the large deletion SVs.
Figure 4.
Methods for detecting variation in a human genome sequence using DNA sequencing technologies. (A) Paired-end reads to detect insertions and deletions. (B) Split read methods for breakpoint idnetification. (C) Read depth analysis to detect CNVs. (D) Local reassembly to reconstruction novel insertions.
Large insertion SVs are identified using a variety of technologies (Fig. 4), such as (1) “read depth,” in which the frequency of reads from a segment of DNA reflects its copy number (Wang et al. 2008; Yoon et al. 2009); (2) paired-end mapping, in which end sequences from DNA fragments of particular sizes are determined and mapped to the genome to find variants (Korbel et al. 2007); and (3) DNA microarrays and comparative genome hybridization in which intensities of hybridization relative to a reference sample reveal copy number (Pinkel et al. 1998; Urban et al. 2006; Wang et al. 2009). These technologies are valuable for detecting large deletions as well. Paired-end mapping can also be used for detecting inversions, which are estimated to comprise ∼10% of SVs >2 kb in size (Korbel et al. 2007).
None of the current methods for detecting SVs are optimal. Many SVs reside in repetitive regions of the genome where reads cannot be unambiguously assigned. Also, SVs are often complex, with multiple events having occurred in close proximity, so that the events cannot be readily deduced using relatively short reads. For the case of transposon insertions, a major class of SVs, specialized algorithms can be used to facilitate their identification (e.g., X Li et al. 2008), although insertions that lie in repetitive regions are still likely to be difficult to identify accurately. Although SVs account for much of the variation in the genome in terms of numbers of base pairs (Korbel et al. 2007; Kidd et al. 2008), it is likely that a substantial number of SVs are missed in most genome sequencing projects. Consistent with this hypothesis, the frequency of large SVs detected in most genome sequences is low compared with that obtained by projects using longer reads, which are better at detecting SVs (e.g., Levy et al. 2007; see below). Many genome sequencing projects have chosen to report very few SVs or ignore them altogether (Table 1).
Methods for determining new sequences are also problematic, particularly when using short reads. In principle, paired-end sequences in which one end lies in a known region and another lies in novel sequence can be used to identify new sequence regions (Kidd et al. 2008), and the new sequences can be assembled into contigs. However, the accuracy of such a method remains to be determined empirically. Currently, very few genome resequencing projects analyze new DNA sequences.
A major complexity for human genome resequencing is the diploid nature of the genome, containing two nearly identical copies of each sequence. This tends to confound many of the structural variant assignment approaches, since reads supporting the variant structure and the “normal” reference genome can be present simultaneously. Moreover, it introduces the problem of “phasing” (determining on which of the two chromosomes a variant is located). Two variants that lie in a gene on the same chromosome “phased variants” are not the same as two variants (i.e., alleles) located in genes on separate chromosomes (Fig. 2). The activity of the two alleles would likely be very different in the two cases. Furthermore, with duplications, there may be three or more copies of a sequence. This can mean that most individuals can have two variants of a gene but a few have three, and phasing of the variant sequences can be quite difficult. Currently, methods for assigning alleles to specific chromosomes require long reads and/or paired-end reads so that alleles can be linked or phased. Alternative approaches are also described below. Presently, very few, if any, individual genome sequencing projects using new high-throughput technologies report phasing of alleles.
Defining performance metrics for genome resequencing
For each of the parameters that may be considered in a genome resequencing project, there should be an estimate of sensitivity (i.e., completeness of coverage) and accuracy (i.e., error rate). For SNPs, these metrics are relatively easy to determine using “SNP microarrays” designed to detect known SNPs (e.g., Affymetrix or Illumina) (Hehir-Kwa et al. 2007) or using other methods (e.g., mass spectrometry) (Gabriel et al. 2009). For Indels, SVs, and new DNA sequences, accuracy can be determined by PCR and/or DNA sequencing across junctions. However, for SVs and new DNA sequences, assessment of accuracy is often easier in principle than in practice, as the affected regions often lie in repetitive sequences or the exact location of the SV breakpoint is not known, making primer design for PCR difficult. Estimates of accuracy for phasing are difficult to determine using measures beyond linked sequencing methods such as paired-end sequencing, and are most readily estimated from a comparison with known haplotype information and an analysis of related individuals.
In spite of these challenges, it is important to define reasonable metrics so that the performance of a resequencing project can be evaluated. Such measures should accurately report the sensitivity and accuracy of detecting variants at different levels (e.g., SNPs, Indels, and SVs). The Archon X Prize Competition (Archon X Prize Foundation 2006) uses a linear performance metric to evaluate the accuracy of resequencing. Their construction errors are counted as the sum of base errors in SNPs and Indels, and the number of wrongly identified rearrangements. Because SNPs, Indels, SVs, new sequences, and phasing are each unique parameters, we suggest each type of event be scored independently. In this fashion, the scoring is similar to that of baseball statistics, in which batting average, home runs, runs batted in, and strikeouts are each maintained as separate parameters. However, if one wants to add them into a single statistic, we suggest picking a simple summation scheme that reflects the total number of base pairs changed—analogous to the way the slugging percentage is computed from a sum of simpler ballplayer statistics. A potential scheme for recording the accuracy of genome sequence parameters is presented in Box 1.
How to sequence a personal genome: technologies and general considerations
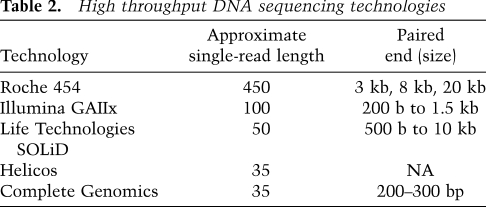
Presently, there are a variety of technologies for high-throughput DNA sequencing, all of which can be used either alone or in combination to help determine a human genome sequence (Table 2). Short-read technologies include Illumina GIIx, Life Technologies SOLiD, Complete Genomics Platform, and Helicos Helicope, and generate up to 400 million uniquely mapped reads of ∼35–120 bp per run (e.g., Bentley et al. 2008; Drmanac et al. 2009; McKernan et al. 2009; Pushkarev et al. 2009). Longer-read technologies from 454 produce ∼1 million reads of 450 bp. Most of these technologies can be used to obtain both single and paired-end reads (e.g., 454, Illumina, and SOLiD). 454 reads are sufficiently long to span Alu repetitive DNA sequences, and therefore are valuable for unambiguous identification. Although longer reads are clearly advantageous, they typically have a significantly higher cost per base.
Table 2.
High throughput DNA sequencing technologies
In principle, most genome sequencing could be performed with paired-end sequencing, because this approach provides both single reads and linkage information simultaneously. The one exception is 454 paired-end technology, for which the length of read from each end is slightly less than half the single-read length (Korbel et al. 2007), so there is benefit to using both single-read runs (to obtain longer reads) and paired-end runs. The paired-end technologies differ considerably in terms of the length of the fragment that can be sequenced from both ends. 454 technology allows for sequencing of ends from 20-kb fragments; Illumina and SOLiD technologies generally allow for sequencing from 1.5 to 3 kb, although claims for longer paired-end reads have been made (see http://www.454.com, http://www.illumina.com, and http://www3.appliedbiosystems.com). Lengths >7 kb are expected to be particularly useful, as these can span common transposon insertions such as L1s, and thereby can identify insertion events in a single read. It is expected that a combination of paired-end reads from different length fragments will provide optimal SV detection.
Box 1. Accuracy calls for genome variations.
The accuracy of SNP, Indel, SV, and haplotype phasing can be scored as follows: The accuracy of SNPs (AccSNP) can be defined by the proper identification and call and a simple 1 (detected) or 0 (miss) scoring system. The accuracy of Indels, SVs, and new sequences can be defined as both the overall identification of the events and the proper call of each inserted/deleted base, both within and associated with the event. Each of these parameters can be reported separately, with the event defined based on the alignment result of the actual variant sequence and the inferred variant sequence. For example, the accuracy of an individual novel insertion is
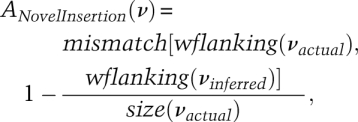 |
where νactual is the actual insertion (in simulations, it is already known; in reality, it will need to be identified in a validation step), νinferred is the insertion sequence inferred by the genome resequencing approach, mismatch returns the number of mismatches of two aligned sequences, wflanking returns a sequence with its flanking sequences on both ends, and size returns the size of a sequence. In this manner, the accuracy of individual insertions (and deletions, inversions, and rearrangements) can be parameterized.If one wants to combine the different assessments into a single score, a weighted approach can be established using a formula such as the one below:
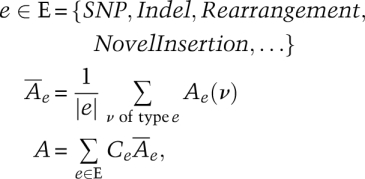 |
where e is a type of variant event (SNP, Indel, etc.), Ae(ν) is the individual detection accuracy for a specific variant ν (which is of type e), 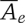 is the average accuracy in the detection of a certain type (e) of variation, |e| computes the number of all of the type e variant events, and A, the overall resequencing accuracy, is a linear combination of these average accuracy values. The Ce coefficients are chosen to reflect various weighting schemes. In the extreme, one can set all of the coefficients to a same normalization factor, and the resulting formula would be equivalent to counting the number of correctly identified variant events. If the coefficients are set according to the average size of the different types of events, the resulting definition will be counting the number of correctly called bases in the variant events. The first weighting scheme obviously puts more emphasis on SNP accuracy, while the later emphasizes SVs. The weighting scheme chosen for the X Prize (discussed in the text) is between these extremes.
is the average accuracy in the detection of a certain type (e) of variation, |e| computes the number of all of the type e variant events, and A, the overall resequencing accuracy, is a linear combination of these average accuracy values. The Ce coefficients are chosen to reflect various weighting schemes. In the extreme, one can set all of the coefficients to a same normalization factor, and the resulting formula would be equivalent to counting the number of correctly identified variant events. If the coefficients are set according to the average size of the different types of events, the resulting definition will be counting the number of correctly called bases in the variant events. The first weighting scheme obviously puts more emphasis on SNP accuracy, while the later emphasizes SVs. The weighting scheme chosen for the X Prize (discussed in the text) is between these extremes.
Accuracy is one of the most important parameters of any genome sequencing project. Technologies that generate 10-fold more sequence but at reduced accuracy may not be advantageous. Indeed, misassignments due to inaccurate short reads will be deleterious to genome sequencing projects. A final genome sequence that contains only one mistake per million bases will still contain 6000 errors! A rigorous comparison of the relative performance of the different high-throughput DNA sequencing technologies for each of the different parameters (SNPs, Indels, SVs, new sequences, and phasing) is sorely needed. Thus far, only one major project has chosen to use each technology on the same DNA (1000 Genomes Project, http://www.1000genomes.org), although an in-depth comparison of technologies was not performed.
Presumably, the optimal way to sequence a human genome will use a combination of technologies. Because each technology has different biases, a combinatorial approach would be expected to increase accuracy and help facilitate genome reconstruction. A reduced bias is particularly important to maximize coverage of both alleles. It has been reported that a DNA sequencing depth of 40-fold average coverage is required to identify 92% of both alleles using Illumina technology (Wang et al. 2008), and Wheeler et al. (2008) have performed a quantitative study on how coverage/sequencing depth affects detection rates of heterozygous SNPs. Du et al. (2009) illustrated how combining technologies would reduce the depth of sequencing required to identify structural variants. The use of different DNA sequence technologies would likely significantly reduce genome sequencing costs while achieving maximum accuracy.
Most individuals and scientific researchers will want to sequence genomes at a fixed budget. Therefore, it will be quite valuable to establish an algorithm to determine which combination of technologies and platforms provide the optimal coverage and accuracy of each sequence parameter—SNPs, Indels, SVs, new sequences, and genotypes/haplotypes—as a function of cost. Such an algorithm would be useful for guiding researchers toward applying technologies based on their research goals. Moreover, as additional technologies become available, it should be possible to incorporate these into the algorithm for evaluation.
A detailed genome resequencing strategy
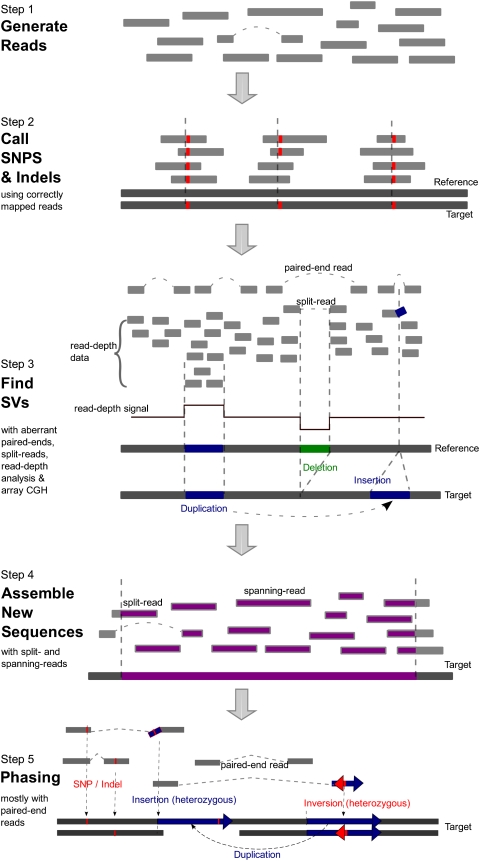
It is likely that most genome sequencing projects for the immediate future will be a hybrid resequencing strategy that incorporates both comparative (Pop et al. 2004) and de novo methods. Because most of the assembly can be done based on the existing reference genome, it is generally unnecessary to perform an experimentally and computationally intensive whole-genome de novo assembly. Local de novo assembly will still be necessary for regions with complex SVs and for new sequences. Perhaps most importantly, different DNA sequencing and array experiments can be integrated to identify different types of variant events (i.e., SNPs, Indels, and SVs) in a cost-effective way. In general, the experimental outputs will be processed in the following ways (Fig. 5):
Figure 5.
A general flow chart for determining a personal genome sequence. The generated reads are first mapped to the reference genome to call high-quality SNPs and small Indels (Step 1) and SVs based on aberrant alignment information (Step 2). The novel insertions can be reconstructed using local de novo assembly algorithms (Step 3), and a final phasing step (Step 4) will be able to deduce the complete diploid genome of the individual.
Mapping the reads back to the reference genome
Short reads will be obtained, and errors will be corrected (Pevzner et al. 2001) and combined into “unipaths” (a maximal unbranched sequence of the genome constructed based on the reads) (Butler et al. 2008). All of the the reads (long/medium/short) from an individual's genome are then mapped to the reference genome, generally allowing for partial matches and a few mismatches within the match—e.g., <6%, or no more than two mismatches—if no other better match exists. This mapping step is usually the most computationally time-consuming part of the project. Thus, one important aspect of the many short-read mapping programs—e.g., ELAND, MAQ (H Li et al. 2008), and Bowtie (Langmead et al. 2009)—is the manner in which they efficiency execute this process. The mapped reads are then divided into three categories according to the alignment results: those with unique best matches, sequences with multiple best matches, and those with no match (but they may contain partial matches). The last category usually corresponds to SVs, breakpoints (Lam et al. 2009), and new sequences with respect to the reference genome.
Identifying small variants (SNPs and Indels)
SNPs can be identified immediately based on results from the sequencing reads with single best matches, and the boundaries of deletions and small insertions will be detected by such reads as well (by allowing gaps in alignment). Algorithms that incorporate both detection frequency and base calling are typically used to ensure that the events called are high confidence and not simply due to DNA sequencing errors (e.g., Wheeler et al. 2008; Li et al. 2009). Results from SNP arrays can also reveal information about SNPs and validate assignments in a cost-effective way, as well as help assess the error frequency.
Finding SVs
Reconstructing large structural variants is the most challenging problem in genome resequencing. It is greatly complicated by the many levels of duplication in the human genome and the often complex nature of the rearrangements. Thus, a variety of approaches should be used to enhance their accurate detection. These approaches include paired-end reads, read depth, and split reads (e.g., Korbel et al. 2007; Wang et al. 2008; Chen et al. 2009; Yoon et al. 2009). All of these can be used to deduce the presence of SVs and their locations, and the split reads and novel sequences can be used to help define the rearrangement breakpoints. Long reads are particularly useful in helping to identify rearrangements and novel sequences located near the breakpoint regions. Because of technical problems detecting SVs (i.e., chimeric clones from ligations used to prepare DNA for sequencing), each event must be detected more than once. Using a combination of the different approaches is valuable for enhancing accuracy in SV identification and assignment of location.
CGH array data can also be integrated into the reconstruction process for such SVs. Incorporating the CGH data can also lower the coverage depth requirement of sequencing experiments, since the inner regions of segmental duplications/deletions not covered by low sequence coverage can still be identified by CGH results. The copy numbers of the genomic regions inferred from CGH array data can be integrated into the rearrangement analysis, and provide additional evidence of the actual SV type. Such CNV results can be correlated with trait-associated SNPs to identify candidate loci for influencing disease susceptibility (Conrad et al. 2009). CGH data can also be used to validate SVs, as well as provide information concerning accuracy.
Additional analyses for mapping SVs should also be applied. First, direct searches can be used to search directly for novel transposon insertions using sequences from the ends of common transposons (L1s, Alus, and retrotransposons). Second, de novo assembly is useful to reconstruct large novel insertions and to help identify complex genomic rearrangement events (e.g., segmental duplication/deletion). Third, SV events can also be identified by comparison with existing SV sequences (Lam et al. 2010). Finally, the heterozygous or homozygous nature of the SVs should be determined based on the existence of reads that map back to the corresponding reference sequences.
Assembling new sequences
De novo assembly of large novel insertions depends primarily on the reads from the new inserted sequences that are linked to known sequences. Misleading reads from elsewhere in the genome (usually highly represented regions) that are also found in the insertion can sometimes hinder the full reconstruction process. In such cases, longer reads that can unambiguously identify new sequences and appropriate assembly strategies are needed to ensure the correct assembly. Paired-end reads with an appropriate gap size can also help the unambiguous mapping of the reads inside novel insertions (Korbel et al. 2007). Simulation is useful in this context to help determine the optimal insert size (Korbel et al. 2009) and to calibrate error rates.
Phasing the variants
The last step in a genome sequencing project involves properly phasing all of the discovered variations. “Haplotype islands” or blocks of properly assigned variants can be extracted based on both the paired-end sequencing information (Lippert et al. 2002; Levy et al. 2007; Bansal et al. 2008) and the existing knowledge of the population haplotype patterns revealed by previous work (The International HapMap Consortium 2005). Other strategies include using information from family trios (Zhang et al. 2006a), in situ genotyping (Zhang et al. 2006a), and “chromosome dilution libraries” in which variants can be assigned to the same chromosome by analyzing dilutions containing individual chromosomes (Zhang et al. 2006b). Although most efforts on phasing have concentrated on SNPs, initial approaches to integrate SVs into the overall phasing scheme have begun recently (Conrad et al. 2009).
Personal genomes: possible uses for the information
The immediate applications of the information derived from a human genome sequence, both on the individual and societal levels, are somewhat limited at present. A reasonably large number of loci that are known to be predictive of disease or physiology (∼1500) (http://www.genetests.org) have been identified, and a number of these are clinically actionable. For the vast majority of these, the frequency of disease-causing alleles is quite rare. Moreover, most common traits are complex and likely involve many different loci and/or may be influenced by environment; how these factors are integrated to produce a given phenotype is poorly understood. Thus, currently, genome sequencing will likely not yet shed light on the genetic basis of these traits. In addition, for many of the known loci that have significant health implications, specific diagnostics tests already exist (e.g., for cystic fibrosis or for EGF receptor) (http://www.cff.org/aboutcf/testing/geneticcarriertest; Cappuzzo et al. 2005). For those interested in using DNA to trace human ancestry, a substantial amount of information may be readily deduced from analysis of a small subset of the genome. Thus, whole-genome sequencing is not necessary for these particular applications.
The promise of personal genomics lies in the future, as we amass a database of personal genomes. Large-scale analyses of multiple genomes are expected to reveal important insights into gene regulation and chromosome structure. Systematic collection of human phenotypic information along with personal genomic information will increase our ability to analyze the genetic basis of development, function, and disease. Integrating genomic information with other types of large-scale molecular data such as proteomics and metabolmics data holds promise for diagnostics in the future (Snyder et al. 2009). We envision a time in the future when personal genomic information is one of the essential tools used to tailor an individual's medical care.
The onus is on the biological community to be good stewards of personal genomics. In principle, and in keeping with the practices in the genomics community (Birney et al. 2009), the data amassed should be free and accessible to all, to ensure rapid scientific progress and hasten any medical advances. At the same time, the biological community has a continuing obligation to educate the general public about what this information can and cannot do, to avoid its misuse. In terms of misuse, personal privacy is of particular concern (Greenbaum et al. 2008). One can imagine many scenarios in which revealed genomic information is mined to determine peoples' personal characteristics without their consent. This is particularly true when one considers that one passes half of their genomic information to their children.
Nevertheless, many people may wish to be open with their personal genomic information (Lunshof et al. 2008). Indeed, as people become more comfortable with their genotype, it is easy to envision a future in which such information is readily shared. For example, at a party, individuals may casually chat about their genetic predispositions, just as they currently often share thoughts on their various ailments or treatments. Regardless of whether genomic information is maintained private or made public, such information, if properly handled, holds enormous value for both science and society.
Acknowledgments
This work was supported by grants from the NIH.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1864110.
References
- Ahn SM, Kim TH, Lee S, Kim D, Ghang H, Kim DS, Kim BC, Kim SY, Kim WY, Kim C, et al. The first Korean genome sequence and analysis: Full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archon X Prize Foundation. Archon X PRIZE competition guidelines. X Prize Foundation. 2006. http://genomics.xprize.org/files/downloads/genomics/Archon_X_PRIZE_for_Genomics_+Competiton_Guidelines.pdf.
- Bansal V, Halpern AL, Axelrod N, Bafna V. An MCMC algorithm for haplotype assembly from whole-genome sequence data. Genome Res. 2008;18:1336–1346. doi: 10.1101/gr.077065.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Hudson TJ, Green ED, Gunter C, Eddy S, Rogers J, Harris JR, Ehrlich SD, Apweiler R, Austin CP, et al. Prepublication data sharing. Nature. 2009;461:168–170. doi: 10.1038/461168a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J, Maccallum I, Kleber M, Shlyakhter IA, Belmonte MK, Lander ES, Nusbaum C, Jaffe DB. ALLPATHS: De novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuzzo F, Hirsc FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. BreakDancer: An algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2009;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Du J, Bjornson RD, Zhang ZD, Kong Y, Snyder M, Gerstein MB. Integrating sequencing technologies in personal genomics: Optimal low cost reconstruction of structural variants. PLoS Comput Biol. 2009;5:e1000432. doi: 10.1371/journal.pbi.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;60:2.12.1–2.12.18. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- Genome Reference Consortium. National Center for Biotechnology Information; 2009. Human genome assembly information. http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/data/index.shtml. [Google Scholar]
- Greenbaum D, Du J, Gerstein M. Genomic anonymity: Have we already lost it? Am J Bioeth. 2008;8:71–74. doi: 10.1080/15265160802478560. [DOI] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Egmont-Petersen M, Janssen IM, Smeets D, van Kessel AG, Veltman JA. Genome-wide copy number profiling on high-density bacterial artificial chromosomes, single-nucleotide polymorphisms, and oligonucleotide microarrays: A platform comparison based on statistical power analysis. DNA Res. 2007;14:1–11. doi: 10.1093/dnares/dsm002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit J, Godwin B, Grubert F, Simons JF, Kim PK, Palejev D, Carriero N, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Abyzov A, Mu XJ, Carriero N, Cayting P, Zhang Z, Snyder M, Gerstein MB. PEMer: A computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol. 2009;10:R23. doi: 10.1186/gb-2009-10-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HYK, Mu XJ, Stutz AM, Tanzer A, Cayting PD, Synder M, Kim PM, Korbel JO, Gerstein MB. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat Biotechnol. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kahveci T, Settles AM. A novel genome-scale repeat finder geared towards transposons. Bioinformatics. 2008;24:468–476. doi: 10.1093/bioinformatics/btm613. [DOI] [PubMed] [Google Scholar]
- Li R, Li Y, Fang X, Yang H, Wang J, Kristiansen K, Wang J. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert R, Schwartz R, Lancia G, Istrail S. Algorithmic strategies for the single nucleotide polymorphism haplotype assembly problem. Brief Bioinform. 2002;3:23–31. doi: 10.1093/bib/3.1.23. [DOI] [PubMed] [Google Scholar]
- Lunshof JE, Chadwick R, Vorhaus DB, Church GM. From genetic privacy to open consent. Nat Rev Genet. 2008;9:406–411. doi: 10.1038/nrg2360. [DOI] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;10:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1525–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010a;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010b;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M, Phillippy A, Delcher AL, Salzberg SL. Comparative genome assembly. Brief Bioinform. 2004;5:237–248. doi: 10.1093/bib/5.3.237. [DOI] [PubMed] [Google Scholar]
- Pushkarev D, Neff N, Quake S. Single-molecule sequencing of an individual human genome. Nat Biotechnol. 2009;27:847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Weissman S, Gerstein M. Personal phenotypes to go with personal genomes. Mol Syst Biol. 2009;5:273. doi: 10.1038/msb.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban AE, Korbel J, Selzer R, Popescu GV, Richmond T, Cubells JF, Green R, Emanuel BS, Gerstein M, Weissman SM, et al. High resolution mapping of DNA copy alterations using high density tiling oligonucleotide arrays. Proc Natl Acad Sci. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LY, Abyzov A, Korbel JO, Snyder M, Gerstein M. MSB: A mean-shift-based approach for the analysis of structural variation in the genome. Genome Res. 2009;19:106–117. doi: 10.1101/gr.080069.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zhu J, Shendure J, Porreca GJ, Aach JD, Mitra RD, Church GM. Long-range polony haplotyping of individual human chromosome molecules. Nat Genet. 2006a;38:382–387. doi: 10.1038/ng1741. [DOI] [PubMed] [Google Scholar]
- Zhang K, Martiny AC, Reppas NB, Barry KW, Malek J, Chisholm SW, Church GM. Sequencing genomes from single cells via polymerase clones. Nat Biotechnol. 2006b;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]