Abstract
Cytoskeletal systems are networks of polymers found in all eukaryotic and many prokaryotic cells. Their purpose is to transmit and integrate information across cellular dimensions and help turn a disorderly mob of macromolecules into a spatially organized, living cell. Information, in this context, includes physical and chemical properties relevant to cellular physiology, including: the number and activity of macromolecules, cell shape, and mechanical force. Most animal cells are 10–50 microns in diameter, whereas the macromolecules that comprise them are 10,000-fold smaller (2–20 nm). To establish long-range order over cellular length scales, individual molecules must, therefore, self-assemble into larger polymers, with lengths (0.1–20 m) comparable to the size of a cell. These polymers must then be cross-linked into organized networks that fill the cytoplasm. Such cell-spanning polymer networks enable different parts of the cytoplasm to communicate directly with each other, either by transmitting forces or by carrying cargo from one spot to another.
Cytoskeletal polymers control cellular architecture and information transfer. Their self-assembly, disassembly, and reorganization underlies processes such as cell polarization and motility.
Because they control the architecture and internal organization of the cell, cytoskeletal systems play uniquely important roles in breaking symmetry and establishing cell polarity (Li and Gundersen, 2008). Broadly speaking, cytoskeletal networks participate in symmetry breaking in three ways: (1) by recording or “freezing” the results of symmetry-breaking events performed by other signaling networks; (2) by providing feedback loops to associated signaling networks that enable them to break symmetry; and (3) by directly generating asymmetry via intrinsic cytoskeletal mechanisms. In this article, I concentrate on symmetry-breaking mechanisms intrinsic to cytoskeletal systems. First, I briefly describe some components and properties of the major eukaryotic cytoskeletal systems. Recent reviews provide more detailed descriptions of the actin (Pollard and Borisy 2003), microtubule (Wittman and Desai 2005), and intermediate filament (Goldman et al. 2008) cytoskeletons. Next, I discuss some basic principles useful for understanding how polymer networks generate asymmetry. Finally, I outline specific examples of cytoskeleton-mediated symmetry breaking.
POLYMERS, PUSHING, AND PULLING
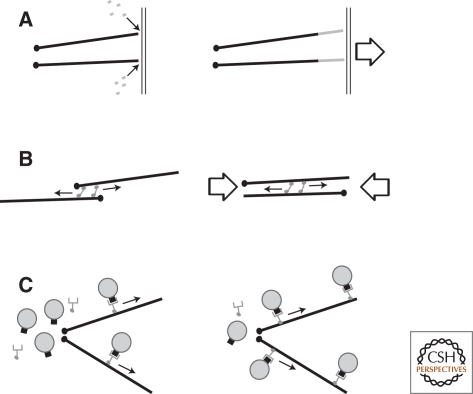
The three main cytoskeletal systems in eukaryotic cells are composed of actin filaments, microtubules, and intermediate filaments. All three systems play a role in determining the shape and mechanics of a cell by resisting and responding to externally applied forces. In addition, networks of actin filaments and microtubules can: (1) generate pushing forces by self-assembly or expansion; (2) transmit pulling forces caused by motor molecules; and (3) provide oriented tracks for directed movement of cargo (Fig. 1). Little is known about how intermediate filaments are assembled or how they contribute to symmetry breaking and cell polarity, so this discussion focuses on actin filaments and microtubules.
Figure 1.
Mechanisms by which cytoskeletal networks integrate information over long distances. (A) Pushing forces generated by polymer assembly or network expansion, (B) pulling forces caused by motor sliding filaments, and (C) oriented tracks for delivery of cargo. Note that pulling forces can also be generated by polymer disassembly (Lombillo et al. 1995) and pushing forces can be generated by motor-mediated filament sliding, but these processes are less common.
Individual actin filaments and microtubules are structurally and kinetically polarized. Structural polarity means that the polymers are not symmetrical. That is, the polymer looks different from its reflection in a mirror or from itself rotated 180°. One consequence of this is that motor molecules move preferentially in one direction or the other on an actin filament or microtubule. In addition, some accessory factors bind specifically to one end of a polymer but not the other. Kinetic polarity means that one end of each polymer grows faster than the other. The fast end of a microtubule is called the plus end and the slow, the minus end. For actin filaments, these are called the barbed (fast) and pointed (slow) ends (Woodrum et al. 1975). Both structural and kinetic polarity are essential for the ability of actin filaments and microtubules to form organized and polarized networks.
The two most important differences between microtubules and actin filaments are mechanical rigidity and assembly dynamics. Microtubules are far more rigid than actin filaments and they have more complex assembly dynamics. One measure of polymer rigidity is a parameter called persistence length, essentially the distance over which the polymer can transmit force. Actin filaments have a persistence length of 10–20 µm, whereas the persistence length of a microtubule is 300–500 times larger (Gittes 1993). Because of their rigidity, individual microtubules can form relatively straight tracks that span the length of a cell. Many animal cells take advantage of this and construct radial arrays of microtubules that function as hubs for intracellular trafficking. These arrays are polarized, with microtubule minus ends collected together at the center and plus ends radiating out to the cell edge. To move toward the center of the cell, cargo molecules must recruit motors that carry them toward minus ends of microtubules. To move to the plasma membrane, they recruit motors that move toward microtubule plus ends. Because they are less rigid, actin filaments that participate in large scale cellular organization are generally cross-linked into ordered bundles or more disordered gels. Stress fibers, for example, are mixed-polarity bundles of actin filaments that connect sites at which cells make intimate contact with the extracellular matrix (Ridley and Hall 1992). They generate contractile forces and enable cells to pull against their external environment. In addition, networks of more randomly crosslinked actin filaments adjacent to the plasma membrane give most animal cells their shape. Filament assembly and disassembly within these networks produce changes in cell shape and amoeboid motility. Other forms of motility, such as swimming driven by the beating of flagella (Riedel-Kruse et al. 2007) or the propagation of kinks in helical spiroplasma (Shaevitz et al. 2005) do not involve such dramatic, global reorganization of the cytoskeleton.
The assembly dynamics of actin filaments and microtubules are adapted to their distinct cellular functions. To a good approximation, the assembly of an actin filament can be described by a single set of rate constants (Pollard 1986) and, under most conditions, the rate of filament assembly depends directly on the concentration of free monomeric subunits available. The steady elongation of actin filaments is well suited to producing sustained forces required to advance the leading edge of a migrating cell (Pollard and Borisy 2003). Microtubules, in contrast, can exist in one of two states: steadily growing or rapidly shortening. In the first state, microtubules are similar to actin filaments and their growth rate depends on the concentration of available tubulin subunits. In the second state, microtubules shorten rapidly, regardless of the concentration of available subunits. Individual microtubules can switch between these two states, growing for a time and then rapidly shrinking. This behavior is known as dynamic instability (Mitchison and Kirschner 1984) and is important for the ability of microtubules to find chromosomes during mitosis (Holy and Leibler 1994; Wollman et al. 2005) and for the ability of radial microtubule arrays to find the center of a cell (Holy et al. 1997).
In addition to polymer assembly and disassembly, motor-driven processes also play important roles in cellular organization and symmetry breaking. Different classes of motor molecules are responsible for sliding filaments and for moving cargo. Actin-based motors are all part of the large myosin family, whose members are adapted to carry out many different cellular tasks. Myosin II, for example, assembles into bipolar minifilaments and induces sliding of anti-parallel actin filaments past each other (Reisler et al. 1980; Pollard 1982). To prevent interference between the multiple motor domains, each subunit of a myosin minifilament interacts only briefly with an actin filament, imparting a brief tug and then letting go (De la Cruz and Ostap 2009). Added together, all the tugs produced by a myosin II minifilament produce a sustained, contractile force in the network. In contrast, other family members, such as myosins V and VI, remain bound through many cycles of motor activity and can move for a considerable distance along a filament before falling off (Mehta et al. 1999; Rock et al. 2001). Single molecules of these “processive” motors can, therefore, transport cargo over long distances. Myosins can also move cargo in both directions along an actin filament. Myosin V transports cargo toward the fast-growing barbed end while myosin VI moves in the opposite direction (Wells et al. 1999). Unlike actin-based motors, microtubule motors appear to have arisen twice in eukaryotic evolution. The newer class of motors, which are not found in primitive eukaryotes such as Giardia labmlia, are called dyneins (Gibbons et al. 1965) and they move from plus end to minus end. Dyneins help construct and power flagella and mitotic spindles and they carry the majority of minus-end-directed cellular cargo (Vaisberg et al. 1996). The older class of motors is a large protein family, called the kinesins (Lawrence et al. 2002; Vale et al. 1985). Some kinesins (e.g., kinesin-1) are processive and move cargo long distances, whereas others (e.g., Eg-5 and other class-five kinesins) are specially adapted to cross-link microtubules or cause them to slide past each other (Miki et al. 2005; Vale and Milligan 2000). Similar to the myosins, the kinesin family contains both plus-end- and minus-end-directed motors.
BASIC PRINCIPLES
I now focus on three principles that appear to be important in the majority of cytoskeleton-mediated symmetry-breaking events: (1) positive feedback, (2) architectural heterogeneity, and (3) network mechanics.
POSITIVE FEEDBACK
Nothing in nature is perfectly symmetrical or homogeneous, so a simple way to generate asymmetry from apparently symmetrical starting conditions is to amplify very small, pre-existing defects or inhomogeneities. Autocatalytic processes, in which the product of a reaction feeds back positively to speed up its own production, can dramatically amplify tiny, spatial fluctuations. This is such a general principle that it is difficult to imagine any form of symmetry breaking in which positive feedback does not play some role. In fact, in some situations, a single positive feedback loop appears to be sufficient to generate cellular asymmetry (Kozubowski et al. 2008; Altschuler et al. 2008). The best experimental evidence for this comes from budding yeast, which determine the position at which a daughter cell forms by localizing the machinery for exocytosis and cell wall synthesis to a single spot on the membrane (see Slaughter et al. 2009). This spot is usually determined by pre-existing cellular landmarks but mutants that lack the landmarks or the ability to recognize them still (randomly) select a single, unique bud site (Chant and Herskowitz 1991). One signaling molecule that is important for this process is a small Rho-family GTPase, Cdc42. Two positive feedback loops have been shown to participate in localization of Cdc42: one involving actin-dependent transport of activated Cdc42 to the membrane (Wedlich-Soldner et al. 2003) and the other involving autocatalytic activation of Cdc42 mediated by a scaffold protein, Bem1 (Kozubowski et al. 2008). Interruption of both feedback loops abolishes polarization but removal of either one alone does not. Mathematical analysis of autocatalytic recruitment of molecules to a membrane poses important constraints on the ability of such a simple mechanism to generate polarity (Altschuler et al. 2008). A unique site on the membrane can be specified only when: (1) the total number of signaling molecules is small and (2) the rate of autocatalytic recruitment is high compared to the rates of spontaneous membrane binding and dissociation. The relative lack of robustness of this simple mechanism may explain why many cellular systems rely on multiple, redundant feedback loops.
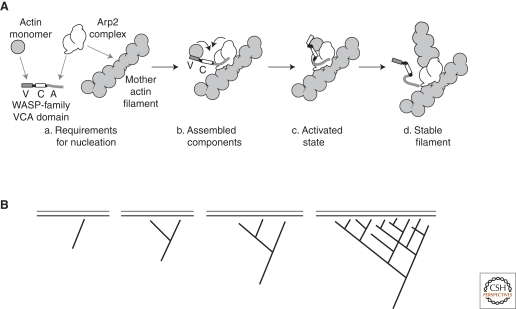
In cytoskeletal networks, positive feedback can include autocatalytic mechanisms for assembling or disassembling component polymers as well as cooperative mechanisms for changing the local architecture of the network. The Arp2/3 complex, for example, is an important nucleator of new actin filaments and its activity is stimulated by the presence of pre-existing filaments (Welch and Mullins 2002). To generate a new filament, the Arp2/3 complex must interact with a pre-existing (or “mother”) filament, an upstream activator, called a nucleation promoting factor (NPF), and an actin monomer bound to the NPF (Fig. 2). After nucleation, the newly formed “daughter” filament remains attached to the mother filament, leading to formation of a cross-linked filament array. As the density of filaments increases, the rate at which the complex is recruited and activated increases. This feedback loop is important for building robust and stable actin networks at the leading edge of motile cells (Svitkina and Borisy 1999; Iwasa and Mullins 2007; Wang 2009) as well as sites of endocytosis (Kaksonen et al. 2006; Orlando and Guo 2009). Obviously, this feedback cycle cannot continue indefinitely, producing an infinite density of actin polymer. Like all positive feedback loops in biology, it is ultimately limited by some physical or biochemical constraint, in this case by the availability of actin monomers required to activate the complex. When the number of growing filaments becomes very large, they act as a sink, competing with the complex for NPF-bound actin (Akin and Mullins 2008). Assembly of microtubules is not as well understood but there does not appear to be an equivalent positive feedback system controlling their nucleation. Instead, a motor-mediated feedback loop controls assembly and stability of the microtubule organizing centers that focus the minus ends of microtubules to discrete spots (discussed later).
Figure 2.
Positive feedback in the activation of the Arp2/3 complex. (A) Molecular mechanism of Arp2/3 activation. (a) Four factors must come together to produce a new filament: An Arp2/3 complex, an actin monomer, a VCA domain, and a pre-existing actin filament. (b) All the components assemble into a preactivation complex. (c) A conformational change in the complex results in formation of a new stable filament (d) attached to the side of the pre-existing (mother) filament. (B) The requirement for a pre-existing filament makes the nucleation reaction auto-catalytic. The existence of a single filament near a surface covered with an Arp2/3 activator leads to nucleation of more filaments, which, in turn, stimulate further Arp2/3-dependent nucleation.
ARCHITECTURAL HETEROGENEITY
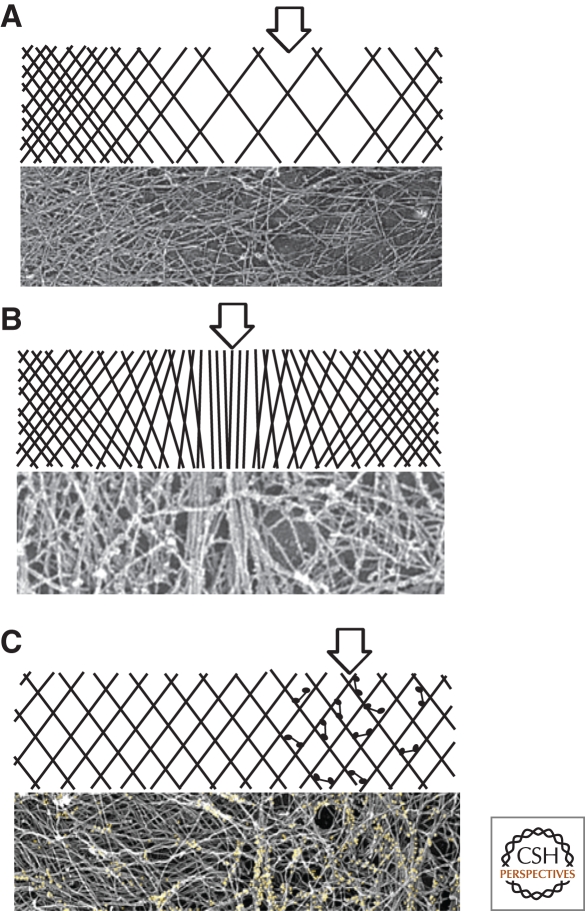
In cell signaling, the most common type of spatial heterogeneity is a fluctuation in the local concentration of an activated signaling molecule, say a kinase or a G-protein. Because cytoskeletal networks are collections of crosslinked, linear polymers, they show additional, often subtle, classes of spatial heterogeneity. These include local differences in: (1) polymer density; (2) polymer alignment and orientation; (3) activity of motor molecules; or (4) number of bound cross-linking proteins (Fig. 3). Each of these architectural heterogeneities can be amplified by positive feedback to produce large-scale variations in network properties. As noted above, local polymer density can affect the activity of the actin-nucleating Arp2/3 complex. In addition, polymer orientation and alignment can affect interaction with motors and cross-linkers. Myosin II, for example, prefers to interact with antiparallel bundles of actin filaments. By recruiting and aligning additional filaments, myosin II motor activity can produce additional myosin II binding sites (Schaub et al. 2007). This positive feedback loop participates in polarization of many motile cells (see the following discussion). Also, in addition to motors, the number of bound cross-linking proteins can determine the degree of filament orientation and the local mechanical properties of a network. At low concentrations, many actin filament cross-linking proteins, including, for example, α actinin, will stabilize filament networks with a variety of geometries, from parallel bundles to orthogonal arrays. At higher concentrations, these cross-linkers cause filaments to zipper together and preferentially form mixed-polarity bundles (Wachsstock et al. 1994). In vivo, the asymmetric localization of the actin-cross-linking protein cortexillin, for example, helps drive shape changes required for cell division, even in the absence of myosin II motor activity (Girard et al. 2004; Weber et al. 1999).
Figure 3.
Typical heterogeneities in the architecture of polymer networks. (A) Variations in polymer density. Arrow denotes region of low density. Lower panel: Electron micrograph of actin filaments in a sea urchin coelomocyte (Henson et al. 1999). (B) Variations in orientation or alignment of polymers. Arrow denotes region of filament alignment in an otherwise random gel. Lower panel: Variations in actin filament alignment in the periphery of a fibroblast cell (Svitkina et al. 2003). (C) Variations in density or activity of accessory factors such as motor or cross-linker molecules. Arrow denotes region of high concentration of motor or cross-linker. Lower panel: distribution of myosin motors (yellow) in the actin network of a fish epidermal keratocyte (Svitkina and Borisy 1997).
NETWORK MECHANICS
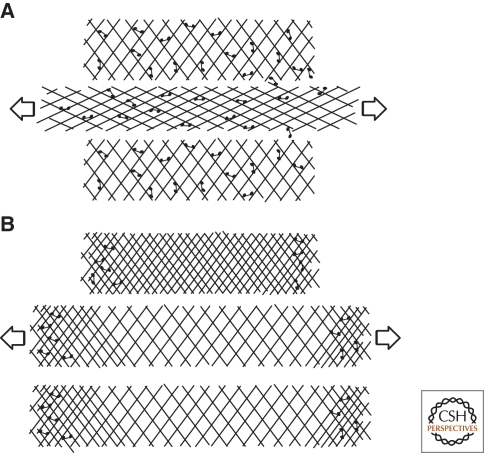
The mechanical properties of a material can be described by how it responds to an applied force. Elastic solids are initially deformed but then return to their original shape after force is removed. Viscous liquids, by contrast, flow in response to force and do not regain their original shape when force is removed. Cytoskeletal networks can behave either like viscous liquids or elastic solids, or combinations of both, depending on the architecture of the network and how force is applied to it (Sato et al. 1987; Wachsstock et al. 1993). Generally, the density and orientation of the polymers and the number of cross-linkers connecting them determine whether a network resists deformation like an elastic solid or flows like viscous liquid (Fig. 4). In addition to forces applied from the outside, cytoskeletal networks can also respond to forces generated within the network by motor molecules. One important class of symmetry-breaking processes makes use of the asymmetrical flow of cortical actin networks caused by nonuniform distributions of motors and cross-linkers. These flows can gather signaling molecules and polarity determinants that are bound to actin filaments and convert a uniform distribution into a highly polarized one. Mechanisms of this type are responsible for polarization of oocytes and fertilized eggs in many species (see the following discussion).
Figure 4.
Elastic versus viscous behavior of cross-linked polymer networks. (A) Some cross-linked cytoskeletal networks respond to some types of applied forces (arrows) like elastic materials. They deform and then return to their original shape after the force is removed. (B) Other networks flow like viscous liquids in response to applied forces and do not return to their original shape after the force is removed. For most networks, the response to applied forces is best described by a combination of these viscous and elastic behaviors.
In addition to deformation and flow, mechanical failure of cytoskeletal networks can also contribute to symmetry breaking. To understand how this works, imagine a breakable, macroscopic object with spherical symmetry—say a balloon. A straightforward way to break the spherical symmetry of this particular object would be to over-inflate it until the skin of the balloon is stretched beyond its elastic limit and fails (i.e., pops). Failure occurs at a specific site, probably determined by micro-heterogeneities in the material. In a similar way, external and internal forces acting on a cytoskeletal network can produce symmetry-breaking ruptures.
The remainder of this article is devoted to discussing specific examples of cytoskeleton-mediated symmetry breaking in light of the general principles discussed previously.
ASTERS AND SPINDLES: THE SPONTANEOUS ASSEMBLY OF MICROTUBULES INTO ORGANIZED STRUCTURES
In many cells, the bipolar mitotic spindle and monopolar interphase microtubule array are organized around well-defined microtubule organizing centers, called centrosomes. The architecture and composition of centrosomes are not completely understood but they contain microtubule nucleating and binding factors that collect and anchor microtubule minus ends. Centrosomes, however, are not essential to life. All plant cells and many meiotic animal cells lack them entirely. In addition, work from several laboratories over the past two decades has shown that cells that normally contain centrosomes can carry out many essential, microtubule-based functions without them. Laser ablation of centrosomes in animal cells, for example, does not prevent assembly of functional mitotic spindles (Khojakov et al. 2000) and fission yeast lacking spindle pole bodies (functional equivalents of centrosomes) assemble functional interphase microtubule arrays, indistinguishable from those normally assembled around spindle pole bodies (Carazo-Salas and Nurse 2006). These results, together with the observation that microtubules and motor proteins can spontaneously self-assemble into radial arrays and bipolar spindles (see following discussion), suggest that centrosomes evolved, in part, to increase the robustness and fidelity of pre-existing pathways for spontaneous assembly of microtubule arrays (Marshall 2009).
Asters
Radially symmetrical arrays of microtubules, like those found in interphase animal cells, are often called asters. It might seem odd to describe assembly of such an array as symmetry breaking, but, from the point of view of a molecule in the cytoplasm, the broken symmetry is obvious. Rather than a linear, anterior–posterior or dorsal–ventral axis, the axis of polarity established by such an array is from inside to outside. Leibler and coworkers showed that radial arrays of microtubules can be spontaneously assembled from mixtures of microtubules and motors (Nedelec et al. 1997). These authors constructed artificial, four-headed kinesins and mixed them with preformed, stabilized microtubules. At certain concentrations, the motors convert the spatially uniform, random distribution of microtubules into a collection of well-separated, radial arrays of approximately the same size. Using plus end-directed kinesins, aster assembly begins with the motors binding to microtubules and moving toward their plus ends. Because they can bind multiple microtubules simultaneously, the motors can draw plus ends together. If a motor binds a pair of antiparallel microtubules, for example, it will force their minus ends in opposite directions and bring their plus ends to the same point. For microtubules that cross each other at an arbitrary angle, a motor that binds both will pull both plus ends to the crossover point. Because each microtubule can bind many motors and contact many other microtubules, many plus ends wind up moving to the same location, producing a radial array, or aster, centered on this point (Fig. 5). The remarkably uniform size of the asters produced in this experiment is determined by the average lengths of the microtubules and the concentration of the motor molecules (Surrey et al. 2001).
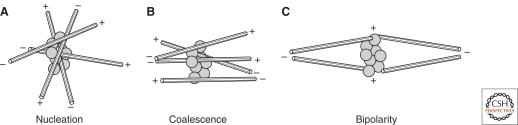
Figure 5.
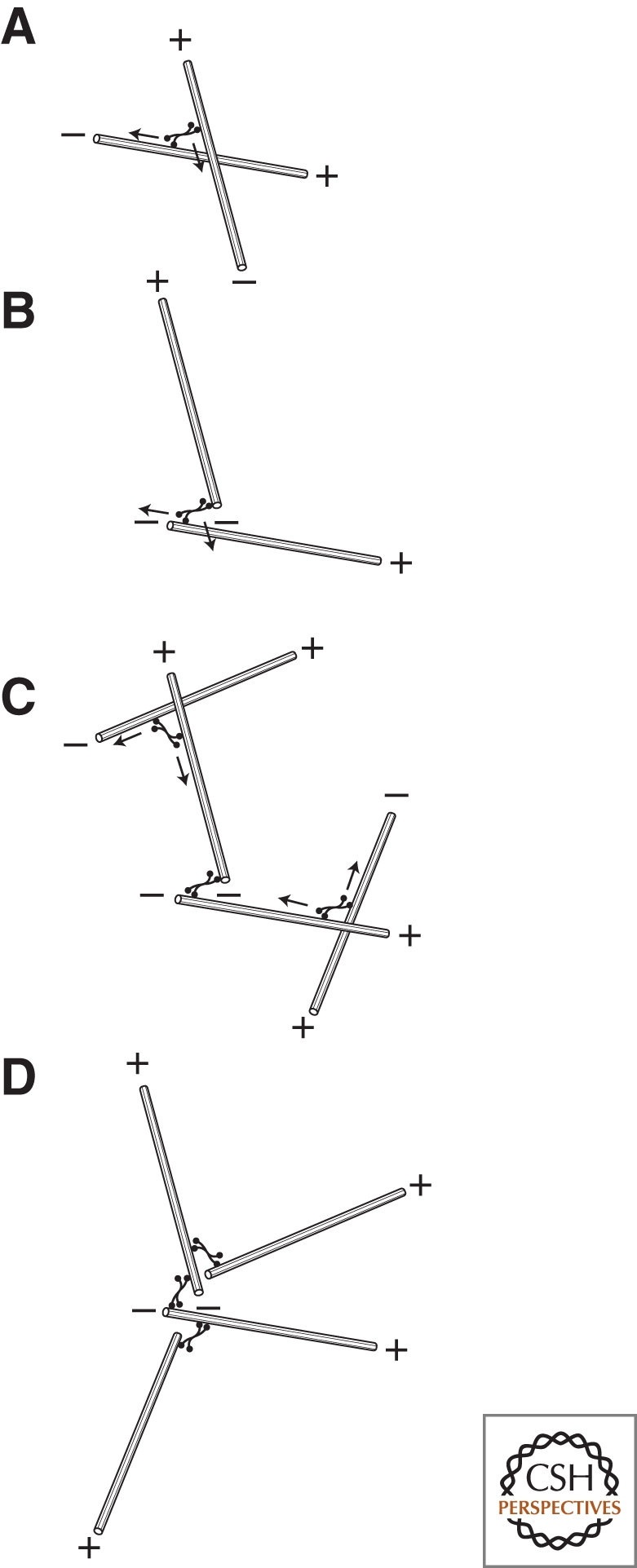
Assembly of polarized, radial arrays of microtubules. (A) Multi-valent microtubule-based motors can cross-link microtubules and cause them to slide past each other. (B) The polarity of the motors (in this case, minus end-directed) causes the minus ends of the microtubules to be pulled into proximity. (C) Additional motor-dependent interactions recruit additional microtubules, eventually forming (D) a polarized radial array.
What about the three principles outlined above? In this system, the architectural heterogeneities required to break symmetry are the randomly occurring points of microtubule overlap, where motors can bind simultaneously to multiple microtubules. As more plus ends are collected together in one spot, the microtubules radiating from this spot will contact and reel in additional microtubules from a larger and larger volume, resulting in positive feedback. Finally, the high mechanical rigidity of the microtubules is required to prevent them from snarling and buckling as they are moved around by motors. If the microtubules were less rigid, they would form a tangled hairball rather than an organized radial array.
In this system, plus end-directed motors produce asters with microtubule minus ends facing out while minus-end-directed motors produce asters of the opposite polarity. A similar mechanism can probably construct radial microtubule arrays in vivo. In cells, dynein motors carry cross-linking factors (e.g., NuMA) to the minus ends of microtubules, promoting their assembly into radial arrays in which plus ends face out. When radial microtubule arrays are placed in a confined volume, they tend to find the center of the space. This self-centering is the result of polymerization-dependent pushing forces and requires dynamic instability of the microtubules (Holy et al. 1997). A similar self-centering of radial arrays is observed in vivo and involves the additional participation of motor proteins located at the cell periphery (Rodionov and Boirsy 1997; Burakov et al. 2003).
Spindles
Microtubules assemble, during mitosis, into a bipolar, DNA-segregating machine called the mitotic spindle. In animal cells, the poles of the spindle are formed by centrosomes and, in addition to aligning and segregating chromosomes, the microtubules between the centrosomes determine the size and morphology of the spindle. Centrosomes, however, are not absolutely required for spindle assembly or DNA segregation. As noted above, plant cells and animal cells undergoing meiosis form spindles without centrosomes and laser ablation of centrosomes in animal cells does not prevent the assembly of functional spindles. Heald et al. (1996) used meiotic Xenopus egg extracts to investigate spindle assembly in vitro and found that neither centrosomes nor intact chromosomes were required for bipolar spindle formation. Addition of micron-sized polystyrene particles coated with plasmid DNA was sufficient to induce localized polymerization of microtubules. These microtubules then spontaneously assembled into bipolar spindles with plus ends facing the DNA-coated particles in the middle and minus ends focused at the poles. The first step in this process, DNA-induced microtubule assembly, involves activation of a small G-protein, Ran, by a DNA-associated GTP-exchange factor, RCC1. Activated Ran then releases microtubule nucleation factors sequestered by importin-family proteins (Zheng 2004). The cloud of microtubules assembled around the DNA-coated beads is then cross-linked by multivalent motors. Multiple cross-links between pairs of microtubules drives their alignment into a mixed-polarity bundle. Formation of a more-or-less linear bundle at this stage ensures that the final spindle has two poles and not, say, three or five. Next, plus end-directed kinesin-family motors bound to DNA push the minus ends of the microtubules out, away from the DNA-coated beads. Finally, dynein motors carry microtubule cross-linking factors to the minus ends and focus them into a tight pole (Fig. 6) (Heald et al. 1997).
Figure 6.
Formation of bipolar spindles in the absence of centrosomes. (A) In cell extracts, DNA-coated particles induce nucleation of microtubules whose polarities are random with respect to the DNA. (B) Microtubule cross-linkers and multivalent motors align the microtubules while minus-end directed motors begin to cluster their minus ends. (C) Plus end-directed microtubule motors associated with DNA push the clustered minus ends away from the DNA-coated particles, forming a bipolar, spindle-shaped structure.
ACTIN-BASED MOTILITY OF PATHOGENS AND PARTICLES: A ROLE FOR MECHANICAL FAILURE
Many micron-sized objects, including some pathogens and newly endocytosed vesicles, harness the assembly of actin networks to generate force and move through the cytoplasm of eukaryotic cells. Motile pathogens, including: Listeria, Shigella, and Rickettsia species, as well as Burkholderia pseudomallei, Mycobacteria marinum, and vaccinia virus, hijack the host cell actin cytoskeleton and use it to power intracellular motility and cell-to-cell spread. Conversely, in many cells, newly endocytosed vesicles appear to be pushed away from the membrane and into the cytoplasm by polarized actin assembly (Merrifield et al. 2002; Kaksonen et al. 2006). A particularly dramatic example of this type of motility is observed in Xenopus eggs, where fertilization produces a global burst of actin-based vesicle motility (Taunton et al. 2000).
Motile pathogens and vesicles express different molecules on their surfaces and interact with different host cell factors but, ultimately, they all recruit and activate the same actin-nucleating Arp2/3 complex (Gouin et al. 2005). In fact, the only molecular requirement for this type of actin-based motility is a surface-attached activator of the Arp2/3 complex. A remarkable feature of this type of motility is that, even when the Arp2/3 activator is uniformly distributed on the surface of the particle, it can robustly form a polarized actin network and move in a vectorial fashion (Cameron et al. 1999). In vitro, polystyrene micro-spheres and lipid vesicles uniformly coated with activators of the Arp2/3 complex, induce formation of spherically symmetrical actin networks which then break symmetry to form motile actin “comet tails.” Several models have been proposed to describe how these symmetrical particles generate asymmetrical actin networks (van Oudenaarden and Theriot 1999; van der Gucht et al. 2005). The most successful is based on treating the crosslinked actin network as an elastic gel with a finite breaking strength (Van der Gucht et al. 2005; Van der Gucht and Sykes 2009). In this model, actin nucleation at the surface of the particle produces a symmetrical actin shell and then continued nucleation and polymerization at the surface of the particle pushes the original shell outward. The network behaves like the skin of a balloon. As more actin polymerizes at the surface, it stretches the network until eventually it ruptures. The energy stored in the elastic deformation causes the expanding network to pull away from the site of rupture, producing a dramatic asymmetry in the distribution of actin around the particle. This asymmetry persists because, even if new actin assembles uniformly over the particle, the new network covering the site of rupture will be thinner and weaker than that on the opposite side, where no rupture has occurred. This asymmetric network will expand and rupture again at the same site (Fig. 7).
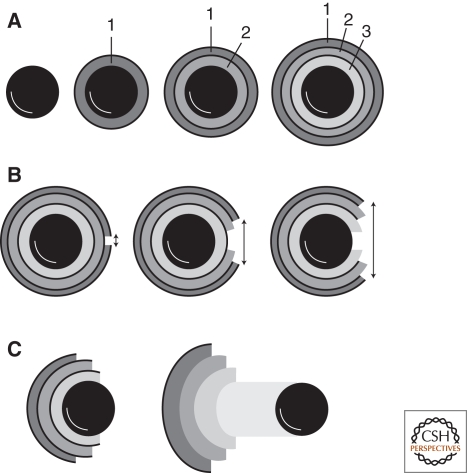
Figure 7.
Polarization of actin networks because of expansion and mechanical rupture. (A) Spherical surfaces coated with activators of the Arp2/3 complex (black circle) direct assembly of spherically symmetrical actin networks (gray rings). Continuous nucleation of new filaments at the surface causes outward displacement and stretching of the initial actin shell (1). (B) Continued expansion eventually stretches the outer portions of the actin network to the point of mechanical failure. The elastic energy stored in the stretched shells is released by contraction. The initial rip propagates because of positive feedback. As the rip progresses, the remaining stress in the network is distributed over a smaller number of cross-links, increasing the probability that that they will fail. (C) Recoil of the actin shell away from the rip results in a polarized distribution of actin on the surface of the particle.
The role of mechanics in this system is obvious. The material properties of the actin network will determine how much force is required to inflate the network and how much stretching it can withstand before it ruptures. The important architectural heterogeneity in the system is a (possibly slight) fluctuation in network density that determines the initial site of rupture. Because the outer network is under tension, the loss of a small number of cross-links results in increased tension on the remaining cross-links. This increased tension increases the probability that some of these cross-links will fail, and so on. This positive feedback means that, once a small rupture occurs, it will tend to propagate, leading to a large scale mechanical failure.
POLARIZATION AND MOTILITY OF CELLS AND CELL FRAGMENTS
To move in a given direction, a cell must first establish an axis of polarity, with functionally distinct regions at the front and back (Xu 2003; Wang 2009). At the front of the cell, the leading membrane is pushed forward by assembly of actin networks similar to those that push pathogens and vesicles through the cytoplasm. At the rear of the cell, the trailing edge is pulled forward by myosin-dependent contraction of the actin network. Adhesion to a substrate is required for some types of amoeboid locomotion but the most basic form of movement through a three-dimensional matrix appears to depend only on actin-dependent protrusion and expansion of the cell’s front and retraction of its rear (Lämmermann et al. 2008).
Cells that crawl toward or away from chemical cues determine direction of motion based on activity of signaling systems linked to cell surface receptors (Parent and Devreotes 1999). In many motile cells, however, the cytoskeleton appears capable of breaking symmetry and determining direction of migration without external cues. Evidence for this comes from the fact that a population of chemotactic cells placed in a spatially uniform concentration of chemoattractant, which activates receptors uniformly over the cell surface, spontaneously break symmetry and move in random directions (Tranquillo et al. 1988; Weiner et al. 1999; Xu 2007). Also, some cells, including fish epidermal keratocytes, move constitutively in the absence of any known external cue. The membrane of these cells appears to be uniformly permissive for assembly or contraction of actin networks and the direction of migration appears to be determined by mechanisms intrinsic to the cytoskeleton (Verkhovsky et al. 1999; Yam et al. 2007; Xu et al. 2007). Recent reviews (e.g., Weiner 2002) provide a detailed discussion of how cytoskeletal assembly is integrated with cellular signaling systems controlling chemotaxis. For purposes of this discussion, I focus on symmetry-breaking mechanisms that appear to be independent of external chemical cues.
Important insight into cytoskeletal processes that establish and maintain polarity has come from the study of motile cell fragments, called cytoplasts. Cytoplasts can be produced from many cell types by several techniques (Malawista et al. 1985). They can be generated, for example, by treatment of fish keratocytes with nonspecific kinase inhibitors, which cause motile regions at the cell front to pull away and detach from the cell body (Verkhovsky et al. 1999). Cytoplasts lack a nucleus, a microtubule organizing center, and contain only a fraction of the cytoplasm of the original cell. Despite this, they maintain a shape similar to intact cells and move for hours in a directed fashion. The organization of cytoplast actin networks is remarkably similar to that of intact cells. The network at the leading edge of both is a gel of cross-linked filaments with a large fraction of barbed ends pointing toward the plasma membrane (Svitkina et al. 1997). Continuous assembly of this network pushes forward the leading edge (Theriot et al. 1991). At the rear of both cells and cytoplasts, actin filaments are collected into large, mixed-polarity bundles. These bundles also contain a large number of myosin II minifilaments that cause them to contract (Anderson and Cross 2009) and pull forward the rear of the cell (Lämmermann et al. 2008; Xu 2003). The combination of polymerization and protrusion at the leading edge and contraction at the trailing edge gives both keratocytes and their cytoplasts a characteristic crescent or canoe shape.
Occasionally, cytoplasts lose polarity and stop moving. Such cytoplasts go from crescent-shaped to round and actin assembly occurs uniformly around their entire circumference. It appears, in these cases, as if the cell fragment has lost its rear end and that the entire periphery is now functionally equivalent to the front of a migrating fragment. These nonpolarized cytoplasts can spontaneously break symmetry and begin moving again. Symmetry can also be broken by small, externally applied perturbations and this type of forced symmetry breaking has provided insight into cytoskeletal mechanisms of generating and maintaining polarity. Verkhovsky et al. (1999) showed that a stream of liquid strong enough to deform the edge of a nonmotile cytoplast can induce polarization and sustained motility. These authors found that induced deformations often propagate along the edge of the cytoplast, becoming a zone of sustained network contraction and defining a new rear. The cytoplast then becomes motile and crawls away from the site of initial perturbation (Verkhovsky et al. 1999). This phenomenon appears to depend on the preference of myosin II minifilaments for antiparallel actin filaments. When the Arp2/3 complex is active all around the periphery, it generates actin networks in which filaments intersect at a variety of angles, all with their barbed ends facing toward the membrane (Svitkina and Borisy 1999; Mullins et al. 1998). This type of network does not contain many favorable binding sites for myosin II. Mechanically deforming the network by crushing (or stretching) it forces some of the filaments into an antiparallel alignment, producing new binding sites for myosin II. Once bound, myosin II pulls additional filaments into the bundle, creating more myosin II binding sites (Anderson et al. 2008). This positive feedback causes an initially small architectural heterogeneity in the network to propagate (Fig. 8). Given the strong positive feedback, why does this architectural change in the network fail to propagate around the entire circumference of the cytoplast? The simplest answer is that it is ultimately limited by cell locomotion. As the zone of contraction propagates, the forces acting on the cytoplast become unbalanced. The pushing force caused by actin assembly on the opposite side combines with the contraction to cause the cell to migrate. Cell migration pushes the leading edge of the cell away from the original actin network. Seen from the perspective of the leading edge itself, the actin network appears to treadmill backwards. When myosin II binds to the network anywhere but the trailing edge, it is swept backward with the flow of actin filaments. This rearward flow continuously sweeps myosin II away from the leading edge and further concentrates it in the rear of the cytoplast (Anderson et al. 2008). This type of flow-induced asymmetry of actin binding proteins also plays a key role in establishing polarity of many types of oocytes and fertilized eggs (see the following discussion). In addition, the myosin-dependent collection and contraction of actin filaments appears to be limited by a poorly understood, myosin-dependent mechanism for disassembling actin filaments (Biron et al. 2005; Medeiros et al. 2006).
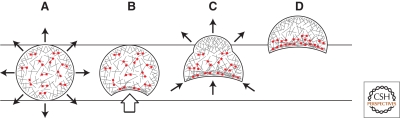
Figure 8.
Polarization and motility of cytoplasts. (A) In cytoplasts that have lost polarization, actin assembly occurs symmetrically around the periphery, producing uniform outward forces (arrows) that sum to zero. Myosin II minifilaments (red symbols) are randomly oriented and do not produce coherent, large-scale pulling forces. (B) Random fluctuations in myosin localization or activity or external mechanical perturbation (large arrow) can produce local alignment of actin filaments. (C) Alignment of filaments produces more favorable binding sites for myosin minifilaments and makes their pulling forces more coherent (arrows at bottom of cell). As the contraction proceeds, more filaments are aligned and recruited into the contractile bundle. This bundle is a poor substrate for the Arp2/3 complex and inhibits production of polymerization-driven outward forces. This imbalance of outward forces enables polymerization on the opposite side of the cell to produce net protrusion. (D) Translocation of the cell in the direction of protrusion results in treadmilling of the actin network away from the leading edge. This treadmilling carries myosin II away from the leading edge and concentrates it in the rear of the cell, further enhancing the polarization of the cell and the spatial separation of protrusive and contractile forces.
So much for cell fragments. What about intact cells? Yam et al. (2007) found that the mechanism by which intact keratocytes break symmetry and establish polarity is remarkably similar to the mechanism described for cytoplasts. These authors watched nonpolarized cells break symmetry and initiate directed motility and found that, in all cases, symmetry breaking began as a contraction in the actin network that propagated to establish a trailing edge for the cell.
POLARIZATION OF OOCYTES AND FERTILIZED EGGS
In most metazoans, the major axes of embryonic development—anterior–posterior and dorsal–ventral—are established at the single cell stage, either during oogenesis or soon after fertilization. Different organisms use different mechanisms to establish polarity but all rely heavily on the actin and microtubule cytoskeletons. One of the most completely understood polarity generating mechanisms occurs in the Caenorhabditis elegans zygote (Munro and Bowerman 2009). In this case, anterior–posterior polarity is determined by sperm entry. Both the actin and microtubule cytoskeletons work together to convert this initial asymmetry into a global cell polarity.
In C. elegans eggs, the determinants that specify posterior cell fates, including the Par-3/Par-6/PKC-3 complex, are uniformly distributed throughout a cortical actin network, just below the plasma membrane. Determinants that specify the anterior fates are found deeper in the cytoplasm. Fertilization induces a dramatic, global activation of myosin-dependent contraction of the actin network, probably via a wave of calcium influx. At the site of fertilization, the sperm nucleus enters the egg, bringing along its centrosome, which nucleates formation of a radial array of microtubules. The microtubules of this sperm aster act as tracks to deliver factors that inhibit myosin contraction to the cortex (Jenkins et al. 2006). The cortex adjacent to the site of sperm entry receives the largest dose of these factors and becomes the site of weakest contraction. Because of the reduced contraction, this zone cannot resist the pulling forces coming from the rest of the cortex and the cortical actin network is, therefore, pulled away from this site, dragging along the associated posterior determinants (Fig. 9).
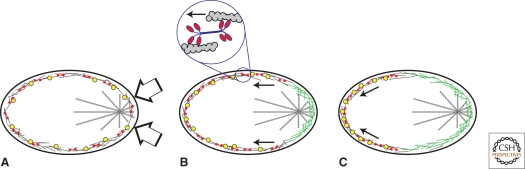
Figure 9.
Contraction-induced polarization of C. elegans zygotes. (A) Before polarization, many posterior determinants (e.g., par6, yellow symbols) are uniformly distributed throughout the cortical actin network of the zygote. Soon after sperm entry, the activity of myosin II minifilaments (red symbols) causes contraction of the cortical actin network. The sperm centrosome (near the right side of the zygote) delivers factors to the cortex that locally inhibit myosin activity. (B) The asymmetric activity of myosin II pulls the cortical actin network away from the sperm centrosome toward the opposite end of the cell. (C) Actin-associated polarity determinants pulled along with the cortical network define the posterior pole of the zygote and, ultimately, the embryo. Cortical actin removed from the future anterior pole is replaced by newly polymerized filaments or filaments that well up to the surface from the underlying cytoplasm (green filaments).
SUMMARY AND PERSPECTIVE
Cytoskeletal systems are organized networks of polymers that control cell shape, organize intracellular spaces, and integrate information over cellular length scales. Establishing cell polarity or otherwise breaking cellular symmetry requires, at the very least, reorganizing the cytoskeleton and, in many cases, is actually driven by cytoskeletal mechanisms. Autocatalytic assembly, disassembly, or reorganization of cytoskeletal networks can amplify small perturbations and help convert subtle external cues into large-scale cellular responses. In the absence of external cues, these feedback loops can often amplify spontaneous fluctuations in network architecture and break symmetry on their own.
In many examples of biological symmetry breaking, the specific contributions of signaling and cytoskeletal systems are difficult to parse. Correct polarization of Drosophila oocytes, for example, is known to require participation of both the actin and microtubule cytoskeletons, as well as multiple cellular signaling systems (Roth and Lynch 2009). At present, however, the specific molecular event that determines the positions of the anterior and posterior poles of the oocyte is unknown. In many cases, a complete understanding of a symmetry-breaking process will require its reconstitution in vitro, either in cell extracts or from purified components. A relatively small number of complex cytoskeletal systems have already been reconstituted in vitro and they have provided deep insight into the pathways by which they are assembled and the mechanisms by which they are regulated. They have also impressed on the field the fact that remarkably complex, and otherwise unpredictable, properties can emerge from the interaction of a small set of molecules. The next phase in cell biological research will be to use such reconstituted systems to bridge the gap between structural and biochemical studies of individual molecules and complex behavior of living cells and organisms.
Footnotes
Editors: Rong Li and Bruce Bowerman
Additional Perspectives on Symmetry Breaking in Biology available at www.cshperspectives.org
REFERENCES
- Akin O, Mullins RD 2008. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133:841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler SJ, Angenent SB, Wang Y, Wu LF 2008. On the spontaneous emergence of cell polarity. Nature 454:886–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KI, Cross R 2009. Contact dynamics during keratocyte motility. Curr Biol 10:253–260 [DOI] [PubMed] [Google Scholar]
- Anderson TW, Vaughan AN, Cramer LP 2008. Retrograde flow and myosin II activity within the leading cell edge deliver F-actin to the lamella to seed the formation of graded polarity actomyosin II filament bundles in migrating fibroblasts. Mol Biol Cell 19:5006–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron D, Alvarez-Lacalle E, Tlusty T, Moses E 2005. Molecular model of the contractile ring. Phys Rev Lettr 95:098102. [DOI] [PubMed] [Google Scholar]
- Burakov A, Nadezhdina E, Slepchenko B, Rodionov V 2003. Centrosome positioning in interphase cells. J Cell Biol 162:963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Sheetz MP 2009. Force propagation across cells: Mechanical coherence of dynamic cytoskeletons. Curr Opin Cell Biol 21:47–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron LA, Footer MJ, van Oudenaarden A, Theriot JA 1999. Motility of ActA protein-coated microspheres driven by actin polymerization. Proc Natl Acad Sci 96:4908–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Nurse P 2006. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol 8:1102–1107 [DOI] [PubMed] [Google Scholar]
- Chant J, Herskowitz I 1991. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65:1203–1212 [DOI] [PubMed] [Google Scholar]
- Co C, Wong DT, Gierke S, Chang V, Taunton J 2007. Mechanism of actin network attachment to moving membranes: Barbed end capture by N-WASP WH2 domains. Cell 128:901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM 2009. Kinetic and equilibrium analysis of the myosin ATPase. Meth Enzymol 455:157–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Rowe AJ 1965. Dynein: A protein with adenosine triphosphatase activity from cilia. Science 149:424–426 [DOI] [PubMed] [Google Scholar]
- Girard KD, Chaney C, Delannoy M, Kuo SC, Robinson DN 2004. Dynacortin contributes to cortical viscoelasticity and helps define the shape changes of cytokinesis. EMBO J 23:1536–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol 120:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Grin B, Mendez MG, Kuczmarski ER 2008. Intermediate filaments: Versatile building blocks of cell structure. Curr Opin Cell Biol 20:28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin E, Welch MD, Cossart P 2005. Actin-based motility of intracellular pathogens. Curr Opin Microbiol 8:35–45 [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382:420–425 [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A 1997. Spindle assembly in Xenopus egg extracts: Respective roles of centrosomes and microtubule self-organization. J Cell Biol 138:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JH, Svitkina TM, Burns AR, Hughes HE, MacPartland KJ, Nazarian R, Borisy GG 1999. Two components of actin-based retrograde flow in sea urchin coelomocytes. Mol Biol Cell 10:4075–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy TE, Leibler S 1994. Dynamic instability of microtubules as an efficient way to search in space. Proc Natl Acad Sci 91:5682–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy TE, Dogterom M, Yurke B, Leibler S 1997. Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci A 94:6228–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD 2007. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol 17:395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE 2006. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science 313:1298–1301 [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 7:404–414 [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL 2000. Centrosome-independent mitotic spindle formation in vertebrates. Current Biology 10:59–67 [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ 2008. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol 18:1719–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Söldner R, Hirsch K, Keller M, Förster R, Critchley DR, Fässler R, et al. 2008. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453:51–55 [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Malmberg RL, Muszynski MG, Dawe RK 2002. Maximum likelihood methods reveal conservation of function among closely related kinesin families. J Mol Evol 54:42–53 [DOI] [PubMed] [Google Scholar]
- Li R, Gundersen GG 2008. Beyond polymer polarity: How the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol 9:860–873 [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR 1995. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature 373:161–164 [DOI] [PubMed] [Google Scholar]
- Malawista SE, Van Blaricom G, Cretella SB 1985. Cytokineplasts from human blood polymorphonuclear leukocytes. Lack of oxidase activity and extended functional longevity. Inflammation 9:99–106 [DOI] [PubMed] [Google Scholar]
- Marshall WF 2009. Centriole evolution. Current Opinion Cell Biol 21:14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nature Cell Biology 8:215–226 [DOI] [PubMed] [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE 1999. Myosin-V is a processive actin-based motor. Nature 400:590–593 [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wah L, Almers W 2002. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol 4:691–698 [DOI] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N 2005. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol 15:467–476 [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M 1984. Dynamic instability of microtubule growth. Nature 312:237–242 [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD 1998. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci 95:6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Bowerman B 2009. Cellular symmetry breaking during C. elegans development. Cold Spring Harb Perspect Biol 1:a003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nédélec FJ, Surrey T, Maggs AC, Leibler S 1997. Self-organization of microtubules and motors. Nature 389:305–308 [DOI] [PubMed] [Google Scholar]
- Orlando K, Guo W 2009. Membrane organization and dynamics in cell polarity. Cold Spring Harb Perspect Biol 1:a001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Devreotes PH 1999. A cell's sense of direction. Science 284:765–770 [DOI] [PubMed] [Google Scholar]
- Pollard TD 1982. Structure and polymerization of Acanthamoeba myosin-II filaments. J Cell Biol 95:816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD 1986. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol 103:2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453–465 [DOI] [PubMed] [Google Scholar]
- Riedel-Kruse IH, Hilfinger A, Howard J, Jülicher F 2007. How molecular motors shape the flagellar beat. HFSP J 1:192–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisler E, Smith C, Seegan G 1980. Myosin minifilaments. J Mol Biol 143:129–145 [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Hall A 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389–399 [DOI] [PubMed] [Google Scholar]
- Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL 2001. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci 98:13655–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Borisy GG 1997. Self-centring activity of cytoplasm. Nature 386:170–173 [DOI] [PubMed] [Google Scholar]
- Roth S, Lynch JA 2009. Symmetry breaking during Drosophila oogenesis. Cold Spring Harb Perspect Biol 1:a001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Schwarz WH, Pollard TD 1987. Dependence of the mechanical properties of actin/α-actinin gels on deformation rate. Nature 325:828–830 [DOI] [PubMed] [Google Scholar]
- Schaub S, Bohnet S, Laurent VM, Meister JJ, Verkhovsky AB 2007. Comparative maps of motion and assembly of filamentous actin and myosin II in migrating cells. Molecular Biology of the Cell 18:3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaevitz JW, Lee JY, Fletcher DA 2005. Spiroplasma swim by a processive change in body helicity. Cell 122:827–828 [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Smith SE, Li R 2009. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb Perspect Biol 1:a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surrey T, Nedelec F, Leibler S, Karsenti E 2001. Physical properties determining self-organization of motors and microtubules. Science 292:1167–1171 [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145:1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG 2003. Mechanism of filopodia initiation by reorganization of a dendritic network. Journal of Cell Biology 160:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG 1997. Analysis of the actin-myosin II system in fish epidermal keratocytes: Mechanism of cell body translocation. J Cell Biol 139:397–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Rowning BA, Coughlin ML, Wu M, Moon RT, Mitchison TJ, Larabell CA 2000. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J Cell Biol 148:519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ 1991. Actin microfilament dynamics in locomoting cells. Nature 352:126–131 [DOI] [PubMed] [Google Scholar]
- Tranquillo RT, Lauffenburger DA, Zigmond SH 1988. A stochastic model for leukocyte random motility and chemotaxis based on receptor binding fluctuations. J Cell Biol 106:303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisberg EA, Grissom PM, McIntosh JR 1996. Mammalian cells express three distinct dynein heavy chains that are localized to different cytoplasmic organelles. J Cell Biol 133:831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Milligan RA 2000. The way things move: Looking under the hood of molecular motor proteins. Science 288:88–95 [DOI] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42:39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Gucht J, Sykes C 2009. Physical model of cellular symmetry breaking. Cold Spring Harb Perspect Biol 1:a001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Gucht J, Paluch E, Plastino J, Sykes C 2005. Stress release drives symmetry breaking for actin-based movement. Proc Natl Acad Sci 102:7847–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oudenaarden A, Theriot JA 1999. Cooperative symmetry-breaking by actin polymerization in a model for cell motility. Nat Cell Biol 1:493–499 [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG 1999. Self-polarization and directional motility of cytoplasm. Curr Biol 9:11–20 [DOI] [PubMed] [Google Scholar]
- Wachsstock DH, Schwartz WH, Pollard TD 1993. Affinity of α-actinin for actin determines the structure and mechanical properties of actin filament gels. Biophys J 65:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F 2009. The signaling mechanisms underlying cell polarity and chemotaxis. Cold Spring Harb Perspect Biol 1:a002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I, Gerisch G, Heizer C, Murphy J, Badelt K, Stock A, Schwartz JM, Faix J 1999. Cytokinesis mediated through the recruitment of cortexillins into the cleavage furrow. EMBO J 18:586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Mullins RD 2002. Cellular control of actin nucleation. Annu Rev Cell Dev Biol 18:247–288 [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Altschuler S, Wu L, Li R 2003. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299:1231–1235 [DOI] [PubMed] [Google Scholar]
- Weiner OD 2002. Regulation of cell polarity during eukaryotic chemotaxis: The chemotactic compass. Curr Opin Cell Biol 14:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Servant G, Welch MD, Mitchison TJ, Sedat JW, Bourne HR 1999. Spatial control of actin polymerization during neutrophil chemotaxis. Nat Cell Biol 1:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL 1999. Myosin VI is an actin-based motor that moves backwards. Nature 401:505–508 [DOI] [PubMed] [Google Scholar]
- Wittmann T, Desai A 2005. Microtubule cytoskeleton: A new twist at the end. Curr Biol 15:R126–129 [DOI] [PubMed] [Google Scholar]
- Woodrum DT, Rich SA, Pollard TD 1975. Evidence for biased bidirectional polymerization of actin filaments using heavy meromyosin prepared by an improved method. J Cell Biol 67:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A 2005. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr Biol 15:828–832 [DOI] [PubMed] [Google Scholar]
- Xu J, Van Keymeulen A, Wakida NM, Carlton P, Berns MW, Bourne HR 2007. Polarity reveals intrinsic cell chirality. Proc Natl Acad Sci 104:9296–9300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang F, Van Keymeulen A, Herzmark P, Straight A, Kelly K, Takuwa Y, Sugimoto N, Mitchison T, Bourne HR 2003. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell 114:201–214 [DOI] [PubMed] [Google Scholar]
- Yam PT, Wilson CA, Ji L, Hebert B, Barnhart EL, Dye NA, Wiseman PW, Danuser G, Theriot JA 2007. Actin-myosin network reorganization breaks symmetry at the cell rear to spontaneously initiate polarized cell motility. J Cell Biol 178:1207–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y 2004. G protein control of microtubule assembly. Annu Rev Cell Dev Biol 20:867–894 [DOI] [PubMed] [Google Scholar]