Abstract
Hypoxic response and inflammation both involve the action of the hypoxia-inducible transcription factors HIF-1α and HIF-2α. Previous studies have revealed that both HIF-α proteins are in a number of aspects similarly regulated post-translationally. However, the functional interrelationship of these two isoforms remains largely unclear. The polarization of macrophages controls functionally divergent processes; one of these is nitric oxide (NO) production, which in turn is controlled in part by HIF factors. We show here that the HIF-α isoforms can be differentially activated: HIF-1α is induced by Th1 cytokines in M1 macrophage polarization, whereas HIF-2α is induced by Th2 cytokines during an M2 response. This differential response was most evident in polarized macrophages through HIF-α isoform-specific regulation of the inducible NO synthase gene by HIF-1α, and the arginase1 gene by HIF-2α. In silico modeling predicted that regulation of overall NO availability is due to differential regulation of HIF-1α versus HIF-2α, acting to, respectively, either increase or suppress NO synthesis. An in vivo model of endotoxin challenge confirmed this; thus, these studies reveal that the two homologous transcription factors, HIF-1α and HIF-2α, can have physiologically antagonistic functions, but that their antiphase regulation allows them to coordinately regulate NO production in a cytokine-induced and transcription-dependent fashion.
Keywords: Hypoxia, HIF, nitric oxide, arginase, macrophage polarization
Functional characterization of myeloid response has allowed macrophage activation to be classified as responsive to Lipopolysaccharide (LPS) or Th1 cytokines such as IFNγ, or Th2 cytokines, including IL-4 and IL-13 (Bonecini-Almeida et al. 1998; Gordon 2003; Mantovani et al. 2004; Mosser and Edwards 2008). Macrophages polarized with Th1 cytokines are called M1 macrophages, and are considered to be classically activated. M1 macrophage polarization is important for the clearance of phagocytosed or intracellular pathogens; this is mediated by production of proinflammatory cytokines, reactive oxygen species, and nitric oxide (NO) (Nathan et al. 1983; Bermudez and Young 1988; Summersgill et al. 1992; Benoit et al. 2008). Macrophages activated by Th2 cytokines are considered M2-polarized, or alternatively activated, and are important in humoral immunity and repair processes (Stein et al. 1992; Mantovani et al. 2002; Martinez et al. 2009).
Macrophages are often present in hypoxic tissues, and hypoxia strongly affects macrophage functions (Murdoch et al. 2005; Nizet and Johnson 2009). Much of the overall transcriptional response to hypoxia is mediated by a group of transcription factors known as hypoxia-inducible factors (HIF) (Wang and Semenza 1993; Weidemann and Johnson 2008). One of these, HIF-1α, is expressed ubiquitously, and is tightly linked to inflammatory response and microbicidal activities of myeloid cells (Cramer et al. 2003; Peyssonnaux et al. 2005). Another oxygen-responsive component of the HIF family, HIF-2α, is expressed in a more limited fashion, although it is also found in myeloid cells (Tian et al. 1997; Talks et al. 2000; Jurgensen et al. 2004). Among the transcriptional targets of HIF-1α, the inducible NO synthase gene (iNOS) is regulated by both hypoxia and a number of other factors (MacNaul and Hutchinson 1993; Melillo et al. 1995; Peyssonnaux et al. 2005). It is also expressed primarily in macrophages that are M1-polarized (Gordon 2003). iNOS produces NO by metabolizing its substrate, the amino acid L-arginine (Bronte and Zanovello 2005). Macrophages also have another arginine-metabolizing enzyme, arginase1, which generates ornithine and urea (Durante et al. 2007). Arginase1 is highly expressed in M2 macrophages, and competes with iNOS for their common substrate, L-arginine (Bansal and Ochoa 2003). Arginase1 activity can thus regulate NO production via the limitation of arginine availability in the extracellular environment (Bronte et al. 2003; El Kasmi et al. 2008). Arginase1 gene expression is also induced by hypoxia (Louis et al. 1998); this raises the question of how these two differing metabolic fates of arginine, NO synthesis and arginase1 activity, are regulated under hypoxia, and how the two transcription factors, HIF-1α and HIF-2α, participate in that regulation.
In this study, we found that HIF-1α and HIF-2α mRNA is expressed differentially in M1- and M2-polarized macrophages, due to differential induction of the two isoforms by Th1 and Th2 cytokines. Through computational analysis of transcription rates, mRNA, and protein half-lives, we determined that these dynamic changes of mRNA levels could strongly influence overall protein levels in the absence of classical effects on post-translational stability. We then found that HIF-1α and HIF-2α act in this way to cooperatively maintain NO homeostasis, and that they act through a functional antagonism to accomplish this by differential action on their two target genes: iNOS and arginase1.
Results
HIF-2α mRNA expression correlates with M2 activation
Primary macrophages from mice are generally from one of two sources: They are bone marrow-derived by culture of bone marrow in M-CSF-containing medium (termed bone marrow-derived macrophages [BMDMs]), or they are elicited from the peritoneum of mice by thioglycollate injection (termed thioglycollate-elicited peritoneal macrophages [TEPMs]). A number of studies have indicated that TEPMs are at least partially activated (Michl et al. 1979; Norton and Munck 1980; Hopper 1986); this includes a recent demonstration that PPARγ, an essential regulator of alternative macrophage activation, is expressed in TEPMs but not in BMDMs (Odegaard et al. 2007).
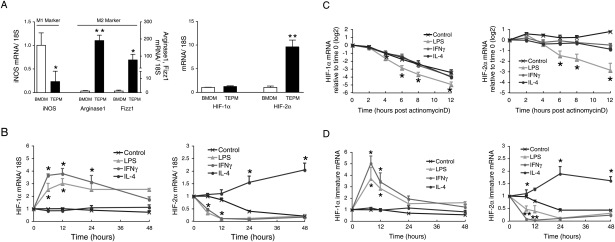
Examination of gene expression in these two types of cells showed that M1- and M2-related gene expression does differ between BMDMs and TEPMs. BMDMs have higher induction of the M1 gene encoding iNOS, whereas TEPMs have higher levels of the M2 genes encoding arginase1 and resistin-like molecule α/found in inflammatory zone (Fizz1) (Fig. 1A). This finding is not universally applicable to M2 genes, as other M2 genes, such as mannose receptor (MR) and chitinase-like lectin ym1 (YM1), are not differentially expressed. Interestingly and unexpectedly, we found that mRNA expression of the HIF-2α, but not the HIF-1α, gene is increased significantly in TEPMs; i.e., partially M2-polarized macrophages (Fig. 1A).
Figure 1.
Th1 and Th2 cytokines oppositely regulate HIF-1α and HIF-2α mRNA expression. (A) TEPM has significantly higher HIF-2α, arginase1, and Fizz1 mRNA, but lower iNOS mRNA expression compared with BMDM (n = 6). (B) BMDMs were treated with LPS (1 μg/mL), IFNγ (20 ng/mL), or IL-4 (20 ng/mL). LPS and IFNγ induced HIF-1α but strongly decreased HIF-2α mRNA levels at 6 and 12 h. IL-4 induced HIF-2α mRNA at 24 or 48 h. (n = 5). (C) BMDMs were treated with actinomycin D (5 μM) together with LPS (1 μg/mL), IFNγ (20 ng/mL), or IL-4 (20 ng/mL). HIF-1α and HIF-2α mRNA abundance after actinomycin D were shown as relative to time 0. Control HIF-1α mRNA was destabilized quickly, whereas HIF-2α mRNA was stable. LPS accelerated the destabilization of both HIF-1α and HIF-2α significantly (n = 3). (D) HIF-1α and HIF-2α immature mRNA synthesis was measured by using pre-mRNA-specific primers. LPS and IFNγ induced HIF-1α mRNA synthesis at 6 and 8 h, whereas both decreased HIF-2α synthesis. IL-4 increased HIF-2α mRNA synthesis at 24 h or 48 h (n = 5). All data represent means ± SEM. (*) P < 0.05; (**) P < 0.01 versus control.
HIF-1α and HIF-2α mRNA expression are differentially regulated by Th1 and Th2 cytokines
To analyze HIF isoform function in polarization, we asked whether HIF-α mRNAs respond differently to differing cytokines, particularly to Th1 and Th2 cytokines. In these experiments, we used BMDMs as representatives of a preactivated state. As can be seen in Figure 1B, IFNγ and LPS increase HIF-1α mRNA expression in BMDMs over 12 h, but significantly repress HIF-2α mRNA expression relative to controls. However, the Th2 cytokine IL-4 acts in almost the opposite fashion; it has no effect on expression of HIF-1α mRNA, but acts to increase HIF-2α mRNA expression. Other Th2 cytokines, such as IL-13, also increased HIF-2α mRNA abundance (data not shown), suggesting Th2 cytokines cause a gradual increase of HIF-2α mRNA, whereas Th1 cytokines suppress HIF-2α mRNA expression. Interestingly, the kinetics of HIF-2α mRNA induction by Th2 cytokines differ from that of HIF-1α mRNA induction by LPS or IFNγ, and the IL-4 induction of HIF-2α mRNA gives rise to a prolonged effect on HIF-2α expression.
Transcriptional regulation of HIF-α by Th1 and Th2 cytokines
To determine how HIF-α mRNA behavior is regulated by cytokines, we first defined mRNA stability in BMDMs; this was done by measuring levels of mRNA over time following treatment with actinomycin D (Fig. 1C). These data demonstrate that HIF-1α mRNA is quite labile, with a decline in abundance of approximately four to five orders of magnitude over 12 h, with a half-life of ∼200 min. On the other hand, HIF-2α mRNA is very stable, and we were unable to determine a conclusive half-life for it in this assay; HIF-2α half-life is affected only by treatment with LPS. LPS’ effects on mRNA stability are also significantly greater in destabilizing HIF-1α mRNA relative to HIF-2α mRNA.
We next measured pre-mRNA by quantitative PCR, using primers that bind at exon 12 and intron 13 in HIF-1α, and at intron 11 and exon 12 in HIF-2α. Amplification of purified nuclear RNA from these sites can be used to determine the rate of HIF-α mRNA synthesis by detecting preprocessed RNA. LPS and IFNγ enhanced the synthesis of HIF-1α mRNA at 6–12 h, whereas they strongly reduced HIF-2α mRNA synthesis (Fig. 1D).
However, IL-4 significantly induced HIF-2α mRNA synthesis, albeit again with a delayed time course relative to the transcriptional effects seen on HIF-1α mRNA stimulated by Th1 cytokines. Collectively, these results demonstrate that HIF-1α mRNA has a high turnover rate, and is controlled primarily by modulating its transcription. In contrast, HIF-2α mRNA is stable, with a lower turnover rate, and its accumulated presence in the cell is affected by both its stability and transcriptional rate.
HIF-1α and HIF-2α protein levels are differentially regulated by Th1 and Th2 cytokines
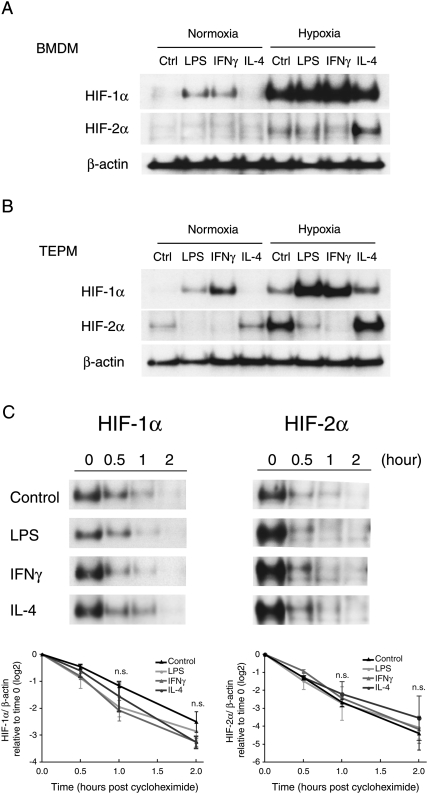
There is great complexity to the role of inflammation in HIF-1α induction, with evidence for transcriptional, translational, and post-translational effects (Sandau et al. 2001; Zhou et al. 2003; Frede et al. 2007; Rius et al. 2008). To determine how Th1 and Th2 cytokines ultimately affect HIF-α protein levels, we first examined HIF-1α and HIF-2α protein abundance during hypoxia. We treated primary macrophages with cytokines for 48 h, and then subjected them to hypoxia for 4 h. Hypoxia alone induced a small amount of HIF-2α protein in BMDMs; however, IL-4 strongly increased HIF-2α abundance (Fig. 2A).
Figure 2.
HIF-1α and HIF-2α protein synthesis were inversely influenced by LPS/Th1 and Th2 cytokines. (A) LPS or IFNγ increased HIF-1α protein expression both in normoxic and hypoxic conditions in BMDM. HIF-2α protein has low-level expression except in the presence of IL-4. (B) TEPM has high HIF-2α protein expression. LPS and IFNγ decreased HIF-2α protein abundance. (C) TEPM from wild-type mice were collected and treated with dipyridyl for 2 h to induce HIF-1α and HIF-2α protein expression. Cells were cultured with cycloheximide (100 μM) together with LPS (1 μg/mL), IFNγ (20 ng/mL), or IL-4 (20 ng/mL), and collected for Western blotting. LPS or IFNγ, and IL-4 did not affect HIF-1α or HIF-2α protein stability. n = 3. (*) P < 0.05 versus control.
TEPMs, which are partially M2-polarized, have much higher HIF-2α expression compared with BMDMs (Fig. 2A,B); this is consistent with their HIF-2α mRNA levels (Fig. 1B). HIF-2α protein expression can be detected even in normoxia in TEPMs. LPS and the Th1 cytokine IFNγ induced HIF-1α protein but strongly decreased HIF-2α protein in both BMDMs and TEPMs. These protein levels are also consistent with the response of HIF-1α or HIF-2α mRNA to LPS or IFNγ (Fig. 1B).
We then sought to determine whether LPS, IFNγ, or IL-4 affect HIF-α protein stability (Fig. 2C). We treated TEPMs with dipyridyl, an inhibitor of proline hydroxylase activity, for 2 h to induce HIF-1α and HIF-2α protein expression, and then administered cycloheximide to inhibit translation. HIF-1α and HIF-2α protein half-lives were 41.5 min and 27.3 min, respectively (Fig. 2C). In the presence of LPS, IFNγ, or IL-4, HIF-1α and HIF-2α protein degradation rates are almost identical to those of non-cytokine-treated controls, indicating that neither LPS, IFNγ, nor IL-4 act to affect HIF-α protein stability (Fig. 2C).
Computational simulation of HIF-1α and HIF-2α expression in macrophages
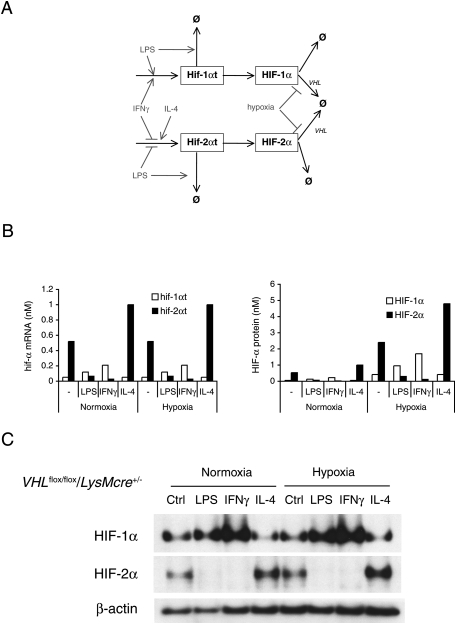
Based on the parameters described above, a mathematical model of HIF-1α and HIF-2α mRNA kinetics under M1 or M2 polarization stimuli was constructed (Fig. 3A). The model was then employed to calculate and predict the net effects of the cytokines on overall levels of HIF-1α or HIF-2α protein (Fig. 3B).
Figure 3.
Computational simulation of HIF-1α and HIF-2α expression demonstrated that HIF-α protein abundances are influenced by changes in their mRNA levels. (A) Network diagram of HIF-1α and HIF-2α regulation by LPS, IFNγ, or IL-4, used to construct a mathematical model. (B) Computational simulation of Th1 or Th2 cytokine acting inversely on HIF-1α and HIF-2α mRNA levels, which are unaffected by hypoxia. Transcription rates were altered to the degree measured experimentally in response to Th1 and Th2 cytokines, and the resulting mRNA and protein levels were calculated. Simulations reveal that HIF-1α and HIF-2α protein levels are regulated by the cytokine-mediated control of mRNA synthesis and stability, even in normoxic conditions; this regulatory effect is enhanced in hypoxic conditions. (C) TEPM from VHLflox/flox/LysMcre+/− mice expressed HIF-1α and HIF-2α protein in normoxia. LPS and IFNγ increased HIF-1α, but decreased HIF-2α protein. IL-4 increased HIF-2α protein in VHL-deficient macrophages.
The in silico model predicts that dynamic changes of HIF-1α and HIF-2α mRNA synthesis and degradation induced by Th1 or Th2 cytokines will strongly affect HI-α protein abundance. To model this experimentally, we analyzed von Hippel-Lindau (VHL)-null TEPMs; these cells lack an essential protein required for post-translational degradation of the HIF-α proteins.
HIF-1α and HIF-2α proteins are present in normoxia in VHL-deficient macrophages (Fig. 3C). However, in keeping with the computed model for effects of transcriptional controls on HIF-α levels, LPS and IFNγ increased HIF-1α but almost completely suppressed HIF-2α protein accumulation. On the other hand, IL-4 increased HIF-2α levels. These results demonstrate that polarizing cytokines affect HIF-1α or HIF-2α protein abundance not by regulating degradation, but through inducing mRNA synthesis.
iNOS expression is controlled by HIF-1α; in contrast, arginase1 expression is regulated mainly by HIF-2α
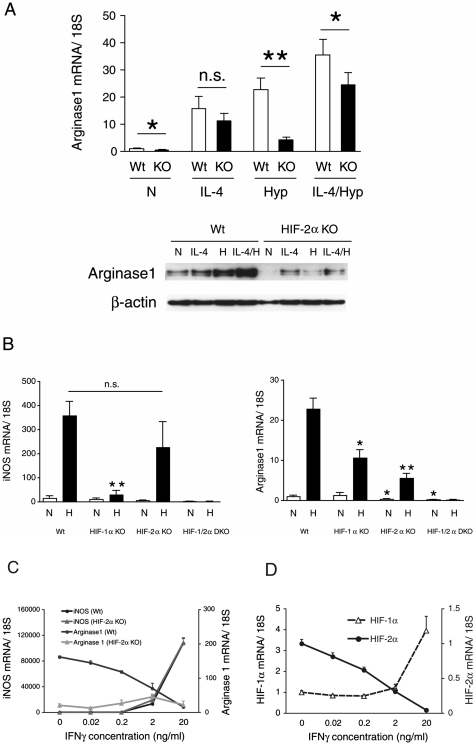
The data above indicate that HIF-2α is exclusively induced by Th2 cytokines. To study this surprising finding further, we collected TEPMs from HIF-2αflox/flox; Tie2cre+/− mice (HIF-2α KO) and used cre recombinase-negative littermates to isolate control TEPMs. Initial analyses demonstrated that loss of HIF-2α did not affect a number of M2 genes, including Fizz1 and Ym-1 (Supplemental Fig. 1). We also found that loss of HIF-2α did not affect hypoxic induction of classical HIF-1α target genes in these cells, including glucose transporter 1 (Glut1) and lactate dehydrogenase-A (LDH-A) (Supplemental Fig. 2). However, arginase1 expression was reduced significantly in both mRNA and protein levels in HIF-2α KO macrophages (Fig. 4A). Basal arginase1 expression as well as hypoxic induction is attenuated in HIF-2α KO macrophages, suggesting that HIF-2α is an essential component of arginase1 induction even in normoxia. Both iNOS and arginase1 are partners in regulating overall NO levels; both are known to be induced by hypoxia, so we hypothesized that the two HIF-α isoforms might act to differentially induce these genes in macrophages. To do this, we generated HIF-1αflox/flox; Tie2cre+/− mice (HIF-1α KO) and HIF-1αflox/flox HIF-2αflox/flox; Tie2cre+/− double-knockout mice (HIF-1/2α DKO) in addition to HIF-2α KO, then collected primary macrophages (TEPMs) and compared hypoxic iNOS and arginase1 induction. iNOS gene induction is reduced significantly in HIF-1α KO, but not in HIF-2α KO macrophages, suggesting hypoxic iNOS gene induction is dependent primarily on HIF-1α (Fig. 4B). In contrast, hypoxic arginase1 induction is reduced to some extent in HIF-1α KO and to a greater extent in HIF-2α KO macrophages, and is completely abrogated in HIF-1/2α DKO (Fig. 4B). These results demonstrated iNOS and arginase1 gene are regulated differently by the two HIF-α isoforms. This notion prompted us to collate dose response profiles of IFNγ stimulation versus HIF-α isoform, iNOS, or arginase1 mRNA expression under hypoxia. The induction of iNOS mRNA expression increased in a manner directly reflective of increasing doses of IFNγ; however, arginase1 gene expression was suppressed (Fig. 4C). Intriguingly, HIF-1α mRNA expression increased with IFNγ, whereas HIF-2α mRNA expression decreased; this dose-dependent response to IFNγ is directly reflective of the respective changes in iNOS and arginase1 (Fig. 4D).
Figure 4.
iNOS expression is controlled by HIF-1α; in contrast, arginase1 expression is regulated mainly by HIF-2α. TEPMs were collected from HIF-1αflox/flox; Tie2cre+/- (HIF-1α KO), HIF-2αflox/flox; Tie2cre+/- (HIF-2α KO), or HIF-1αflox/flox HIF-2αflox/flox; Tie2cre+/- (HIF-1/2α DKO) mice, or wild-type mice as a control. (A) Wild-type (Wt) and HIF-2KO (KO) TEPMs were treated with IL-4 (20 ng/mL) or hypoxia (1%) (H) for 14 h. IL-4 induced arginase1 expression in HIF-2α KO TEPMs normally. However, hypoxic induction of arginase1 mRNA and protein were reduced significantly in HIF-2α KO TEPMs. (B) TEPMs were exposed to hypoxia (1%) (H) for 14 h and collected for gene expression analysis. Hypoxic induction of iNOS was abrogated in HIF-1α KO, but not in HIF-2α KO TEPMs. Hypoxic induction of arginase1 was suppressed in HIF-1α KO TEPMs, but was more strongly suppressed in HIF-2α KO TEPMs. Both iNOS and arginase1 induction with hypoxia were completely inhibited in HIF-1/2α DKO TEPMs (n = 4). (*) P < 0.05; (**) P < 0.01 versus wild type. (C) Wild-type TEPMs were treated with different doses of IFNγ (0.02–20 ng/mL) for 12 h, then incubated in hypoxia (1%) for 16 h. iNOS expression was increased, while arginase1 expression was reduced with increasing doses of IFNγ. (D) HIF-1α expression was also induced, while HIF-2α diminished after increasing doses of IFNγ. Arginase1 but not iNOS expression was affected in the HIF-2α KO macrophage. The decrement of arginase1 expression is abrogated in HIF-2α KO TEPMs.
NO homeostasis and HIF-α dynamic response
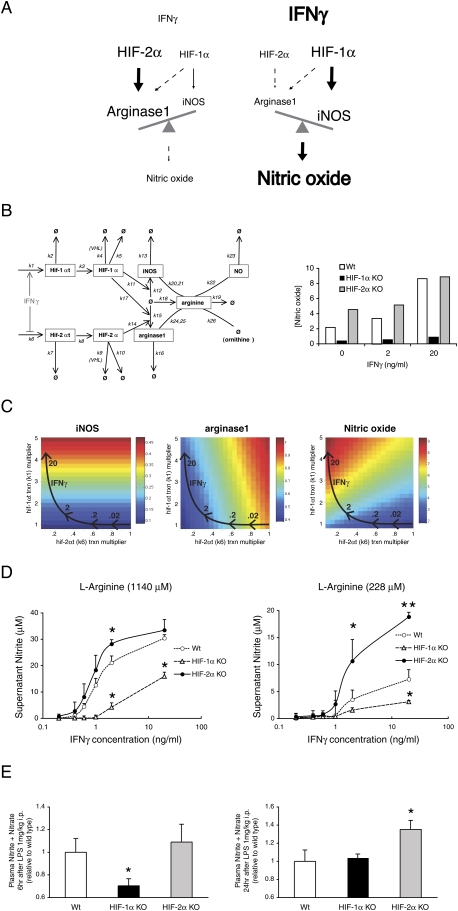
The data presented to this point support the hypothetical model for NO control shown in Figure 5A, which argues that cytokine induction of NO acts differentially through Th1 and Th2 cytokines, and uses the two HIF-α isoforms to accomplish this; in short, elevated Th1 cytokines act to induce HIF-1α, and this in turn increases iNOS levels, which drive NO synthesis. Increases in Th2 cytokine levels conversely increase HIF-2α levels, which drive arginase1 expression, which metabolizes L-arginine and suppresses NO synthesis.
Figure 5.
HIF-1α and HIF-2α acts antagonistically in terms of NO production by inducing iNOS and arginase1 expression. (A) Hypothesized model of NO production under low or high concentration of IFNγ. Under low IFNγ condition, HIF-2α is present and induces arginase1 expression, resulting in the suppression of NO production. Under high IFNγ condition, HIF-2α is diminished and iNOS uses L-arginine for the production of NO. (B) Network diagram for a mathematical model of L-arginine metabolism through iNOS and arginase1 (values for the rate constants k1–k26 are listed in Table 1). HIF-1α and HIF-2α act antagonistically in terms of NO production in macrophages. (C) Heat maps of computational simulation results of iNOS, arginase1, and NO levels in response to altered rates of HIF-1α and HIF-2α transcription under IFNγ stimulation revealed that both L-arginine concentration and the balance of HIF-1α/HIF-2α could affect NO secretion from macrophages. Arrows indicate the responses to different doses of IFNγ. (D) Nitrite productions from TEPMs were measured in supernatant of cultured macrophages using Griess reaction systems. TEPMs were incubated in hypoxia (1%) for 12 h, and were treated with IFNγ under normoxia for 36 h. HIF-2α KO TEPMs produced higher NO than wild type in low–middle concentration of IFNγ under an excess amount of L-arginine (1140 mM). Nitrite production was decreased in HIF-1α KO TEPMs. Production of nitrite was strikingly increased in HIF-2α KO TEPMs under physiological concentration of L-arginine (228 mM) (n = 6). (E) Measurement of plasma nitrite and nitrate levels 6 h or 24 h after LPS 1 mg/kg i.p. of HIF-1αflox/flox; LysM+/− (HIF-1α KO) or HIF-2αflox/flox; LysM+/− (HIF-2α KO) mice. NO level was decreased in 6 h after LPS in HIF-1α KO; however, NO level was increased at 24 h in HIF-2α KO mice (n = 8).
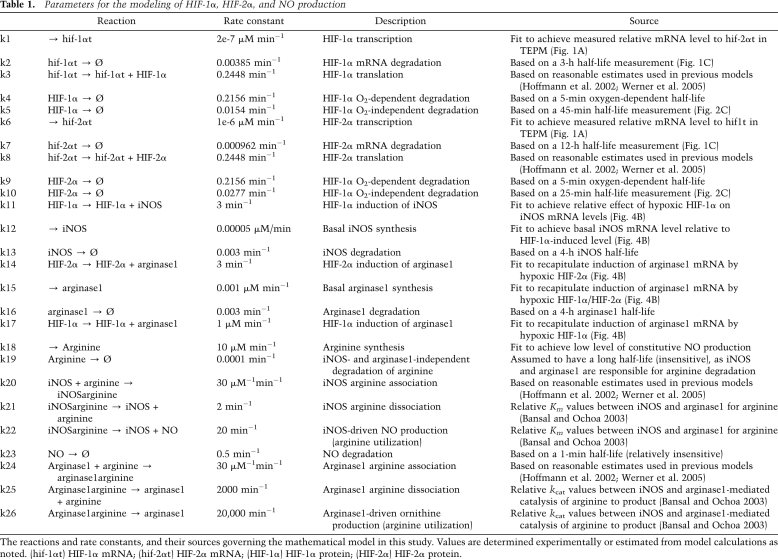
To test this hypothesis, we extended our mathematical model to allow us to predict the net effects of these differential levels of HIF-α function across Th1 and Th2 stimulus gradients on NO synthesis (Fig. 5B). Figure 5B is a graphic representation of the equations described in Table 1; the algorithm encompasses our observed rates of HIF-α mRNA and protein expression and degradation and iNOS and arginase1 synthesis, and the readout of arginine metabolism expressed as NO synthesis. The bar graph in Figure 5B represents a quantitative prediction of NO production to differing doses of IFNγ, which is calculated based on the parameters shown in Table 1. As shown, it predicts that loss of HIF-1α at all doses should lower NO production, but surprisingly, that loss of HIF-2α should cause increased levels of NO, most strikingly at lower doses of Th1 cytokines. As can be seen in Figure 5C, a heat map predicts that iNOS synthesis is essentially a product of HIF-1α transcription, whereas arginase1 has mixed HIF-1α and HIF-2α inputs; the net result, however, as seen in the heat map in the last panel in Figure 5C, is predicted to result in the highest NO levels when HIF-1α mRNA levels are high and HIF-2α mRNA levels are suppressed.
Table 1.
Parameters for the modeling of HIF-1α, HIF-2α, and NO production
The reactions and rate constants, and their sources governing the mathematical model in this study. Values are determined experimentally or estimated from model calculations as noted. (hif-1αt) HIF-1α mRNA; (hif-2αt) HIF-2α mRNA; (HIF-1α) HIF-1α protein; (HIF-2α) HIF-2α protein.
Given a model, we were now able to ascertain whether the predictions it made reflect findings in vivo, and, in particular, to test the surprising supposition that loss of HIF-2α would increase NO levels. To test this, we examined NO production from macrophages cultured in normal or L-arginine-reduced RPMI medium by assaying for NO metabolite levels. As can be seen in Figure 5D, when L-arginine is abundant (1140 mM), NO production is abrogated in HIF-1α-null macrophages. In contrast, significant elevation of NO production is observed in HIF-2α-null cells, suggesting validity of our model. It is noteworthy that the elevation of NO production in HIF-2α-null macrophages is more prominent when cells are cultured in more physiological levels of L-arginine (228 mM; normal L-arginine levels in plasma are predicted to be 80–200 mM, although are much lower in wounds or infections) (Albina et al. 1990; Suh et al. 1997; Wu and Morris 1998; Kamada et al. 2001).
A critical question was whether these results and this model have relevance to in vivo HIF-α function and NO metabolism. To ascertain this, we analyzed plasma NO induced by LPS by using myeloid-specific HIF-1α knockout (HIF-1αflox/flox; LysMcre+/−) and HIF-2α knockout (HIF-2αflox/flox; LysMcre+/−) mice. We measured plasma nitrite + nitrate levels 6 h or 24 h after LPS injection. We found that plasma NO was reduced at 6 h in myeloid-specific HIF-1α knockout mice; in contrast, it increased at 24 h in myeloid-specific HIF-2α knockout mice compared with controls (Fig. 5E). Collectively, these data validate the hypothesis that the HIF-α isoforms regulate NO metabolism in inflammatory cells through a functional antagonism, where HIF-2α suppresses NO production through the induction of arginase1, and HIF-1α induces NO production by induction of iNOS, both acting in response to different polarizing cytokines.
Discussion
The differential functions of the HIF-α isoforms have been difficult to determine. Although loss of either isoform has clear-cut effects on development and function in a range of physiological processes, there is nonetheless a wide overlap in putative HIF targets and binding to hypoxic response elements. In addition, an understanding of differential functions of the two isoforms is limited by their susceptibility to similar mechanisms for post-translational degradation through the VHL-mediated ubiquitin–proteasome pathway (Maxwell et al. 1999; Krieg et al. 2000; Weidemann and Johnson 2008).
One of the more puzzling questions in this regard is how HIF-α protein abundance is adjusted in response to nonhypoxic stimuli. With regard to inflammation, a number of reports have demonstrated that NF-κB signaling can influence HIF-1α mRNA levels (Frede et al. 2006; Rius et al. 2008; van Uden et al. 2008). In the work presented here, we analyzed HIF-1α and HIF-2α mRNA expression in primary macrophages, and established a simulation model of HIF-1α and HIF-2α behavior to ascertain whether, given specific input effects on transcription, regulation at the mRNA level could influence differential protein levels of the two isoforms in the presence of post-translational controls that acted similarly on the two molecules. Our algorithm indicates that Th1 and Th2 cytokines could affect HIF-1α and HIF-2α protein levels by regulating their transcript levels. This was confirmed by an analysis of protein levels in cells with and without VHL, a key component of post-translational regulation of the HIF-α factors (Maxwell et al. 1999; Weidemann and Johnson 2008). These data argue, in turn, that transcriptionally differentiated induction of the HIF-α gene could be an important means for regulating HIF-α expression in a number of other tissues and systems, and could possibly be an explanation for differential roles of the factors in physiological induction and response to nonhypoxic stress.
It is also clear that HIF-2α is expressed exclusively in M2-polarized macrophages, and that this is in contrast to abundant HIF-1α expression in M1 macrophages. This is the first demonstration as well of a cytokine that specifically induces HIF-2α as opposed to HIF-1α. Given that a number of HIF-2α-specific targets or functions have been identified (e.g., erythropoietin, Oct4, c-Myc, and p53) (Covello et al. 2006; Gordan et al. 2007; Rankin et al. 2007; Bertout et al. 2009), but that the mechanisms to explain HIF-2α specificity of these genes is still unclear, it will be useful to investigate the possible transcriptional control of HIF-2α in cells in which these target genes are induced in a HIF-2α-specific manner.
Interestingly, HIF-1α and HIF-2α mRNA showed quite distinctive kinetic profiles; HIF-1α mRNA has a relatively short half-life, while HIF-2α RNA has a remarkably longer half-life. HIF-1α mRNA increased rapidly in response to IFNγ, whereas HIF-2α mRNA responded slowly to IL-4. These data suggest that HIF-1α has a role in transient, acute phase reactions, whereas HIF-2α is working in long-term responses. This is consistent with data described previously for the HIF-α proteins and their kinetic profiles (Holmquist-Mengelbier et al. 2006; Kong et al. 2007); interestingly, in terms of targets, iNOS mRNA also has a short half-life, and arginase1 mRNA has a long half-life (Supplemental Fig. 3). These data would argue for a coevolution of acute and chronic responses via the HIF-α proteins that would suit homeostatic regulation; in the case of NO, the acute phase acts to synthesize the effector, and the chronic phase acts to resolve induction and restore preinduction levels.
In addition to IL-4, we saw that the Th2 cytokine IL-13 also induces HIF-2α mRNA (data not shown). IL-4 and IL-13 both bind to the common receptor IL-4Rα, which signals through the JAK/STAT6 pathway (Chatila 2004). It should be noted that the murine HIF-2α promoter sequence has a putative STAT6-binding site at −448 nucleotides upstream of its transcription start site (data not shown). Interestingly, and in contrast, IFNγ and LPS are known to suppress IL-4/STAT6 signaling by inducing the expression of SOCS (suppressor of cytokine signaling) proteins (Dickensheets et al. 1999; Losman et al. 1999). This could represent one mechanism for IFNγ-mediated suppression of HIF-2α mRNA synthesis.
NO is a powerful bactericidal agent; however, it can potentially cause tissue damage and accelerate inflammation. The homeostatic regulation of NO during inflammation is a primary concern in the activation of the innate immune system. Here, we showed that the HIF-α isoforms have a previously unanticipated cooperative role in temporally regulating the enzymes responsible for both induction and suppression of NO synthesis. That the two HIF-α isoforms act under the control of differing polarizing cytokines argues that these responses are, in turn, controlled by Th1 and Th2 signaling, and that this signaling, unexpectedly using the HIF transcription factors, is key to regulating NO homeostasis.
Materials and methods
Transgenic mice
All procedures involving animals were approved by the University of California at San Diego Animal Care Committee, which serves to ensure that all federal guidelines concerning animal experimentation are met. Generation of mice carrying the loxP-flanked conditional alleles of HIF-1α, HIF-2α, and VHL was described previously (Ryan et al. 1998; Haase et al. 2001; Gruber et al. 2007). Hematopoietic/endothelial or myeloid cell-specific inactivation was achieved by cross-breeding those mice to Tie2-Cre, or LysM-Cre transgenic mice (Clausen et al. 1999; Kisanuki et al. 2001) in C57BL/6 background. Cre-negative homozygous littermates for the conditional alleles were used as controls. Animals were between 8 and 12 wk at the time of the experiments.
Harvest of BMDM and TEPM
To isolate BMDMs, the marrow of femurs and tibias of wild-type mice were collected. Cells were plated in DMEM supplemented with 10% heat-inactivated FBS and 30% conditioned medium (a 7-d supernatant of M-CSF expressing L-929 cells). Mature adherent BM cells were harvested by incubation with mixture A (1:1 mixture of 10 mM EDTA in PBS and RPMI 1640 medium containing 20% FBS) for 15 min and gentle scraping after 7 d in culture. TEPMs were isolated from the peritoneal cavity 4 d after injection of thioglycollate. LPS (L3012), actinomycin D, and cycloheximide were obtained from Sigma. IFNγ and IL-4 were obtained from eBioscience.
RNA analysis by reverse transcription and real-time quantitative PCR
Total RNA was isolated from cultured cells using RNeasy method (Qiagen, Inc.) as described previously (Takeda et al. 2007). First strand synthesis was performed with 0.5 or 1 μg of total RNA by the SuperScript System (Invitrogen) according to the manufacturer's protocol. Fot real-time PCR analysis, cDNA was amplified in SYBR Green or TaqMan Universal Master Mix (Applied Biosystems) with an ABI Prism 7700 sequence detection system (Applied Biosystems). Expression levels were related to 18 sec using the ΔCt method. Primer sequences are in the Supplemental Material.
Western blot analysis
Cells were collected in buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, all chemicals from Sigma) and complete PI tablets (Roche) and incubated for 10 min on ice. After incubation with 0.6% NP-40 for 30 sec and vortex for 30 sec, the lysates were spun down at 12,000g for 30 sec, and the supernatant was collected as cytoplasmic fraction. The pellet was resuspend in buffer C (20 mM HEPES at pH 7.9, 420 mM NaCl, 1.5 mL MgCl2, 0.2 mM EDTA, 25% glycerol, 1 mM DTT, 1 mM PMSF, complete PI), and incubated for 30 min on ice. After centrifugation at 14,000g for 15 min, the supernatant was collected as nuclear extract. Ten micrograms of nuclear protein were separated on 4%–8% Tris-acetate gels (Invitrogen) followed by immunoblotting. Primary antibodies for HIF-1α (B1049-49, Novus Biologicals), HIF-2α (AF2997, R&D), and β-actin (AC-15, Sigma) were probed with secondary antibodies from Amersham. Chemiluminescence was visualized with ECL Plus (GE Healthcare).
Griess assay
The measurement of NO in the supernatant of cultured macrophages was carried out using Griess Reagent System (Promega), and was performed according to the manufacturer's protocol. Before the experiments, cells were plated onto 96-well plates at a plating density of 2 × 105 per well. To measure plasma NO levels, we converted nitrate into nitrite via nitrate reductase prior to the reaction with Griess reagent, since nitrite is rapidly oxidized by oxyhemoglobin to nitrate. Following the reduction, samples were deproteinizated using a 10-kDa cutoff filter (Millipore). Measurement of nitrate + nitrite was performed using NO colorimetric assay kit (BioVision).
Computational simulations
A mass action, ODE-based model was constructed based on measurements made in this study as well as in previous studies (Table 1; Hoffmann et al. 2002; Bansal and Ochoa 2003; Werner et al. 2005). Simulations were done in Matlab version 2008b (Mathworks) with the built-in ode15s silver at default settings. Simulations involving cytokine- or LPS-induced changes to HIF-1α and HIF-2α transcription rates were done based on measured experimental results of the LPS and cytokine effect on HIF-1α and HIF-2α mRNA levels.
Statistical analyses
All statistic analyses were performed using Prism (GraphPad Software). If not otherwise noted, a two-sided unpaired Student's t-test was used for the analysis of differences in mean values between groups.
Acknowledgments
We acknowledge Thorsten Cramer, Carole Peyssonnaux, Ananda Goldrath, Victor Nizet, and Michael Karin for helpful discussions. N.T. carried out experimental work, A.D. initiated design and analysis of tissue-specific deletions, E.L.O. and A.H. carried out modeling studies, M.C.S. provided HIF-2α conditional knockout mice, and all coauthors participated in analysis of data and writing of the manuscript. We acknowledge support for N.T. and M.A. from the Japanese Society for the Promotion of Science; this work was supported by NIH grants CA082515 and CA118165 and Komen grant KG081021.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1881410.
Supplemental material is available at http://www.genesdev.org.
References
- Albina JE, Mills CD, Henry WL, Jr, Caldwell MD. Temporal expression of different pathways of 1-arginine metabolism in healing wounds. J Immunol. 1990;144:3877–3880. [PubMed] [Google Scholar]
- Bansal V, Ochoa JB. Arginine availability, arginase, and the immune response. Curr Opin Clin Nutr Metab Care. 2003;6:223–228. doi: 10.1097/00075197-200303000-00012. [DOI] [PubMed] [Google Scholar]
- Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- Bermudez LE, Young LS. Tumor necrosis factor, alone or in combination with IL-2, but not IFN-γ, is associated with macrophage killing of Mycobacterium avium complex. J Immunol. 1988;140:3006–3013. [PubMed] [Google Scholar]
- Bertout JA, Majmundar AJ, Gordan JD, Lam JC, Ditsworth D, Keith B, Brown EJ, Nathanson KL, Simon MC. HIF2α inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci. 2009;106:14391–14396. doi: 10.1073/pnas.0907357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonecini-Almeida MG, Chitale S, Boutsikakis I, Geng J, Doo H, He S, Ho JL. Induction of in vitro human macrophage anti-Mycobacterium tuberculosis activity: Requirement for IFN-γ and primed lymphocytes. J Immunol. 1998;160:4490–4499. [PubMed] [Google Scholar]
- Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- Bronte V, Serafini P, Mazzoni A, Segal DM, Zanovello P. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends Immunol. 2003;24:302–306. doi: 10.1016/s1471-4906(03)00132-7. [DOI] [PubMed] [Google Scholar]
- Chatila TA. Interleukin-4 receptor signaling pathways in asthma pathogenesis. Trends Mol Med. 2004;10:493–499. doi: 10.1016/j.molmed.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B. HIF-2α regulates Oct-4: Effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes & Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T, Yamanishi Y, Clausen BE, Forster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickensheets HL, Venkataraman C, Schindler U, Donnelly RP. Interferons inhibit activation of STAT6 by interleukin 4 in human monocytes by inducing SOCS-1 gene expression. Proc Natl Acad Sci. 1999;96:10800–10805. doi: 10.1073/pnas.96.19.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante W, Johnson FK, Johnson RA. Arginase: A critical regulator of nitric oxide synthesis and vascular function. Clin Exp Pharmacol Physiol. 2007;34:906–911. doi: 10.1111/j.1440-1681.2007.04638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, Konig T, Schleicher U, Koo MS, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9:1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-κB. Biochem J. 2006;396:517–527. doi: 10.1042/BJ20051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frede S, Berchner-Pfannschmidt U, Fandrey J. Regulation of hypoxia-inducible factors during inflammation. Methods Enzymol. 2007;435:405–419. doi: 10.1016/S0076-6879(07)35021-0. [DOI] [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: Sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Gruber M, Hu CJ, Johnson RS, Brown EJ, Keith B, Simon MC. Acute postnatal ablation of Hif-2α results in anemia. Proc Natl Acad Sci. 2007;104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase VH, Glickman JN, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Natl Acad Sci. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IκB–NF-κB signaling module: Temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, et al. Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Hopper KE. Kinetics of macrophage recruitment and turnover in peritoneal inflammatory exudates induced by Salmonella or thioglycollate broth. J Leukoc Biol. 1986;39:435–446. doi: 10.1002/jlb.39.4.435. [DOI] [PubMed] [Google Scholar]
- Jurgensen JS, Rosenberger C, Wiesener MS, Warnecke C, Horstrup JH, Grafe M, Philipp S, Griethe W, Maxwell PH, Frei U, et al. Persistent induction of HIF-1α and -2α in cardiomyocytes and stromal cells of ischemic myocardium. FASEB J. 2004;18:1415–1417. doi: 10.1096/fj.04-1605fje. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Nagaretani H, Tamura S, Ohama T, Maruyama T, Hiraoka H, Yamashita S, Yamada A, Kiso S, Inui Y, et al. Vascular endothelial dysfunction resulting from L-arginine deficiency in a patient with lysinuric protein intolerance. J Clin Invest. 2001;108:717–724. doi: 10.1172/JCI11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Kong X, Alvarez-Castelao B, Lin Z, Castano JG, Caro J. Constitutive/hypoxic degradation of HIF-α proteins by the proteasome is independent of von Hippel Lindau protein ubiquitylation and the transactivation activity of the protein. J Biol Chem. 2007;282:15498–15505. doi: 10.1074/jbc.M700704200. [DOI] [PubMed] [Google Scholar]
- Krieg M, Haas R, Brauch H, Acker T, Flamme I, Plate KH. Up-regulation of hypoxia-inducible factors HIF-1α and HIF-2α under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene. 2000;19:5435–5443. doi: 10.1038/sj.onc.1203938. [DOI] [PubMed] [Google Scholar]
- Losman JA, Chen XP, Hilton D, Rothman P. Cutting edge: SOCS-1 is a potent inhibitor of IL-4 signal transduction. J Immunol. 1999;162:3770–3774. [PMC free article] [PubMed] [Google Scholar]
- Louis CA, Reichner JS, Henry WL, Jr, Mastrofrancesco B, Gotoh T, Mori M, Albina JE. Distinct arginase isoforms expressed in primary and transformed macrophages: Regulation by oxygen tension. Am J Physiol. 1998;274:R775–R782. doi: 10.1152/ajpregu.1998.274.3.R775. [DOI] [PubMed] [Google Scholar]
- MacNaul KL, Hutchinson NI. Differential expression of iNOS and cNOS mRNA in human vascular smooth muscle cells and endothelial cells under normal and inflammatory conditions. Biochem Biophys Res Commun. 1993;196:1330–1334. doi: 10.1006/bbrc.1993.2398. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: An immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Melillo G, Musso T, Sica A, Taylor LS, Cox GW, Varesio L. A hypoxia-responsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. J Exp Med. 1995;182:1683–1693. doi: 10.1084/jem.182.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl J, Pieczonka MM, Unkeless JC, Silverstein SC. Effects of immobilized immune complexes on Fc- and complement-receptor function in resident and thioglycollate-elicited mouse peritoneal macrophages. J Exp Med. 1979;150:607–621. doi: 10.1084/jem.150.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch C, Muthana M, Lewis CE. Hypoxia regulates macrophage functions in inflammation. J Immunol. 2005;175:6257–6263. doi: 10.4049/jimmunol.175.10.6257. [DOI] [PubMed] [Google Scholar]
- Nathan CF, Murray HW, Wiebe ME, Rubin BY. Identification of interferon-γ as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med. 1983;158:670–689. doi: 10.1084/jem.158.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Johnson RS. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol. 2009;9:609–617. doi: 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JM, Munck A. Glucose transport in murine macrophages: In vitro characterization of the monosaccharide transport system of the thioglycollate-elicited mouse peritoneal macrophage. J Immunol. 1980;125:252–258. [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. HIF-1α expression regulates the bactericidal capacity of phagocytes. J Clin Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin EB, Biju MP, Liu Q, Unger TL, Rha J, Johnson RS, Simon MC, Keith B, Haase VH. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117:1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandau KB, Fandrey J, Brune B. Accumulation of HIF-1α under the influence of nitric oxide. Blood. 2001;97:1009–1015. doi: 10.1182/blood.v97.4.1009. [DOI] [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Peresleni T, Wadhwa N, McNurlan M, Garlick P, Goligorsky MS. Amino acid profile and nitric oxide pathway in patients on continuous ambulatory peritoneal dialysis: L-arginine depletion in acute peritonitis. Am J Kidney Dis. 1997;29:712–719. doi: 10.1016/s0272-6386(97)90124-4. [DOI] [PubMed] [Google Scholar]
- Summersgill JT, Powell LA, Buster BL, Miller RD, Ramirez JA. Killing of Legionella pneumophila by nitric oxide in γ-interferon-activated macrophages. J Leukoc Biol. 1992;52:625–629. doi: 10.1002/jlb.52.6.625. [DOI] [PubMed] [Google Scholar]
- Takeda N, Maemura K, Horie S, Oishi K, Imai Y, Harada T, Saito T, Shiga T, Amiya E, Manabe I, et al. Thrombomodulin is a clock-controlled gene in vascular endothelial cells. J Biol Chem. 2007;282:32561–32567. doi: 10.1074/jbc.M705692200. [DOI] [PubMed] [Google Scholar]
- Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes & Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- van Uden P, Kenneth NS, Rocha S. Regulation of hypoxia-inducible factor-1α by NF-κB. Biochem J. 2008;412:477–484. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidemann A, Johnson RS. Biology of HIF-1α. Cell Death Differ. 2008;15:621–627. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- Wu G, Morris SM., Jr Arginine metabolism: Nitric oxide and beyond. Biochem J. 1998;336:1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Fandrey J, Schumann J, Tiegs G, Brune B. NO and TNF-α released from activated macrophages stabilize HIF-1α in resting tubular LLC-PK1 cells. Am J Physiol Cell Physiol. 2003;284:C439–C446. doi: 10.1152/ajpcell.00294.2002. [DOI] [PubMed] [Google Scholar]