Abstract
Spinal muscular atrophy (SMA) is caused by homozygous survival of motor neurons 1 (SMN1) gene deletions, leaving a duplicate gene, SMN2, as the sole source of SMN protein. However, most of the mRNA produced from SMN2 pre-mRNA is exon 7-skipped (∼80%), resulting in a highly unstable and almost undetectable protein (SMNΔ7). We show that this splicing defect creates a potent degradation signal (degron; SMNΔ7-DEG) at SMNΔ7's C-terminal 15 amino acids. The S270A mutation inactivates SMNΔ7-DEG, generating a stable SMNΔ7 that rescues viability of SMN-deleted cells. These findings explain a key aspect of the SMA disease mechanism, and suggest new treatment approaches based on interference with SMNΔ7-DEG activity.
Keywords: Survival of motor neurons (SMN), spinal muscular atrophy (SMA), motor neuron degenerative disease, protein degradation signal (degron), protein stability, pre-mRNA splicing
Spinal muscular atrophy (SMA) is a common and often fatal motor neuron degenerative disease, and a leading genetic cause of infant mortality (Talbot and Davies 2001; Wirth et al. 2006a; Burnett et al. 2009a). SMA severity corresponds to the degree of functional survival of motor neurons (SMN) protein deficiency. SMN is a ubiquitously expressed protein that plays a critical role in RNA metabolism, and is essential for viability of all cells in eukaryotes (Yong et al. 2004; Neuenkirchen et al. 2008). As part of a large multiprotein complex, the SMN complex, SMN functions in the biogenesis of small nuclear ribonucleoproteins (snRNPs), the major subunits of the spliceosome (Fischer et al. 1997; Liu et al. 1997; Meister et al. 2001; Pellizzoni et al. 2002). Although SMN deficiency manifests itself as a motor neuron disease, its molecular consequences are evident as profound disruptions in RNA metabolism in all tissues tested in an SMA mouse model (Gabanella et al. 2007; Zhang et al. 2008). There are two SMN genes in humans, SMN1 and SMN2, both encoding the same ORF. The vast majority of SMA patients have homozygous SMN1 deletions and are sustained by one or more copies of SMN2. However, due to a C/T substitution at position 6 of exon 7 that does not change the encoded amino acid, the splicing of the SMN2 pre-mRNA incurs frequent (∼80%) exon 7 skipping. This produces an SMN protein (SMNΔ7) that lacks the normal C-terminal 16 amino acids and acquires instead four amino acids, EMLA, encoded by exon 8 (Le et al. 2005). Thus, SMN1 deletions expose the splicing defect of SMN2 and its ineffectiveness in producing full-length normal SMN protein (Wirth et al. 2006a; Cooper et al. 2009). Biochemical experiments in vitro suggested that SMNΔ7 is not fully functional compared with normal SMN protein, including a diminished oligomerization and binding to protein substrates such as the snRNP Sm proteins (Lorson et al. 1998; Pellizzoni et al. 1999). However, as SMNΔ7 is extremely unstable and is generally undetectable, a definitive measure of its functional deficit in cells has not been possible. Nevertheless, increased SMN2 copy number correlates with a milder clinical phenotype in SMA patients (Wirth et al. 2006b). For example, severe SMA (type I) patients typically have one or two SMN2 copies, intermediate severity SMA (type II) patients usually have three SMN2 copies, and patients with mild SMA (type III) mostly have three or four SMN2 copies (Feldkotter et al. 2002; Cusco et al. 2006). Furthermore, studies in cells (Wang and Dreyfuss 2001b) suggested—and experiments in SMN-deficient mice demonstrated—that expression of an increasing copy number of SMNΔ7 cDNA transgenes proportionately lessens SMA severity (Le et al. 2005). This suggests that even a modest SMNΔ7 increase is beneficial in SMA. With this in mind, our experiments here were designed to determine the cause of SMNΔ7 instability.
Results and Discussion
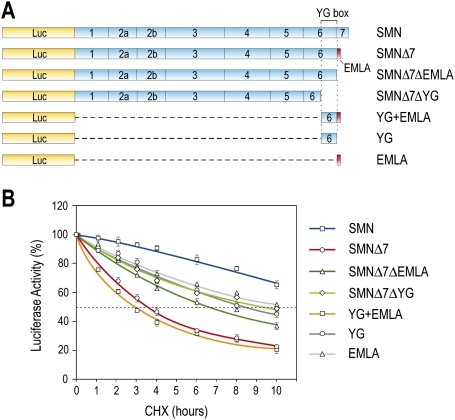
We first established a reporter system that recapitulates the differential stability of full-length SMN and SMNΔ7 and allows quantitative assessment of SMNΔ7's instability determinants. Luciferase (Luc) reporter proteins consisting of normal SMN or SMNΔ7 fused to the C terminus of Luc were produced by transfection of the corresponding cDNA constructs in 293T cells (Fig. 1). Forty-eight hours after transfection, cells were treated with the protein synthesis inhibitor cycloheximide (CHX), and Luc activity was measured at time intervals of up to 10 h. Consistent with previous reports (Lorson and Androphy 2000), SMN has a half-life of >8 h, whereas SMNΔ7 has a half-life of ∼3 h. After 10 h of CHX chase, there was three times more SMN than SMNΔ7. Several constructs were prepared to determine the role of the C-terminal sequence of SMNΔ7 in this protein's instability. Deletion of the C-terminal EMLA from SMNΔ7 (SMNΔ7ΔEMLA) increased the half-life of SMNΔ7 by twofold (Fig. 1B), and a further deletion of the YG box (SMNΔ7ΔYG), a conserved tyrosine/glycine-rich motif in divergent SMNs (Talbot et al. 1997) that is essential for SMN oligomerization (Pellizzoni et al. 1999), also had the same effect. These results suggest that EMLA and the YG box are major contributors to SMNΔ7's instability. Importantly, YG + EMLA alone was sufficient to cause dramatic instability of Luc that is similar to that of SMNΔ7. Neither YG nor EMLA alone was sufficient for full destabilization activity (Fig. 1B). N-terminal deletions in the YG box decreased the destabilizing activity of YG + EMLA (data not shown). These data indicate that YG + EMLA, corresponding to SMNΔ7 amino acids 268–282, is the minimal sequence required for full SMNΔ7 destabilization, and is both necessary and sufficient to trigger rapid degradation of a heterologous protein.
Figure 1.
Delineation of YG + EMLA as a protein destabilization sequence in SMNΔ7. (A) Schematic diagram of Luc-fused SMN and a series of deletion constructs used for quantitative measurement of protein stability. Shown are SMN exon structures. YG box denotes the tyrosine/glycine (YG)-rich sequences in exon 6 of SMN. The EMLA sequence encoded by exon 8 is depicted by the red box at the C-terminal end of SMNΔ7. (B) 293T cells were transfected with plasmids expressing Luc-SMN, Luc-SMNΔ7, and the indicated deletion constructs. Forty-eight hours after transfection, the cells were treated with CHX (0.1 mg/mL) for various times as indicated, and then assayed for luciferase activity. Luc activity at each time point was calculated by comparison with those at time 0, which was set to 100%. Fifty percent activity is indicated by the gray dotted line. Error bars represent standard deviation (SD) from three independent experiments.
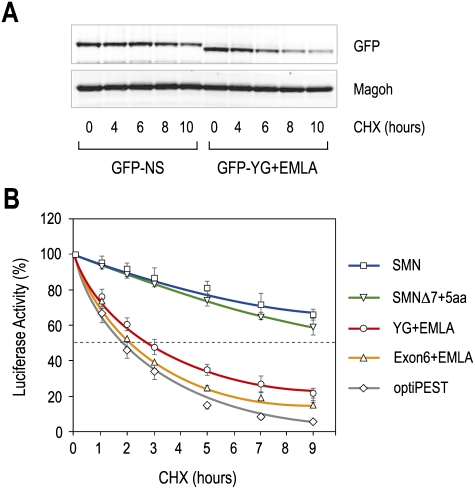
As a further test of this conclusion, we fused YG + EMLA to another reporter, GFP, and expressed this protein as well as GFP as a control in 293T cells. The GFP signal from GFP-YG + EMLA, as determined by Western blots, showed a gradual decrease after treatment with CHX (Fig. 2A). GFP-YG + EMLA protein decreased faster than GFP-NS (nonspecific sequence), and the half-life of GFP-YG + EMLA was about half that of GFP-NS. These results suggest that YG + EMLA functions as a protein degradation sequence. For comparison, we tested the destabilizing activity on the same reporter of YG + EMLA and an optimized PEST, a potent and well-characterized protein destabilizing signal (Li et al. 1998). YG + EMLA had a similar effect to that of the genetically improved PEST (Fig. 2B). Furthermore, SMN Exon6 + EMLA, which is the same size as the 41-amino-acid PEST sequence, conferred similar instability. As this optimized PEST sequence has about half the half-life of the natural one (Li et al. 1998), YG + EMLA could be estimated to have similar or stronger destabilization activity than that of the natural PEST sequence, and Exon6 + EMLA is about twice as strong. These data demonstrate that YG + EMLA is a highly potent and transferable protein degradation signal (degron), which we term SMNΔ7-DEG, for SMNΔ7 degron. Addition of five amino acids to the C-terminal end of EMLA (SMNΔ7 + 5aa) caused SMNΔ7 stabilization, indicating that SMNΔ7-DEG must be exposed at the C terminus of the protein for activity (Fig. 2B). This is consistent with the observation that several additional amino acids, which can be effected by aminoglycoside-forced translational read-through, enhanced SMNΔ7 stability and functionality (Mattis et al. 2008; Heier and DiDonato 2009).
Figure 2.
The C terminus of SMNΔ7, YG + EMLA, is a strong protein destabilizing signal (degron). (A) Plasmids expressing GFP-YG + EMLA or GFP-NS (nonspecific sequence) were transfected into 293T cells. Twenty-four hours after transfection, the cells were treated with CHX (0.1 mg/mL) for various times as indicated. GFP fusion proteins were detected by Western blot using anti-GFP antibody, and Magoh was used as a loading control. (B) Comparison of YG + EMLA and Exon6 + EMLA of SMNΔ7 with an optimized protein-destabilizing element (optiPEST). Shown also are Luc-SMN and Luc-SMNΔ7 containing an additional five amino acids at the C-terminal end (SMNΔ7 + 5aa). Luc activities were measured as in Figure 1. Error bars represent SDs from three independent experiments.
SMN was shown previously to be degraded by the proteasome (Chang et al. 2004; Burnett et al. 2009b). To determine if SMNΔ7 is also degraded by this system, cells expressing Luc-SMNΔ7 were treated with proteasome inhibitors (MG132 and Lactacystin) for 5 h in the presence of CHX. CHX treatment alone resulted in a 60% decrease in signal, but a much smaller decrease was seen in the presence of proteasome inhibitors MG132 and Lactacystin (Supplemental Fig. 1). Inhibitors of other proteolytic activities—such as lysosomal proteases, autophagy, and calpain (NH4Cl, 3-methyladenine, and calpeptin, respectively)—had no effect. These data demonstrate that SMNΔ7 is degraded by the proteasome.
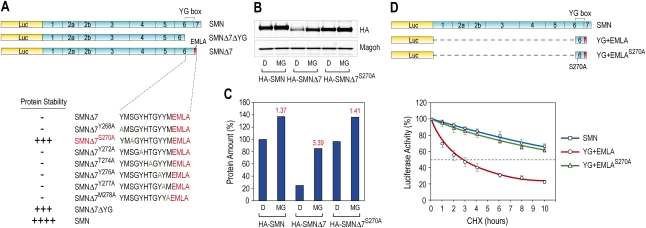
To identify specific residues in the SMNΔ7-DEG that are important for its activity, we performed mutagenesis of the YG box, converting every second residue to alanine in the context of full-length SMNΔ7, and determined the half-life of each in 293T cells. Of seven mutations tested, S270A produced the most striking effect, reversing the destabilizing activity of the SMNΔ7-DEG (Fig. 3A). To confirm that S270A stabilizes SMNΔ7, HA-tagged proteins SMN, SMNΔ7, and SMNΔ7S270A were expressed in 293T cells for 24 h, and then treated with the proteasome inhibitor MG132 for 16 h. The levels of the tagged SMN proteins were then monitored by Western blots using anti-HA antibody (Fig. 3B). As expected, the amount of SMNΔ7 without MG132 treatment was much lower than that of SMN. However, the amount of SMNΔ7S270A was similar to that of normal SMN, indicating an almost complete restoration of stability by S270A mutation. MG132 caused a dramatic increase in the amount of SMNΔ7 (∼3.4-fold), but only a moderate effect on SMN and SMNΔ7S270A (Fig. 3C). Therefore, the S270A mutation limits the proteosome degradation of SMNΔ7 and increases its stability very significantly. We further tested the effect of S270A in the context of SMNΔ7-DEG alone. S270A mutation strongly increased the stability of Luc-YG + EMLA to a level similar to that of SMN (Fig. 3D). These data indicate that the enhancement of stability of SMNΔ7 by the S270A mutation occurs through SMNΔ7-DEG.
Figure 3.
S270 is critical for the activity of the SMNΔ7-DEG through YG + EMLA. (A) Seven residues in the YG box were each mutated to alanine as indicated. All constructs had Luc fusions, and Luc activity was assayed as in Figure 1. (B) HA-tagged SMN, SMNΔ7, and SMNΔ7S270A were expressed in 293T cells for 24 h, and then cells were treated with DMSO (D) or 10 μM MG132 (MG) for 16 h. Fusion proteins were monitored by Western blot using an anti-HA tag antibody, and Magoh was used as a loading control. (C) HA-tagged proteins in B were quantified and compared with HA-SMN without MG132 treatment, which was set to 100%. The fold change of each fusion protein amount upon MG132 treatment is indicated in red above the column. (D) All constructs had Luc fusions, and Luc activity was assayed as in Figure 1. Error bars represent SDs from three independent experiments.
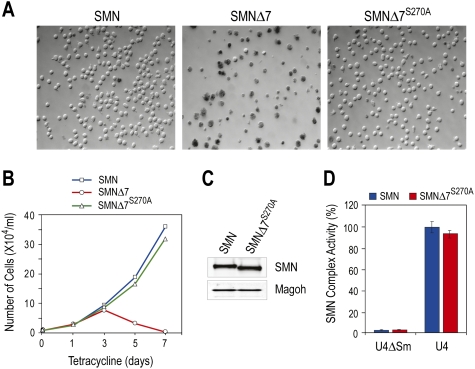
To determine whether SMNΔ7S270A is a functional SMN protein, we used a previously established cell system, the S5 cell line, to ask if it could rescue the viability of SMN-depleted cells. S5 is derived from chicken DT40 cells in which the endogenous chicken SMN gene is disrupted by homologous recombination and SMN protein is expressed exogenously from a cDNA under a tetracycline-repressible promoter (Wang and Dreyfuss 2001a). Upon depletion of chicken SMN (cSMN), S5 cell growth arrests at 72 h and cell death occurs. It is therefore useful to assess the physiological functionality of SMN mutants in this cell system by monitoring cell viability after turning off cSMN cDNA expression and simultaneously expressing exogenous SMN mutants of interest. To do so, we constructed recombinant retroviruses expressing SMN, SMNΔ7, or SMNΔ7S270A and transduced S5 cells. One week after repression of cSMN expression by tetracycline (1 μg/mL), there was a very clear difference in viable cell number among three samples (Fig. 4A,B). As expected (Wang and Dreyfuss 2001b), while SMN rescued the viability of S5 cells, SMNΔ7 did not. Importantly, SMNΔ7S270A also rescued S5 cells to a similar extent as SMN. The two rescued cell lines expressed a similar level of SMN protein (Fig. 4C). Since the deficiency in functional SMN protein is correlated directly with snRNP assembly defects in cells of SMA patients (Wan et al. 2005), we next examined whether SMNΔ7S270A is active in snRNP assembly as a further measure of functionality. Extracts from cells expressing SMN and SMNΔ7S270A were prepared, and their snRNP assembly activity was determined by measuring the amount of Sm protein cores that form on biotinylated snRNA substrate captured on streptavidin beads (Wan et al. 2005). Sm cores are the major constituents of snRNPs whose assembly on snRNAs depends on the SMN complex. As shown in Figure 4D, both cell lines showed similar activity. These data indicate that SMNΔ7 S270A is a functional protein similar to normal SMN in S5 cells. We conclude that the instability of SMNΔ7 conferred by SMNΔ7-DEG is a principal contributor to the deleterious phenotype of exon 7 skipping, and that S270A substitution in SMNΔ7 abrogates the degron activity, thereby restoring the function of SMN.
Figure 4.
SMNΔ7S270A rescues SMN-deficient cells and is functional in snRNP assembly. (A) S5 cells were cultured in the presence of tetracycline (1 μg/mL) to deplete endogenous SMN, and were infected with retroviruses expressing SMN, SMNΔ7, or SMNΔ7S270A. One week after tetracycline addition, cells were stained with Trypan blue and visualized by DIC microscopy. (B) Cell growth as in A was measured by monitoring the number of live cells at the indicated time points following tetracycline addition. (C) Western blots of SMN protein in rescued cells (10 d after tetracycline addition). (D) Cytoplasmic extracts from rescued cells were assayed for snRNP assembly on U4 snRNA in vitro, using U4ΔSm RNA as a control.
Several diverse classes of degrons that target proteins to various degradation pathways have been described. Most noted are N degrons comprised of destabilizing N-terminal residues, C-terminal determinants containing relatively unstructured hydrophobic residues, and phospho-degrons that are modulated by the phosphorylation status of their serine/threonine residues in response to cell signaling (Parsell et al. 1990; Ravid and Hochstrasser 2008). The short-lived tumor suppressor protein PTEN's stability depends on a 50-amino-acid C-terminal tail that is phosphorylated at specific serine/threonine residues (Vazquez et al. 2000). Interestingly, while many of the residues of the SMNΔ7-DEG could be substituted by alanines without loss of degron function, S270 is critical for the destabilizing function. It is therefore possible that S270 is phosphorylated, and that this regulates the SMNΔ7-DEG activity. However, phosphorylation site analysis by NetPhos did not reveal strong candidate kinases for it. SMNΔ7-DEG has no obvious sequence similarity with the known degrons and, thus, represents a novel protein-destabilizing element. Protein database searches did not identify other known proteins containing sequences highly similar to SMNΔ7-DEG.
The reduced oligomerization efficiency of SMNΔ7 has been suggested recently to account for its instability (Burnett et al. 2009b). Indeed, intermolecular SMN oxidative cross-linking provided direct evidence that SMN is oligomeric in cells (Wan et al. 2008). Oligomerization is likely to be important for SMN function, and also to contribute to its stability. However, although SMN oligomerization correlated with its stability, this did not explain the intrinsic instability of SMNΔ7. Our findings show that attachment of SMNΔ7-DEG to monomeric protein reporters (Luc and GFP) triggered their rapid degradation, indicating that lack of oligomerization is not the major cause of SMNΔ7's instability. Loss of oligomerization capacity and other possible deficits as a result of deletion of the peptide encoded by exon 7 may result in an SMN protein that is functionally suboptimal. However, the detrimental effect of exon 7 skipping does not arise primarily from deletion of a functionally essential domain, but from the creation of a positively acting and potent degron that causes severe deficiency of SMNΔ7 protein.
Given the ability of S270A mutation to restore SMNΔ7's stability and complement SMN loss of function, it is reasonable to predict that polymorphisms that inactivate SMNΔ7-DEG, such as at S270, would result in a milder SMA phenotype than the genotype predicts based on SMN2 copy number in SMN1-deleted individuals. Our finding with SMNΔ7S270A indicates that SMNΔ7 is a functional SMN protein, and that its stabilization could prevent or lessen SMA severity. We suggest that interfering with SMNΔ7-DEG activity could be an effective approach for mitigating its deficiency as a potential treatment for SMA. Although the inhibitor studies suggest that the degradation of SMNΔ7 likely occurs in the proteasome, general inhibition of proteasome activity would be very toxic, particularly in the long-term treatment that SMA would be expected to require. A targeted inhibition of the factors that mediate the SMNΔ7-DEG-dependent degradation should provide a more specific therapeutic approach, and their identification will be of great interest for SMA therapy.
SMA is thus the result of a fateful chain of events. Homozygous SMN1 deletion is a cause of SMA only because it exposes the splicing defect of SMN2. We argue that the splicing defect in SMN2 causes SMN deficiency because it fortuitously creates a degron. The degron is a key to SMA, as it is the most direct cause of SMN deficiency, which then results in major perturbations in RNA metabolism.
Materials and methods
Plasmid construction and generation of mutations
To construct plasmids expressing Luc-fused proteins, the Luc gene was cloned into pcDNA3.1 vector at HindIII/KpnI sites, and then DNA fragments encoding full-length wild-type human SMN, SMNΔ7, several deletion mutants of SMNΔ7, and optiPEST were inserted into the KpnI/XhoI sites. SMNΔ7 mutants with a single amino acid change were generated by mutating residues in YG + EMLA to alanine by QuickChange site-directed mutagenesis kit (Stratagene). Plasmid expressing GFP-YG + EMLA was constructed by inserting a DNA fragment encoding YG + EMLA into pEGFP vector (Clontech) at KpnI/BamHI sites. Plasmids expressing HA-SMNs were constructed by inserting DNA fragments encoding HA-tagged SMN, SMNΔ7, and SMNΔ7S270A into the BamHI/XhoI sites of pcDNA3 vector. To generate retroviral plasmids to express SMNs in S5 cells, DNA fragments encoding SMN, SMNΔ7, and SMNΔ7S270A were cloned into the EcoRI/XhoI sites of pMX vector as described (Wang and Dreyfuss 2001a).
Assays for protein stability
Luc- and GFP-based assays were performed as described in the legends for Figures 1 and 2A, respectively. Luc activities were measured using One-Glo reagent (Promega).
Rescue of S5 cell viability
S5 cells were maintained and infected with retroviruses expressing SMN, SMNΔ7, and SMNΔ7S270A as described (Wang and Dreyfuss 2001a).
SMN complex activity assay
Cytoplasmic extracts from rescued S5 cells were prepared and assayed for snRNP assembly in vitro as described (Wan et al. 2005).
Antibodies
Mouse monoclonal antibodies anti-SMN (62E7) and anti-Magoh (18G12) were used as described previously (Wan et al. 2005). Rabbit polyclonal antibodies anti-HA (Santa Cruz Biotechnologies) and anti-GFP (Santa Cruz Biotechnologies) were used as recommended by the manufacturer.
Acknowledgments
We thank the members of our laboratory, especially Dr. Lili Wan, for helpful discussions and comments on this manuscript. This work was supported by the Association Française Contre les Myopathies (AFM). G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1884910.
Supplemental material is available at http://www.genesdev.org.
References
- Burnett BG, Crawford TO, Sumner CJ. Emerging treatment options for spinal muscular atrophy. Curr Treat Options Neurol. 2009a;11:90–101. doi: 10.1007/s11940-009-0012-x. [DOI] [PubMed] [Google Scholar]
- Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009b;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Hung WC, Chuang YJ, Jong YJ. Degradation of survival motor neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem Int. 2004;45:1107–1112. doi: 10.1016/j.neuint.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusco I, Barcelo MJ, Rojas-Garcia R, Illa I, Gamez J, Cervera C, Pou A, Izquierdo G, Baiget M, Tizzano EF. SMN2 copy number predicts acute or chronic spinal muscular atrophy but does not account for intrafamilial variability in siblings. J Neurol. 2006;253:21–25. doi: 10.1007/s00415-005-0912-y. [DOI] [PubMed] [Google Scholar]
- Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier CR, DiDonato CJ. Translational readthrough by the aminoglycoside geneticin (G418) modulates SMN stability in vitro and improves motor function in SMA mice in vivo. Hum Mol Genet. 2009;18:1310–1322. doi: 10.1093/hmg/ddp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNΔ7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Androphy EJ. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AH, Androphy EJ. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- Mattis VB, Bowerman M, Kothary R, Lorson CL. A SMNΔ7 read-through product confers functionality to the SMNΔ7 protein. Neurosci Lett. 2008;442:54–58. doi: 10.1016/j.neulet.2008.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Silber KR, Sauer RT. Carboxy-terminal determinants of intracellular protein degradation. Genes & Dev. 1990;4:277–286. doi: 10.1101/gad.4.2.277. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Davies KE. Spinal muscular atrophy. Semin Neurol. 2001;21:189–197. doi: 10.1055/s-2001-15264. [DOI] [PubMed] [Google Scholar]
- Talbot K, Ponting CP, Theodosiou AM, Rodrigues NR, Surtees R, Mountford R, Davies KE. Missense mutation clustering in the survival motor neuron gene: A role for a conserved tyrosine and glycine rich region of the protein in RNA metabolism? Hum Mol Genet. 1997;6:497–500. doi: 10.1093/hmg/6.3.497. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: Biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Ottinger E, Cho S, Dreyfuss G. Inactivation of the SMN complex by oxidative stress. Mol Cell. 2008;31:244–254. doi: 10.1016/j.molcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dreyfuss G. A cell system with targeted disruption of the SMN gene: Functional conservation of the SMN protein and dependence of Gemin2 on SMN. J Biol Chem. 2001a;276:9599–9605. doi: 10.1074/jbc.M009162200. [DOI] [PubMed] [Google Scholar]
- Wang J, Dreyfuss G. Characterization of functional domains of the SMN protein in vivo. J Biol Chem. 2001b;276:45387–45393. doi: 10.1074/jbc.M105059200. [DOI] [PubMed] [Google Scholar]
- Wirth B, Brichta L, Hahnen E. Spinal muscular atrophy: From gene to therapy. Semin Pediatr Neurol. 2006a;13:121–131. doi: 10.1016/j.spen.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Wirth B, Brichta L, Schrank B, Lochmuller H, Blick S, Baasner A, Heller R. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet. 2006b;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- Yong J, Wan L, Dreyfuss G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 2004;14:226–232. doi: 10.1016/j.tcb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]