Abstract
Methylation of histone H3 Lys 9 and Lys 27 (H3K9 and H3K27) is associated with transcriptional silencing. Here we show that KDM7, a JmjC domain-containing protein, catalyzes demethylation of both mono- or dimethylated H3K9 and H3K27. Inhibition of KDM7 orthologs in zebrafish resulted in developmental brain defects. KDM7 interacts with the follistatin gene locus, and KDM7 depletion in mammalian neuronal cells suppressed follistatin gene transcription in association with increased levels of dimethylated H3K9 and H3K27. Our findings identify KDM7 as a dual demethylase for H3K9 and H3K27 that functions as an eraser of silencing marks on chromatin during brain development.
Keywords: Demethylase, JmjC, methylation, histone, chromatin, epigenetics
Histone methylation status defines the epigenetic program of a cell by determining chromatin structure and thereby regulating DNA-dependent processes such as transcription (Strahl and Allis 2000; Lachner et al. 2003; Margueron et al. 2005; Martin and Zhang 2005). Histone methylation has also been linked to regulation of neuronal function (Iwase et al. 2007). The recent discovery of histone demethylases revealed that histone methylation is a more dynamic process than previously recognized, and that most identified demethylases show a strict substrate specificity limited to a single methylation site (Bannister et al. 2002; Shi et al. 2004; Klose et al. 2006a; Tsukada et al. 2006; Shi and Whetstine 2007; Cloos et al. 2008; Lan et al. 2008). A number of histone demethylases contain a JmjC domain, and a subfamily of JmjC domain-containing proteins (comprising KIAA1718, PHF8, and PHF2) is evolutionarily conserved from Caenorhabditis elegans to humans and is characterized by the presence of a PHD-type zinc finger motif in addition to the JmjC domain (Supplemental Fig. S1A). Whereas the human genes for PHF8 and PHF2 are associated with X-linked mental retardation and hereditary sensory neuropathy type I, respectively (Hasenpusch-Theil et al. 1999; Laumonnier et al. 2005; Abidi et al. 2007; Koivisto et al. 2007), little is known about KIAA1718. Bioinformatic analysis of the JmjC domains of KIAA1718, PHF8, and PHF2 indicated that predicted Fe(II)- and α-ketoglutarate (α-KG)-binding sites are conserved, with the exception of the former in PHF2, and that they share extensive similarity with the JmjC domain of JHDM1/KDM2 (Supplemental Fig. S1B). Conservation of residues within the putative cofactor-binding sites of KIAA1718 suggested that this protein might possess histone demethylase activity, and therefore might also contribute to transcriptional regulation of genes in the nervous system.
Results and Discussion
KIAA1718 possesses histone demethylase activity
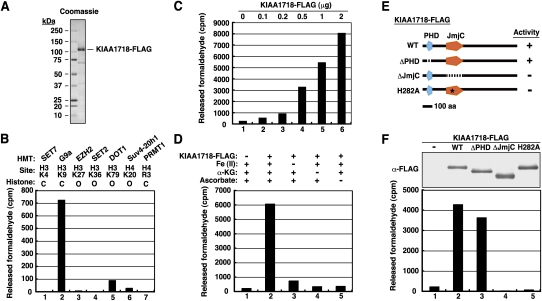
To examine whether KIAA1718 indeed possesses histone demethylase activity, we generated the mouse protein tagged with the Flag epitope at its C terminus in insect cells (Fig. 1A), and incubated the recombinant protein with histone substrates labeled with 3H at various characterized methyl-lysine or methyl-arginine sites by corresponding histone methyltransferases (HMTs). Histone demethylase activity was monitored by measurement of the release of the labeled demethylation product, formaldehyde. Substantial release of labeled formaldehyde was observed in the reaction mixture containing histone H3 labeled on Lys 9 (H3K9) by G9a, but not in those containing histone substrates modified by other HMTs (Fig. 1B). Consistent with the notion that the observed enzymatic activity was intrinsic to KIAA1718, formaldehyde release from G9a-labeled H3 was dependent on KIAA1718 concentration (Fig. 1C). To ascertain whether the demethylation mediated by KIAA1718 is oxidative in nature, with Fe(II) and α-KG as cofactors, we examined whether the enzymatic activity of KIAA1718 is dependent on these cofactors. The release of formaldehyde mediated by KIAA1718 was indeed found to require both Fe(II) and α-KG (Fig. 1D). Ascorbate was also required for the enzymatic activity, presumably as a result of its ability to regenerate Fe(II) from Fe(III). To verify further that the observed enzymatic activity is attributable to a genuine demethylase, we generated recombinant forms of KIAA1718 that either lack the JmjC or PHD domains or contain a mutation (H282A) in the Fe(II)-binding site in insect cells (Fig. 1E). Analysis of similar amounts of the mutant proteins for histone demethylase activity revealed that deletion of the JmjC domain or mutation of His282 abolished the activity of KIAA1718, whereas the PHD domain appeared to be dispensable for such activity (Fig. 1E,F). Together, these results showed that KIAA1718 is a histone demethylase capable of removing methyl groups from H3K9. Given that histone demethylase activity is the first function attributed to KIAA1718, we named this protein KDM7 on the basis of the previously described nomenclature (Allis et al. 2007).
Figure 1.
KIAA1718 is a histone demethylase that targets H3K9. (A, right lane) SDS–polyacrylamide gel electrophoresis with Coomassie blue staining of a C-terminally Flag-tagged recombinant KIAA1718 protein. Molecular size standards are shown in the left lane. (B) Histone demethylase activity of purified KIAA1718-Flag with various methylated histone substrates. The HMTs used to generate the various substrates and their sites of methylation are indicated. The methylated substrates were generated with the indicated forms of histone ([C] core histone octamer; [O] oligonucleosome) on the basis of the substrate preference of each HMT. The presented counts have been corrected for control counts. (C) Histone demethylase activity of the indicated amounts of KIAA1718-Flag. (D) Effects of removal of Fe(II), α-KG, or ascorbate from the reaction mixture on the histone demethylase activity of KIAA1718-Flag with G9a-methylated histone substrate. (E) Schematic representation of wild-type (WT) and mutant forms of KIAA1718 showing whether they are active (+) or inactive (−) as H3K9 demethylases. (F, bottom panel) Demethylase activity of purified wild-type or mutant forms of KIAA1718-Flag with G9a-methylated histone substrate. (Top panel) The similar amounts of KIAA1718-Flag proteins used in the demethylase assay are revealed by immunoblot analysis with antibodies to Flag (α-Flag).
KDM7 is a dual demethylase for dimethylated and monomethylated H3K9 (H3K9me2/me1) and H3K27me2/me1
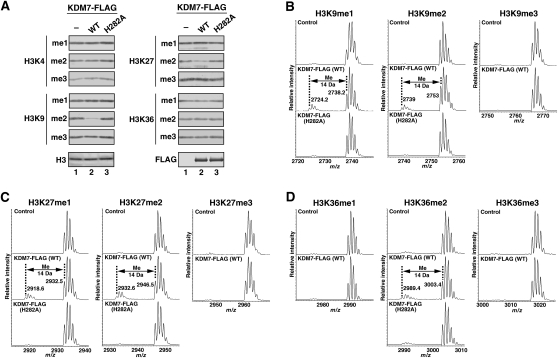
To define the substrate and modification state specificity of KDM7, we included core histones as substrates in demethylation reaction mixtures and examined the modification status of individual methylation sites by immunoblot analysis with a series of methylation-specific antibodies. Wild-type KDM7 mediated a marked decrease in the methylation level of both H3K9me2 and H3K27me2, without affecting that of other histone methylation sites (Fig. 2A; Supplemental Fig. S2A). In addition, KDM7 efficiently removed methyl groups from H3K9me2 and H3K27me2 in core histones, but not from those in mono- or oligonucleosomes (Supplemental Fig. S2B,C). These results suggested that KDM7 prefers core histones rather than mono- or oligonucleosomes as substrates, and explain why demethylase activity was not detected by the radioactive formaldehyde release assay with nucleosomal histones methylated by the HMT EZH2 (Fig. 1B). They also suggested that the low level of reactivity apparent in the formaldehyde release assay with oligonucleosomes modified by DOT1 or Suv4-20h1 as substrates does not reflect demethylase activity of KDM7. To refine further the specificity of KDM7, we used methylated peptides as substrates in demethylation reactions and analyzed the removal of methyl groups from the peptides by mass spectrometry. This assay showed that KDM7 removed methyl groups from both H3K9me2 and H3K27me2 (Fig. 2B,C), eliminating the possibility of cross-reaction of antibodies between methylated H3K9 and H3K27 in the immunoblot analysis. KDM7 also demethylated both H3K9me1 and H3K27me1 (Fig. 2B,C), activity that was not apparent by immunoblot analysis (Fig. 2A), probably because demethylation by KDM7 is not highly processive, so that a reduction in the level of monomethylation is masked by production of monomethylated histone from dimethylated histone in a reaction with core histones that contain all three states of methylation, as compared with peptides containing a single monomethylation state. No demethylation was detected with trimethylated H3K9 (H3K9me3) or H3K27me3 peptides. A low level of demethylation activity was also apparent with an H3K36me2 peptide, but no demethylation of H3K36me1 or H3K36me3 was detected (Fig. 2D). The decrease in mass corresponding to a methyl group was not detected in reaction mixtures containing other methylated histone peptides (Supplemental Fig. S3). Together, these data suggested that KDM7 is an authentic histone demethylase with the ability to mediate the direct removal of methyl groups from H3K9me2/me1 and H3K27me2/me1.
Figure 2.
KDM7 is a histone demethylase specific for dimethylated or monomethylated H3K9 or H3K27. (A) Calf thymus core histones were incubated in the absence or presence of 5 μg of wild-type or H282A mutant forms of KDM7-Flag, after which histone demethylation was evaluated by immunoblot analysis with antibodies to specific modified histones, as indicated on the left. (B–D) Mass spectrometric analysis of the demethylase activity of 4 μg of wild-type or H282A mutant forms of KDM7-Flag with methylated H3K9, H3K27, or H3K36 peptide substrates. Numbers represent the masses of the peptide substrates and products.
Zebrafish KDM7 orthologs possess histone demethylase activity for H3K9/K27 and are expressed predominantly in the brain
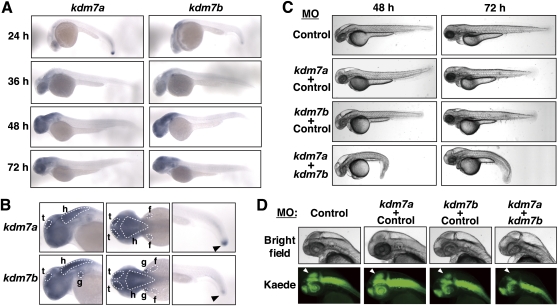
To explore the biological function of KDM7 in vivo, we characterized the KDM7 orthologs in zebrafish—LOC321248 and LOC558416, hereafter designated drKDM7a and drKDM7b, respectively (Supplemental Fig. S1A)—both of which were also found to manifest histone demethylase activity toward H3K9me2 and H3K27me2 in core histones (Supplemental Fig. S4A). We first examined the expression patterns of kdm7a and kdm7b during development by in situ hybridization. Transcripts corresponding to kdm7a and kdm7b were detected as early as the post-somitogenesis stage at 24 h post-fertilization (hpf) in the brain and tail bud (Fig. 3A,B), whereas no signals were observed in embryos at 6 or 12 hpf (data not shown). The expression of both kdm7 genes became prominent in the tectum, hindbrain, fin bud, and gill at 48 hpf (Fig. 3A,B). Corresponding sense probes did not yield any signals at these various stages of development (Supplemental Fig. S4B), indicating that the signals attributed to kdm7 transcripts were specific.
Figure 3.
Zebrafish kdm7 genes are expressed predominantly in brain and are required for tectum development. (A) In situ hybridization of whole-mount zebrafish embryos at the indicated stages (hpf) with antisense kdm7a or kdm7b RNA probes. (B) In situ hybridization of whole-mount embryos at 48 hpf (left and middle panels) or 24 hpf (right panels) with antisense kdm7a or kdm7b RNA probes. Lowercase letters indicate the tectum (t), hindbrain (h), fin bud (f), and gill (g). Arrowheads indicate the tail bud. (C) The Tg(HuC:Kaede) embryos were injected at the one-cell stage with antisense MOs for kdm7a (5 ng) or kdm7b (5 ng) or with a control MO (5 or 10 ng for a total of 10 ng of MO) in the indicated combinations. The morphology of the embryos at the indicated times (hpf) was examined by bright-field microscopy. (D) Morphology of the head region of embryos at 48 hpf that were injected with MOs as in C. (Bottom panels) Neurons expressing Kaede under the control of the HuC gene promoter were visualized by fluorescence microscopy. Arrowheads indicate the tectum or presumptive tectal region. Bright-field images are shown in the top panels.
Zebrafish KDM7 orthologs are required for tectum development
Given that both kdm7 genes are expressed predominantly in the brain, we examined whether drKDM7 might function in brain development. To examine this possibility, we inhibited the function of drKDM7 with the use of two splicing-blocking antisense morpholino oligonucleotides (MOs) that independently target kdm7a or kdm7b. We also studied the stable transgenic line Tg(HuC:Kaede), which expresses the fluorescent protein Kaede in neurons under the control of the vertebrate neuron-specific promoter of the HuC gene, in order to visualize neurons (Sato et al. 2006). The level of mature mRNAs derived from the two kdm7 genes was reduced specifically in embryos injected with the corresponding MO, but not in those injected with a control MO (Supplemental Fig. S5A). At 48 hpf, embryos that had been subjected to simultaneous injection of both kdm7 MOs at the one-cell stage manifested a curly tail and marked decrease in size of the tectum (Fig. 3C,D), consistent with the observed expression of kdm7 in the brain and tail bud. Importantly, the reduction in tectum size was accompanied by the loss of neurons from this region, although neurons in the spinal cord and other regions of the brain were unaffected (Fig. 3D; Supplemental Fig. S5B,C). In contrast, injection of MOs specific for each kdm7 gene alone elicited only marginal effects compared with those of the control MO, a finding likely attributable to functional redundancy of the two kdm7 genes. The persistence at 72 hpf of the phenotypes of the embryos injected with both Kdm7 MOs eliminates the possibility that they were attributable to developmental delay (Fig. 3C; Supplemental Fig. S5B,C). Embryos that had been subjected to simultaneous injection of another set of MOs that independently target kdm7a and kdm7b also manifested phenotypes (Supplemental Fig. S6) similar to those observed with the original set (Fig. 3C,D), suggesting that the phenotypes were the specific consequence of depletion of kdm7 transcripts. Coinjection of a validated MO for p53 (Robu et al. 2007) with either of the two independent sets of MOs for kdm7 did not affect the phenotypes induced by MO-mediated depletion of kdm7 transcripts (Supplemental Fig. S7), eliminating the possibility that the phenotypes were the result of p53 activation. Given that the phenotypes were not attributable to cell death (Supplemental Fig. S8), drKDM7 might regulate the proliferation or differentiation of neurons. On the other hand, ectopic overexpression of a fragment of drKDM7a comprising amino acids 1–480 (which manifested demethylase activity similar to that of the full-length protein) achieved by mRNA injection at the one-cell stage resulted in severe developmental defects in zebrafish embryos (Supplemental Fig. S9), suggesting that spatially and temporally regulated expression of drKDM7 is necessary for proper development. Together, these results thus indicated that KDM7 plays an important role in brain development.
KDM7 directly regulates transcription and H3K9me2 and H3K27me2 levels of the follistatin gene
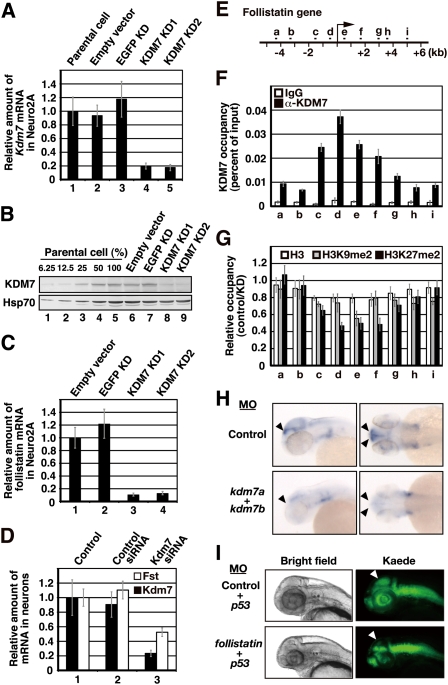
Quantitative RT–PCR analysis revealed that Kdm7 mRNA was more abundant specifically in the cerebrum and cerebellum than in other mouse tissues, although low levels of Kdm7 expression were apparent in a wide spectrum of tissues in the mouse (Supplemental Fig. S10A). To investigate the molecular basis for the abnormal brain development in zebrafish embryos depleted of drKDM7, as well as the function of KDM7 in neurons, we examined the effect of KDM7 depletion on the mRNA profile of the mouse neuroblastoma cell line Neuro2A, in which the abundance of KDM7 was found to be greater than that in other cell lines originating from various tissues (Supplemental Fig. S10B). To this end, we stably transfected Neuro2A cells with vectors for two shRNAs—KD1 and KD2—that target two different regions of mouse Kdm7 mRNA. Quantitative RT–PCR and immunoblot analyses revealed that the amounts of Kdm7 mRNA and KDM7 protein were markedly decreased in cells transfected with either of these vectors compared with those in parental cells or in cells transfected with a vector for a control shRNA (Fig. 4A,B). Microarray analysis of mRNAs in control cells and in those depleted of KDM7 resulted in the identification of genes whose expression was affected by KDM7 depletion (Supplemental Fig. S10C). One of these genes whose expression was markedly decreased by loss of KDM7 was that for follistatin, on which we initially focused, given that follistatin functions as an endogenous inhibitor of members of the transforming growth factor (TGF)-β superfamily, including activin, which plays an important role in brain development (Hemmati-Brivanlou et al. 1994; Lin et al. 2003; Zhu et al. 2008). We confirmed by quantitative RT–PCR analysis that the abundance of follistatin mRNA was decreased in Neuro2A cells depleted of KDM7 (Fig. 4C). Consistent with the results obtained with Neuro2A cells, depletion of KDM7 by RNAi in primary cultured mouse neurons also resulted in down-regulation of follistatin mRNA (Fig. 4D).
Figure 4.
KDM7 directly regulates transcription and both H3K9me2 and H3K27me2 levels of the mouse follistatin gene. (A) Quantitative RT–PCR analysis of Kdm7 mRNA in Neuro2A cell lines stably transfected with vectors for one of two KDM7 shRNAs (KD1 or KD2), with a vector for a control (EGFP) shRNA, or with the empty vector. The amount of Kdm7 mRNA was normalized by that of Gapdh mRNA, and the normalized values are presented relative to that for the parental cells. Data are means ± SD. (B) Immunoblot analysis of KDM7 and Hsp70 (loading control) in the cell lines described in A. (C) Quantitative RT–PCR analysis of follistatin mRNA in the cell lines described in A. The amount of follistatin mRNA was normalized by that of Gapdh mRNA, and the normalized values are presented relative to that for the cells transfected with the empty vector. Data are means ± SD. (D) Quantitative RT–PCR analysis of Kdm7 and follistatin (Fst) mRNAs in primary cultured mouse neurons treated with control or Kdm7 siRNAs. The amounts of Kdm7 and follistatin mRNAs were normalized by that of Gapdh mRNA, and the normalized values are presented relative to that for control cells. Data are means ± SD. (E) Schematic representation of the mouse follistatin genomic locus. The region from a to i was analyzed by ChIP experiments. The transcription start site is indicated by the arrow. (F,G) ChIP analysis of the relative occupancy of the sites in the follistatin genomic region indicated in E with KMD7 (F), as well as with H3 (white bars), H3K9me2 (gray bars), and H3K27me2 (black bars) (G). The analysis was performed with cells stably transfected with the vector for EGFP shRNA (control) and with cells stably expressing the KD1 shRNA for KDM7 (KD), and the results are presented as the percent of input for control cells (F) or the control/KD ratio (G). All data are means ± SD. (H) In situ hybridization of whole-mount zebrafish embryos at 48 hpf with an antisense follistatin RNA probe. Arrowheads indicate regions expressing the follistatin gene in embryos injected at the one-cell stage with antisense MOs for kdm7a (5 ng) and kdm7b (5 ng) or with a control MO (10 ng). (I) The Tg(HuC:Kaede) embryos were injected at the one-cell stage with antisense MOs for follistatin (5 ng) or p53 (5 ng) genes or with a control MO (5 ng) in the indicated combinations. The morphology of the embryos at 48 hpf was examined by bright-field (left panels) or fluorescence (right panels) microscopy. Arrowheads indicate the tectum or presumptive tectal region.
To determine whether the follistatin gene is a direct target of KDM7, we performed a series of chromatin immunoprecipitation (ChIP) experiments to examine its promoter and coding regions in Neuro2A cells (Fig. 4E). This analysis revealed the association of KDM7 with the follistatin gene, predominantly around the transcription start site (Fig. 4F). To investigate the consequences of this association, we analyzed H3K9me2 and H3K27me2 levels in the promoter and coding regions of the gene. Depletion of KDM7 resulted in an increase in both H3K9me2 and H3K27me2 levels around the transcription start site of the gene that appeared to correlate with KDM7 occupancy (Fig. 4G). Depletion of KDM7 by RNAi in Neuro2A cells did not markedly affect H3K9me3 levels of the follistatin gene (Supplemental Fig. S11A). In contrast, depletion of KDM7 paradoxically increased H3K27me3 levels in the entire region of the gene (Supplemental Fig. S11B), although KDM7 showed no activity toward H3K27me3 in vitro. Given that the regions in which H3K27me3 levels were increased did not correlate with KDM7 occupancy, the observed changes in H3K27me3 levels were most likely an indirect effect of KDM7 depletion. These results suggested that the follistatin gene is a direct target of KDM7-mediated transcriptional activation.
We thus examined whether the zebrafish follistatin gene is dysregulated in kdm7 morphants with the use of in situ hybridization. Whereas control morphants showed expression of the follistatin gene in an anterior edge region of the tectum at 48 hpf, kdm7 morphants manifested a substantial decrease in such expression (Fig. 4H). The down-regulation of follistatin gene expression in this particular region was sustained at 60 and 72 hpf (Supplemental Fig. S12A). The corresponding sense probe did not yield any signals at the corresponding stages (Supplemental Fig. S12B). These results thus suggested that KDM7 is recruited to specific regions of the genome, and there functions as an H3K9 and H3K27 demethylase in vivo. To investigate whether follistatin contributes to brain development in zebrafish, we inhibited the function of follistatin with the use of two independent MOs that target the follistatin gene. At 48 hpf, embryos that had been injected with either follistatin MO at the one-cell stage manifested a loss of neurons from the tectum region, although neurons in the spinal cord and other regions of the brain were unaffected (Supplemental Fig. S13). Coinjection of the validated MO for p53 did not affect this phenotype (Fig. 4I; Supplemental Fig. S13C). Together, these results indicated that KDM7 contributes to brain development at least in part through regulation of follistatin gene expression.
Methylation of H3K9 and H3K27 is linked to formation of tightly packed chromatin (heterochromatin) and transcriptional silencing (Martin and Zhang 2005). We showed here that KDM7 is a histone demethylase that catalyzes demethylation at both H3K9 and H3K27. Among the JmjC domain-containing histone demethylases, only JHDM3/JMJD2 has been shown to act as a dual demethylase, targeting both H3K9 and H3K36 and functioning as a transcriptional repressor of the ASCL2 gene, although the consequence of simultaneous methylation of these sites remains unclear (Klose et al. 2006b; Whetstine et al. 2006). We therefore propose that KDM7 functions as an eraser of silencing marks on chromatin to unlock gene silencing. Consistent with this notion, we found that removal of methyl groups from H3K9 and H3K27 by KDM7 is associated with transcriptional activation of the follistatin gene. KDM7 belongs to the subfamily of JmjC domain-containing proteins composed of PHF2 and PHF8 in addition to KDM7. The expression of Phf2 is concentrated in the embryonic neural tube and root ganglia in mice, and mutation of human PHF8 causes inherited X-linked mental retardation (Hasenpusch-Theil et al. 1999; Laumonnier et al. 2005; Abidi et al. 2007; Koivisto et al. 2007). Our results showed that KDM7 is expressed predominantly in the brain of fish and mice as well as in mammalian neuronal cells and is essential for development of the fish brain, suggesting that transcriptional regulation of the follistatin gene by KDM7 may be evolutionarily conserved. Functions in neuronal development based on their demethylase activity may thus be common to this class of JmjC domain-containing proteins.
Materials and methods
In vitro histone demethylase assays
We performed in vitro demethylation assays with the use of purified recombinant proteins and various forms of histone substrates. Demethylase activity was detected by measurement of formaldehyde release, immunoblot analysis with a series of methylation-specific antibodies (Supplemental Table S1), or matrix-assisted laser desorption ionization (MALDI)–time-of-flight (TOF) mass spectrometry, as detailed in the Supplemental Material.
In situ hybridization
Whole-mount zebrafish embryos were subjected to in situ hybridization under standard conditions with digoxigenin-labeled antisense RNA probes prepared from zebrafish kdm7a (XM_687822), kdm7b (XM_681621), or follistatin 1 (DQ317968) genes.
Other methods
Details of other procedures are provided in the Supplemental Material.
Acknowledgments
We thank B.D. Strahl for providing pCAL-SpSet2, National BioResource Project ZEBRAFISH for providing HuC:Kaede transgenic fish, and members of our laboratories for discussion. This work was supported in part by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1864410.
Supplemental material is available at http://www.genesdev.org.
References
- Abidi FE, Miano MG, Murray JC, Schwartz CE. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin Genet. 2007;72:19–22. doi: 10.1111/j.1399-0004.2007.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T. Histone methylation: Dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes & Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenpusch-Theil K, Chadwick BP, Theil T, Heath SK, Wilkinson DG, Frischauf AM. PHF2, a novel PHD finger gene located on human chromosome 9q22. Mamm Genome. 1999;10:294–298. doi: 10.1007/s003359900989. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006a;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006b;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Koivisto AM, Ala-Mello S, Lemmela S, Komu HA, Rautio J, Jarvela I. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin Genet. 2007;72:145–149. doi: 10.1111/j.1399-0004.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE, Lespinasse J, Van Esch H, Lacombe D, Goizet C, et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Morrison JR, Phillips DJ, de Kretser DM. Regulation of ovarian function by the TGF-β superfamily and follistatin. Reproduction. 2003;126:133–148. doi: 10.1530/rep.0.1260133. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: Interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44:136–142. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Zhu CC, Boone JQ, Jensen PA, Hanna S, Podemski L, Locke J, Doe CQ, O'Connor MB. Drosophila Activin- and the Activin-like product Dawdle function redundantly to regulate proliferation in the larval brain. Development. 2008;135:513–521. doi: 10.1242/dev.010876. [DOI] [PubMed] [Google Scholar]