Abstract
Somatic mutations in the TP53 gene are one of the most frequent alterations in human cancers, and germline mutations are the underlying cause of Li-Fraumeni syndrome, which predisposes to a wide spectrum of early-onset cancers. Most mutations are single-base substitutions distributed throughout the coding sequence. Their diverse types and positions may inform on the nature of mutagenic mechanisms involved in cancer etiology. TP53 mutations are also potential prognostic and predictive markers, as well as targets for pharmacological intervention. All mutations found in human cancers are compiled in the IARC TP53 Database (http://www-p53.iarc.fr/). A human TP53 knockin mouse model (Hupki mouse) provides an experimental model to study mutagenesis in the context of a human TP53 sequence. Here, we summarize current knowledge on TP53 gene variations observed in human cancers and populations, and current clinical applications derived from this knowledge.
Various mutations in the p53 tumor suppressor can occur in cancer. These have different “fingerprints” and may serve as biomarkers for distinct tumors and targeted therapies.
Genetic variations in the tumor suppressor gene TP53 (OMIM #191117) contribute to human cancers in different ways. First, somatic mutations are frequent in most cancers (Hollstein et al. 1991). The antiproliferative role of p53 protein in response to various stresses and during physiological processes such as senescence makes it a primary target for inactivation in cancer (Levine 1997). The main modes of TP53 inactivation are single-base substitution and loss of alleles, with inactivation by viral or cellular proteins playing a major role in specific cancers (Tommasino et al. 2003). Second, inheritance of a TP53 mutation causes predisposition to early-onset cancers including breast carcinomas, sarcomas, brain tumors, and adrenal cortical carcinomas, defining the Li-Fraumeni (LFS) and Li-Fraumeni-like (LFL) syndromes (Li et al. 1988; Olivier et al. 2003). Third, TP53 is highly polymorphic in coding and noncoding regions and some of these polymorphisms have been shown to increase cancer susceptibility and to modify cancer phenotypes in TP53 mutation carriers (Whibley et al. 2009).
Whereas tumor suppressors are commonly inactivated by frameshift or nonsense mutations, most TP53 mutations are missense and cause single amino-acid changes at many different positions. Mutations are thus diverse in their type, sequence context, position, and structural impact, making it possible to identify mutation patterns in relation with cancer type and etiology. The occurrence of special mutation patterns may inform on the nature of the mutagens that have caused them, making TP53 an interesting gene to analyze in the realm of molecular epidemiology.
Data on mutation prevalence in human cancer can be conveniently accessed through the IARC TP53 database (http://www-p53.iarc.fr/), a resource that compiles all TP53 gene variations reported in human cancers with annotations on tumor phenotype, patient characteristics, and structural and functional impact of mutations (Petitjean et al. 2007b). Recently, it has become possible to confront these observations with experimental data generated in a novel mouse model, the HupKi mouse, that contains a human TP53 sequence at the mouse TP53 locus and recapitulates the effects of environmental mutagens in a human sequence context (Luo et al. 2001). In this article, we review the current knowledge on the origin, causes, and consequences of TP53 variations and mutations in cancer and we discuss their significance as biomarkers in epidemiology and in the clinics.
TP53 VARIATION LANDSCAPES IN HUMAN CANCERS AND POPULATIONS
Somatic Mutations
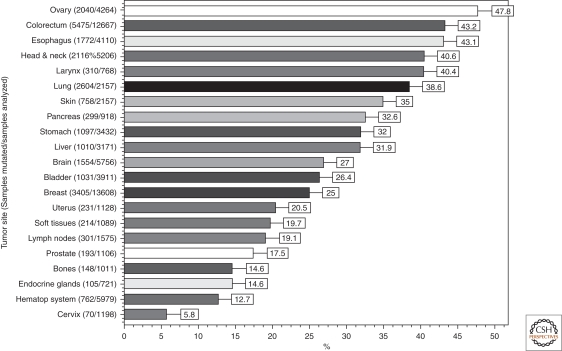
Somatic TP53 mutations occur in almost every type of cancer at rates from 38%–50% in ovarian, esophageal, colorectal, head and neck, larynx, and lung cancers to about 5% in primary leukemia, sarcoma, testicular cancer, malignant melanoma, and cervical cancer (Fig. 1). Mutations are more frequent in advanced stage or in cancer subtypes with aggressive behavior (such as triple negative or HER2-amplified breast cancers) (Wang et al. 2004a; Wang et al. 2004b; Langerod et al. 2007). In cancers with low mutation rates, p53 is often inactivated by alternative mechanisms. This is the case for cervical cancer in which p53 is targeted for degradation by HPV E6 (Tommasino et al. 2003) or for sarcoma that overexpress amplified HDM2.
Figure 1.
TP53 mutations prevalence in sporadic cancers. The proportion of tumors with somatic TP53 mutations is indicated. Data from IARC TP53 Database (R13, November 2008)(Petitjean et al. 2007b).
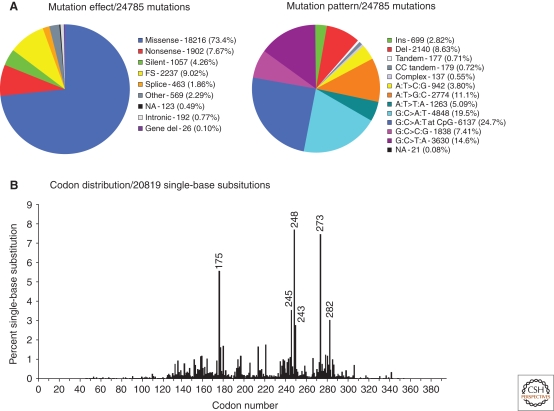
Figure 2.
Type of somatic TP53 mutations in human cancers. (A) Pie charts showing the proportion of the different types of TP53 somatic mutations found in all human cancers. (B) Histogram displaying the position of somatic point mutations in the coding sequence of the TP53 gene. Data from the IARC TP53 Database (R13, November 2008)(Petitjean et al. 2007b).
Based on studies that examined the whole coding sequence, 86% of mutations cluster between codons 125 and 300, corresponding mainly to the DNA binding domain (Fig. 2). Most mutations in this region are missense (87.9%). In contrast, outside this region, missense mutations represent only about 40%, the majority of mutations being nonsense or frameshift. Among single-base substitutions, about 25% are C:G>T:A substitutions at CpG sites. CpG dinucleotides mutate at a rate 10 times higher than other nucleotides, generating transitions (Jones et al. 1992). About 3%–5% of cytosines in the human genome are methylated at position 5′ by a postreplicative mechanism that is restricted to CpG dinucleotides and is catalyzed by DNA methyltransferases. The 5′methylcytosine (5mC) is less stable than cytosine and undergoes spontaneous deamination into thymine at a rate five times higher than the unmethylated base. This process is enhanced by oxygen and nitrogen radicals, leading to a higher load of CpG transitions in cancers arising from inflammatory precursors such as Barrett's mucosa or ulcerative colitis (Schmutte et al. 1996; Ambs et al. 1999; Vaninetti et al. 2008). Among the 22 CpG of the DNA-binding domain (DBD), three hotspot codons (175, 248, and 273) represent 60% of CpG mutations and another five residues (196, 213, 245, 282, and 306) account for 26% of these mutations. The lack of mutations at other CpG sites reflects the fact that substitution at these residues does not generate a dysfunctional protein. Although the same CpG hotspot mutations occur in many cancer types, other types of mutations tend to show differences from one cancer to the other. Some of these differences have been linked to the effect of specific mutagens. Geographic differences have also been reported in relation with environmental exposures. These aspects have been extensively discussed in other reviews (Hainaut et al. 2000; Olivier et al. 2004) and some examples are briefly discussed later in this article.
Germline Mutations: Li-Fraumeni Syndrome
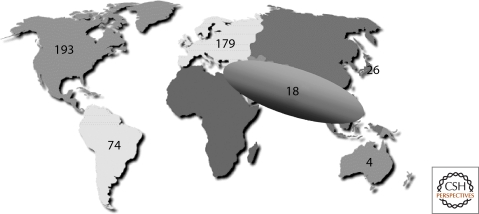
T53 germline mutations are the underlying cause of LFS, a familial clustering of early onset tumors including sarcomas, breast cancers, brain tumors, and adrenal cortical carcinomas (Li et al. 1988; Malkin et al. 1990). Over the past 20 years, TP53 germline mutations have been detected in about 500 families or individuals with complete or partial LFS features (the latter defined as Li-Fraumeni-like, LFL) (Olivier et al. 2003). LFS/LFL has been generally considered as a rare syndrome (Eeles 1995). However, screening for TP53 germline mutation in patients with early onset breast cancer and unselected for familial history has shown TP53 mutations in 2%–3% of the cases (Lalloo et al. 2006), whereas screening of 525 patients with any kind of cancer family history has identified 91 (17.3%) TP53 mutations (Gonzalez et al. 2009). Based on these results, TP53 mutation may contribute to up to 17% of all familial cancer cases. Studies in southern Brazil have identified many families with a founder mutation (R337H) (Fig. 3). Thus, TP53 germline mutations may be more common than previously recognized, occurring in about 1 in 5,000 to 1 in 20,000 births (Lalloo et al. 2006) (Gonzalez et al. 2009).
Figure 3.
Geographic distribution of germline TP53 mutations. Number of TP53 germline mutation carrier families in each world region. Data from the IARC TP53 Database (R13, November 2008) (Petitjean et al. 2007b).
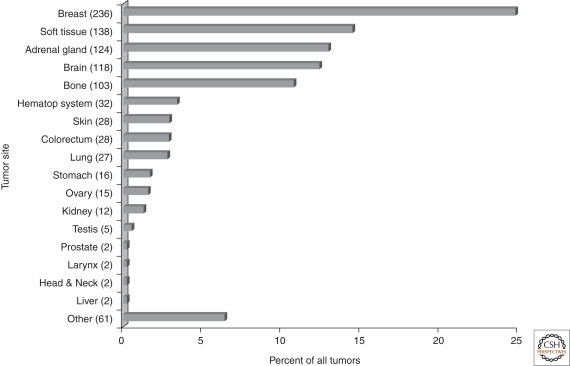
Breast cancer and soft tissue and bone sarcoma account for over 50% of tumors in TP53 mutation carriers, followed by adrenocortical carcinomas and brain tumors (Fig. 4). Other cancers include hematological malignancies, gastric, colorectal, and ovarian cancers, occurring at earlier ages than in the general population (Olivier et al. 2003). Rarer cancers associated with TP53 germline mutation are choroid plexus carcinoma or papilloma before the age of 15, Wilms' tumor, and malignant phyllodes tumors (Birch et al. 2001; Gonzalez et al. 2009).
Figure 4.
Tumor spectrum in individuals with a germline TP53 mutation. The proportion of specific tumor types among all tumors reported in confirmed TP53 germline mutation carriers is indicated. Data from IARC TP53 Germline Database (R13, November 2008, http://www-p53.iarc.fr/Germline.html).
The distribution of germline mutations is similar to somatic mutations, with mostly missense mutations (77%) located at the same hotspots. The proportion of CpG mutations (54% vs. 25% in somatic mutations) may reflect the spontaneous nature of germline mutations. Genotype–phenotype correlations suggest that the most significant defect is loss of function because large deletions encompassing the whole TP53 gene have been found in LFS families with aggressive features (Bougeard et al. 2003).
TP53 Polymorphisms
Over 80 TP53 polymorphisms have been identified and validated in human populations (IARC TP53 Database, R13). The majority (90%) are located in introns, outside splice sites, or in noncoding exons. Few of them have been tested in functional assays or studied for effects on cancer risk. Among 18 exonic SNPs (Table 1), five are silent and seven are located after the stop codon in exon 11. Four exonic polymorphisms alter the protein sequence and have only subtle effects on transactivation capacity as measured in yeast-based assays (Kato et al. 2003).
Table 1.
Selected polymorphisms in TP53 gene
| ExonIntron | g_Description | c_Description | AvgHet | SNPlink | Effect |
|---|---|---|---|---|---|
| 4-exon | g.11333C>A | c.102C>A | - | 11575998 | silent |
| 4-exon | g.11339G>A | c.108G>A | 0.012738 | 1800370 | silent |
| 4-exon | g.11370C>T | c.139C>T (P47S) | 0.029329 | 1800371 | missense |
| 4-exon | g.11446C>G | c.215C>G (R72P) | 0.492248 | 1042522 | missense |
| 6-exon | g.12708A>G | c.639A>G | 0.023526 | 1800372 | silent |
| 6-exon | g.12718G>A | c.649G>A (V217M) | 0.0098 | 35163653 | missense |
| 10-exon | g.16970G>C | c.1079G>C (G360A) | 0.009892 | 35993958 | missense |
| 11-exon | g.18096C>T | c.1182+105C>T | 0.019761 | 35919705 | silent |
| 11-exon | g.18196G>A | c.1182+205G>A | 0.024239 | 16956880 | silent |
| 11-exon | g.18305G>A | c.1182+314G>A | 0.019569 | 34486624 | noncoding |
| 11-exon | g.18319G>A | c.1182+328G>A | 0.010246 | 17881366 | noncoding |
| 11-exon | g.18476G>A | c.1182+485G>A | 0.091723 | 4968187 | noncoding |
| 11-exon | g.18560_18561del2 | c.1182+569_1182+570del2 | 0.436246 | 17886358 | noncoding |
| 11-exon | g.18604C>A | c.1182+613C>A | 0.023872 | 17879353 | noncoding |
| 11-exon | g.18817G>A | c.1182+826G>A | 0.136576 | 17884306 | noncoding |
| 11-exon | g.18877G>A | c.1182+886G>A | 0.009892 | 35659787 | noncoding |
| 11-exon | g.19166A>C | c.1182+1175A>C | 0.00555 | - | intronic |
| 3-intron | g.11259_11274del16 | c.96+41_96+56del16 | 0.5 | 17878362 | intronic |
| 2-intron | g.11117C>G | c.74+38C>G | 0.468059 | 1642785 | Intronic |
| 9-intron | g.16143G>A | c.994-742G>A | 0.401235 | 1641549 | Intronic |
| 9-intron | g.14496C>T | c.993+431C>T | 0.3432 | 1642791 | Intronic |
| 7-intron | g.13511T>G | c.782+92T>G | 0.310766 | 12951053 | Intronic |
| 9-intron | g.15185T>C | c.993+1120T>C | 0.256705 | 12949655 | Intronic |
| 7-intron | g.13491C>T | c.782+72C>T | 0.246362 | 12947788 | Intronic |
| 4-intron | g.12273G>A | c.376-91G>A | 0.212093 | 2909430 | Intronic |
| 6-intron | g.12803A>G | c.672+62A>G | 0.210458 | 1625895 | Intronic |
| 4-intron | g.12239T>C | c.376-125T>C | 0.157451 | 9895829 | Intronic |
| 3-intron | g.11299C>A | c.97-29C>A | 0.15648 | 17883323 | intronic |
Complete list available at http://www-p53.iarc.fr/PolymorphismView.asp.
Missense polymorphisms are highlighted (amino-acid substitution and codon into brackets).
V217M is the only nonsilent polymorphism in the DBD. In yeast functional assays, the rare variant shows increased transactivation of some p53 response elements (CDKN1A, BAX, and PMAIP1) and may thus be protective against cancer. G360A, located next to the tetramerization domain (TET) showed slightly reduced activity that may result in an increased cancer risk. However, the impact of these SNP has not been investigated in clinical or epidemiological studies.
P47S has been reported in African populations only, at a frequency of approximately 5% (Felley-Bosco et al. 1993). This polymorphism may affect phosphorylation at S46, which enhances p53-mediated apoptosis (Pistritto et al. 2007; Oda et al. 2000). hosphorylation of S46 by p38 and homeodomain-interacting protein kinase 2 (HIPK2) requires a proline adjacent to S46. Presence of a serine may thus alter kinase recognition. However, functional data in different experimental systems have proved inconsistent. In mice, blocking S46 phosphorylation has only modest phenotypical consequences (Feng et al. 2006; Toledo et al. 2006). It is possible that this SNP may impact on p53 function only under specific stress conditions. Its impact on cancer susceptibility remains to be elucidated.
The P72R polymorphism is the most extensively studied both in experimental and population studies. Sharp ethnic differences have been observed, the P72 allele showing a north–south gradient from 0.17 in Swedish Saamis to 0.63 in Africans (Yorubas). A recent report has shown an association of this SNP with latitude within Asian populations (Shi et al. 2009) and has proposed that allele frequency may be an adaptation to differences in winter temperature. R72 is more active than P72 in transactivating the expression of leukaemia-inhibitory factor (Kang et al. 2009), which is critical for blastocyst implantation, leading to the conjecture that R72 could thus provide a selective advantage by reducing the risk of implantation failure in colder climates.
Codon 72 is located within a proline-rich region and may affect a putative SH3-binding domain. The current consensus from a large number of studies is that R72 is more effective in inducing apoptosis than P72. However, the relevance of these observations for cancer susceptibility is not understood. An early suggestion that individuals homozygous for R72 may be at greatly higher risk for HPV-related cervical cancers (Storey et al. 1998) has not been substantiated by extensive population-based studies (Klug et al. 2001). Many studies have reported associations between the R72P SNP and risk of different cancers, with inconsistent results. Meta-analyses for breast (Schmidt et al. 2007), lung (Matakidou et al. 2003), and other cancers do not support a significant role for this polymorphism in susceptibility.
The P72R SNP has been studied for its impact on prognosis or response to treatment. Many of these studies must be interpreted cautiously as they lack power and rigorous design. There is, however, evidence of a modifier effect on TP53 mutations, with a bias toward expression of mutant R72 allele in tumors of heterozygous patients with LOH affecting wild-type P72 (Langerod et al. 2002; Bergamaschi et al. 2003; Bonafe et al. 2003; Vikhanskaya et al. 2005; Nelson et al. 2005; Zawlik et al. 2009). Furthermore, experimental studies showed that R72 mutants were more potent in cooperating with EJ-Ras in transforming primary cells and in neutralizing p73 activity (Marin et al. 2000; Bergamaschi et al. 2003). A modifier effect of the R72P SNP has also been reported in germline TP53 mutation carriers, in whom R72 was associated with earlier age at first diagnosis of cancer (Bougeard et al. 2006).
P72R is in linkage disequilibrium with a polymorphism in intron 3 (PIN3), consisting of a duplication of a 16 bp, GC rich sequence (allele frequency: 0.2 in Caucasians). PIN3 has been associated with increased risk of several cancers (Wang-Gohrke et al. 1998; Wang-Gohrke et al. 1999; Gemignani et al. 2004). The duplicated PIN3 allele is associated with reduced expression of TP53 mRNA in lymphoblastoid cell lines (Gemignani et al. 2004). Recently, TP53 germline mutation carriers with duplicated PIN3 were shown to develop cancer, on average, 20 years later than carriers with nonduplicated alleles (Marcel et al. 2009).
Given the complex polymorphic structure of TP53, haplotypes may provide more relevant information than individual polymorphisms. In addition, polymorphisms in gene regulating p53 have a critical impact, as for example MDM2 SNP309 polymorphism (See Grochola et al. 2010).
STRUCTURAL AND FUNCTIONAL IMPACT OF MUTATIONS
Impact on Protein Structure
The p53 DBD is made of an immunoglobulin-like β-sandwich of two antiparallel β-sheets, providing a scaffold for a flexible DNA-binding surface (Cho et al. 1994). This surface is formed by two large loops (L2 and L3) stabilized by a zinc atom and a loop–sheet–helix motif (loop L1). Zinc binding (coordinated by H179, C176, C238, and C242) is critical for correct folding, and requires reduction of thiol groups on cysteines. Hotspot mutations are at residues involved either in making contacts with DNA or in supporting the structure of the DNA-binding surface. Mutant proteins have thus been classified as “contact” (e.g., R248 and R273) or “structural” (e.g., R175, G245, R249, and R282) (Joerger et al. 2007). The structure of several common mutants has been elucidated by NMR spectroscopy or X-ray crystallography (Joerger et al. 2005; Joerger et al. 2006). DNA-contact mutants retain the overall architecture of the DBD with loss of a critical DNA contact. They may actively prevent DNA binding if a large hydrophobic side chain is introduced (e.g., S241F, R248W, and C277F). Zinc-binding mutants affect the zinc coordination sphere (e.g., C176F, H179R, and C242F). This category includes R175H, the most frequent hotspot mutant, because introduction of histidine residue causes distortions that directly interfere with zinc binding. Substitutions introducing smaller residues at this position have been shown to be less destabilizing with partial retention of function (Bullock et al. 2000). Structural mutants cause distortions that create internal cavities or surface crevices in the protein scaffold, inducing conformational changes in the DNA binding surface. Overall, these studies highlight the structural heterogeneity of mutant proteins, with possible consequences on their biochemical and biological properties.
Impact on Transcriptional Activities
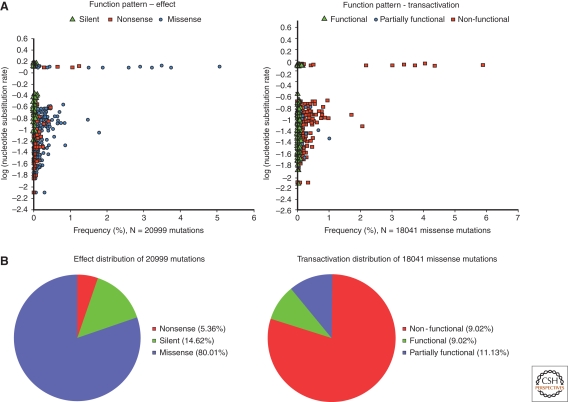
p53 regulates transcription through specific binding to response elements in the promoters or introns of target genes (Riley et al. 2008). p53REs are variations of a consensus constituted by two decamers (PuPuPuC(A/T)(T/A) GPyPyPy) separated by a spacer of variable length (El-Deiry et al. 1992). These sequence variations affect p53 binding affinity and may contribute to shape the repertoire of genes activated by p53 in response to specific signals (Inga et al. 2002). Recent genome-wide analyses of p53 binding sites suggest that hundreds of genes may be up- or down-regulated by p53 (Smeenk et al. 2008). Kato et al. (2003) have used a yeast-based functional assay to analyze the transactivation of 2314 different missense mutants on eight p53REs. All hotspot mutants displayed loss of function on all tested p53REs (Fig. 5). However, several rare mutants retained partial activity on all REs or even full activity on some REs. A small category of mutants even showed increased activity compared with wild-type p53 (so-called “supertrans mutants,” often occurring in loop L1, which is not a frequent target for cancer-related mutations). Using a different type of yeast based assay, Inga et al. (2002) further showed that differential transactivation by wild-type and mutant proteins depends on both protein levels and target sequence.
Figure 5.
Functional impact of somatic TP53 mutations in human cancers. (A) Scatter plots showing the frequency (x-axis) of single amino-acid substitutions in relation to their effect (left panel) or functional impacts on transactivation (right panel), and expected substitution rates (y-axis in log). Each point represents a single amino-acid substitution that is shaped and colored according to the mutation effect or functional impact. (B) Pie charts displaying the proportion of all somatic single amino-acid substitutions according to their effect (left panel) or functional impacts on transactivation (right panel). Data from the IARC TP53 Database (R13, November 2008)(Petitjean et al. 2007b).
Dominant–Negative and Gain-of-Function Effects
Mutant p53 proteins often accumulate in the nucleus of in situ and metastatic cancer cells, suggesting an oncogenic effect in addition to loss of wild-type suppressor function. Experimentally, several hotspot mutants have been shown to cooperate with oncogenes for cellular transformation (Hinds et al. 1990), setting the concept of gain-of-function (GOF). Alternatively, mutant p53 proteins may exert dominant–negative effects (DNE) over wild-type p53. Indeed, transcriptional activity relies on the formation of p53 tetramers (dimers of dimers) and incorporation of mutant p53 in these structures may preclude their activity.
DNE has been experimentally studied in yeast and human cell assays, generating data on the DNE effects for over 200 mutants (Brachmann et al. 1996). Analysis of the IARC TP53 database showed that mutants with DNE toward p53-RE from WAF-1 and RGC promoters were over-represented in cancer (Petitjean et al. 2007b), suggesting that DNE plays a role in selecting for mutations during carcinogenesis.
Only a few mutants have been studied for GOF activities and there is no consensus on the molecular definition of such activities (Petitjean et al. 2007b). Knockin mice with R175H and R273H mutants (Lang et al. 2004; Olive et al. 2004) showed increased metastases and different cancer distribution compared with p53 knockout mice, providing in vivo evidence of GOF. Various molecular mechanisms have been proposed as the basis of GOF, such as interference with the p53-family proteins p63 and p73, interference with the ATM DNA repair pathway (Song et al. 2007), activation of genes normally unaffected or repressed by wild-type p53, interference with other transcription factors, and resistance to drugs (see Oren and Rotter 2010). However, no systematic study has been performed to distinguish mutants with GOF properties. Thus, there is so far no definite evidence that GOF contribute to mutant p53 selection in human cancers.
It should be noted that, when compared with expected numbers, if all mutations were equal, missense mutations in p53 DBD are only slightly overrepresented in cancer (87.1% observed vs. 72.9% expected), whereas nonsense mutations are overrepresented by a factor of two (8% vs. 3.6%) and silent mutations underrepresented by a factor of four (4.8% vs. 22.9%) (Table 2). These observations further support that loss of function is the critical factor for the selection of mutations in cancer. This is particularly striking when considering mutations at hypermutable CpG sites. Of 34 possible missense mutations at CpG sites in the DBD, only seven are frequently observed in cancers. These seven mutants show complete loss of transactivation activities, whereas the 27 rare mutants retain significant transactivational activity on one or more p53RE.
Table 2.
Observed and expected frequency of single-nucleotide substitutions within exons 5–8 of TP53 gene
| Mutation type | Expected mutations | Observed mutations* |
|---|---|---|
| Missense | 1150 (73.4%) | 17,191 (87.9%) |
| Nonsense | 58 (3.7%) | 1435 (7.3%) |
| Silent | 359 (22.9%) | 932 (4.8%) |
| Total | 1567 | 19,558 |
*Somatic mutations reported in the IARC TP53 Database (R13, November 2008).
MUTAGEN-INDUCED MUTATION SPECTRA
Human Data
In several types of cancers, mutation patterns bear the hallmarks of chemical damage induced by particular mutagens, leading to the concept of “mutagen fingerprints” (Table 3). These fingerprints are defined by the relative frequencies, types of base change, strand orientation, and location of base substitutions. The factors shaping mutation patterns have been studied in detail in various organisms and experimental models. They include (1) the nature of the environmental agent that chemically modifies a base, (2) the DNA sequence itself (base context), (3) epigenetic modifications at the target sequence (e.g., base methylation), (4) DNA repair and replication processes, (5) transcription activity at the locus, and (6) biological selection (as discussed previously).
Table 3.
Human carcinogen TP53 signature mutations
| TP53 Signature | |||||
|---|---|---|---|---|---|
| Base substitution | Distinctive | ||||
| Carcinogen/(exposure) | Target organ | Metabolic activation | IARC DB | HUF assay | Hotspots (codons) |
| PAH (B[a]P) (smoking) | Lung | Epoxidation | G to T with strand bias | G to T with strand bias | 157, 158, 273 |
| AA (dietary contam.) | Urothelium | Nitroreduction | A to T with strand bias | A to T with strand bias | 131, 209, (280) |
| Aflatoxin B1 (dietary contam.) | Liver | Epoxidation | G to T | n.i. | 249 (3rd base) |
| UV radiation (sunlight) | Skin | n.a. | CC to TT | CC to TT | 248, 278 |
| 3-NBA (diesel) | Lung | Nitroreduction | n.i. | G to T | n.i. |
| Vinyl chloride | Liver | Epoxidation | A:T to T:A with strand bias | n.i. | |
Although in many cancers transitions at CpG sites predominate, a complex picture emerges when concentrating on rarer types of mutations. The patterns of these mutations can vary dramatically between two types of cancers, or two patient populations with a given cancer type. In some instances, the mutations reveal characteristics expected from the effects of environmental mutagens on DNA. There are four well-documented examples of associations between an etiologic agent and a tumor mutation fingerprint. Three of these have been reviewed in detail: exposure to sunlight and the presence of tandem CC to TT transitions in nonmelanoma skin cancers (Giglia-Mari et al. 2003), tobacco smoking and PAH-induced G to T transversions at specific G:C base pairs oriented with the (premutated) guanine on the nontranscribed strand (codons 157, 158, 245, 248, and 273)(Pfeifer et al. 2002; Pfeifer et al. 2003), and exposure to dietary aflatoxinB1 and G to T transversions at the third base of codon 249 (AGG to AGT) in tumors from HBV carriers (Hussain et al. 2007). The fourth and most recent example links TP53 A to T transversions with crops contaminated with seeds of Aristolochia sp, and is worth mentioning even though the estimated number of cancer patients exposed worldwide to aristolochic acids (AA), the carcinogenic agent, is expected to be small in comparison to the three exposures just cited (Arlt et al. 2007). Premutagenic AA-specific DNA adducts have been detected in individuals suffering from Balkan endemic nephropathy (BEN), a form of nephropathy associated with high risk of urothelial cancer that is historically common in some parts of southeastern Europe. TP53 mutations in urothelial tumors of these patients are predominantly (74%) A to T transversions (Grollman et al. 2007), a form of mutation that is rare in urothelial tumors from non-BEN patients (4%) as well as in human tumors overall (5%). Moreover, BEN tumor mutations show a strong strand bias, with the premutated adenine base residing on the nontranscribed strand in 93% of the instances reported so far. This bias suggests preferential (transcription-coupled) repair of bulky lesions on the DNA strand that serves as template for mRNA synthesis. Reminiscent of the first discoveries of AFB1-associated codon 249 G to T transversions in two small sets of liver tumors (Hsu et al. 1991; Bressac et al. 1991), the AA-associated mutations suggest a mutation fingerprint even though only 19 mutations in this cohort of BEN patients have been reported thus far.
Recent large-scale sequencing studies are having a profound impact on our understanding of mutation load in human cancers (Ding et al. 2008; Stratton et al. 2009). In keeping with the smokers' lung tumor TP53 mutation data, a screen of 623 genes in 188 lung adenocarcinomas confirmed the higher mutation load and higher fraction of strand-biased G to T transversions in lung tumor of smokers when compared with nonsmokers (Ding et al. 2008). Genome-wide studies highlighted an unexpected and statistically significant elevation in G:C to C:G transversions in breast cancers (28% of all substitutions) compared with colorectal, pancreatic, and brain cancers (7–10%) (Stephens et al. 2005; Sjoblom et al. 2006; Greenman et al. 2007; Jones et al. 2008). Oddly, this is not seen in breast cancer TP53 mutations (9% of which are G:C to C:G transversions) (see Pfeifer and Besaratinia 2009 for discussion of G:C to C:G mutations). Next generation DNA sequencing technologies will offer new opportunities for studying factors that govern mutation load in normal and pre-neoplastic cell populations, including stem cells, which may be especially pertinent to elucidating origins of critical cancer-causing mutations.
Experimental Systems
Experimental mutagenesis studies in lower organisms and mammalian cells have provided evidence linking the four human TP53 tumor mutation signatures discussed above to the suspected carcinogens (Hainaut et al. 2000; vom Brocke J. et al. 2006). However, this approach is difficult to extrapolate to more complex situations involving multiple suspected carcinogens and where etiologic clues are needed. Classical experimental systems to generate mutation spectra are based on mutations in reporter genes that are not cancer related, but that allow efficient selection and recovery of mutants (e.g., the microbial lacI and LacZ genes, or the mammalian TK and HPRT genes). However, a reporter gene will differ from TP53 or any other genes of interest in base sequence context, transcriptional level (thus transcription-coupled repair), and biological selection of mutants, three key factors that shape human TP53 tumor mutation patterns. Until recently, the human TP53 gene was considered a “nonselectable” gene in an experimental setting, which would preclude its use as a reporter of experimentally induced mutations. However, innovative approaches have been developed to map experimental DNA damage along the human TP53 sequence as a first approximation of the sites vulnerable to accumulation of mutations. This strategy was used successfully to show the correlation between human tumor TP53 mutations and the positions of the main DNA photoproducts induced by UV light or those of adducts formed by metabolites of PAH (Denissenko et al. 1996). Another powerful approach was devised to score mutations in human TP53 sequences cloned into a vector by assessing loss of p53 transcriptional transactivation function when the plasmid is then introduced into specially designed yeast strains (Frebourg et al. 1992; Ishioka et al. 1993).
A direct method to induce and select TP53 mutations in mammalian cells would be to conduct a rodent cancer assay and sequence the murine TP53 gene in the carcinogen-induced tumors (Zielinski et al. 2002). Although this approach has been used successfully to examine UV-induced mutations in murine skin tumors, it has limited applicability considering the unexplained paucity of TP53 mutations in most mouse tumor types other than skin. Furthermore, murine and human TP53 genes, although highly homologous, differ in DNA sequence at critical hotspot codons, posttranslational modification sites, and intragenic suppressor sites (Hergenhahn et al. 2004). An in vivo carcinogenesis assay in mouse harboring human TP53 gene sequences would bypass this latter difficulty. However, its applicability may still be limited to studies of skin lesions induced by topical application of suspected agents. The HUF Assay (Hupki fibroblast assay) is a novel mutagenesis test in primary cells from “humanized” mice that was designed to circumvent the difficulties discussed above (Liu et al. 2004). HupKi (Human p53-Knockin) mice harbor human TP53 sequences encoding both the polyproline and the DNA binding domain, where most human tumor mutations arise (Luo et al. 2001; Reinbold et al. 2008), replacing the homologous murine gene segments within the mouse TP53 locus. Hupki p53 is functional and regulated normally in mice. The “domain-swap” strategy is necessary, rather than replacement of the entire murine p53 by its human counterpart, because in a complete swap the TP53 gene loses wild-type function in mice, due to aberrant human p53 N-terminal interaction with the murine major negative regulator Mdm2 (Dudgeon et al. 2006).
The HUF assay takes advantage of the fact that p53 inactivation is the critical rate-limiting event in escape of replicative senescence and spontaneous immortalization of mouse embryo fibroblasts (MEF) in culture (Hahn et al. 2002). Fibroblasts are explanted from Hupki embryos, exposed to carcinogens, and cultured until immortalization has occurred. TP53 gene is then sequenced in MEF cell lines in search of mutations induced by carcinogens in a human sequence context. Several human carcinogens have been tested in the HUF assay and shown to induce TP53 mutations that are consistent with signatures identified in human tumors (Pfeifer and Besaratinia 2009; vom Brocke et al. 2006; Reinbold et al. 2008). Of note, aristolochic acid-induced A to T mutations in HUF cells correspond remarkably well with the TP53 tumor mutations in cancer patients with BEN (Nedelko et al. 2009). The HupKi mouse concept is now being extended to a whole family of mice carrying humanized TP53 sequence variants (recapitulating human TP53 polymorphisms, hotspot TP53 mutations, or mutations abrogating critical post-translational modification sites). By crossing Hupki mice with other genetically modified mouse strains (e.g., mice with humanized metabolic or DNA repair enzymes, or knockout strains with deficiencies in specific defense systems), many outstanding questions on the mechanisms of mutagenesis pertinent to human tumorigenesis could be addressed.
EPIDEMIOLOGICAL, DIAGNOSTIC, AND THERAPEUTIC UTILITY OF TP53 MUTATIONS
Biomarkers in Molecular Epidemiology
As discussed above, specific TP53 mutations are observed in some types of cancer that are likely to have arisen from exposure to known carcinogens. The existence of important variations in mutation patterns between different groups of patients with the same cancer suggests that further mutation fingerprints related to environmental exposures are still to be discovered. This is particularly the case in squamous cell carcinomas of the head and neck and of the esophagus, where variations have been described in relation with geographic origin of the patients, environmental risk factors, and lifestyle. However, in the natural history of human cancers, there is often no evidence for high exposure to genotoxic substances and most mutations may arise through spontaneous replication errors, endogenous pro-mutagenic states (induced by reactive oxygen species), spontaneous deamination, or other mechanisms contributing to the spontaneous decay of DNA (Lindahl 1993). Furthermore, the association between characteristic mutation type and carcinogen needs support from experimental systems and other lines of evidence to gain plausibility. Moreover, any characteristic single-base “fingerprint” mutation induced by an exogenous risk factor may also arise spontaneously, i.e., in the absence of exposure (albeit perhaps rarely), calling for corroborative studies to substantiate links between exposures and cancer mutations. In the realm of molecular epidemiology, where large numbers, control groups, and robust statistics are mandatory, such studies are facilitated by technical advances allowing the identification of mutations in DNA extracted from surrogate biological samples such as plasma, urine, sputum, or exfoliated cells from bronchus, bladder, oral cavity, and esophagus (Kirk et al. 2000). The use of such surrogate materials is compatible with the development of studies that follow consistent epidemiological designs, e.g., case-control and prospective cohort studies.
Biomarkers in the Clinics
Mutations in TP53 are useful markers of tumor clonality to compare, in individual patients, separate clusters of tumor cells from the same lesion, or multiple lesions arising in the same tissue (Dix et al. 1995; Ponten et al. 1997; Franklin et al. 1997). Mutations are also useful for the follow-up of minimal residual disease, for comparison between primary and recurrent tumors and for tracing the origin of distant metastases (Franklin et al. 1997). However, the finding of different TP53 mutations in separate clusters of a tumor does not exclude a clonal origin with occurrence of TP53 mutations at a late stage. Detection of TP53 mutations may also help to identify early lesions at a high risk of evolution. For example, in esophageal and endometrial cancers, detection of a mutation in low-grade dysplasia should be considered as an indicator of high risk of malignant evolution (Montesano et al. 1998; Jia et al. 2008).
TP53 mutations, but not p53 positive immunohistochemistry (IHC), have been consistently associated with poor prognosis in cancers such as breast, colorectal, head and neck, and leukemia (Petitjean et al. 2007a). In breast cancers, there is now clear evidence that TP53 mutation is an independent marker of poor prognosis, in particular in hormone receptor-positive cases. A large number of studies that have used IHC as a surrogate marker for mutation status have failed to provide such consistent results. IHC leads to an unacceptable number of misclassified cases (false-positive and false-negative) and to a large interstudy variability. TP53 gene status has also been associated with response to specific treatment regimens in breast cancer (Bergh et al. 1995; Aas et al. 1996; Olivier et al. 2006). However, it should be noted that the prognositic and predictive significance of TP53 mutations is extremely variable according to tumor type and/or treatment (Bertheau et al. 2008), and there is therefore no simple, universal clinical message that can be delivered by TP53 mutation analysis.
Mutations may also serve as biomarkers for targeted therapy. Several strategies have been developed to reactivate normal p53 functions in p53 mutated tumors. They include molecules that target broad classes of mutants to reactivate suppressive functions in tumor cells (PRIMA, RITA, scFv) (Caron de Fromentel et al. 1999; Issaeva et al. 2004; Bykov et al. 2005), or compounds that specifically target particular missense mutants to restore wild-type-like structure (Phikan059 targeting R220C)(Boeckler et al. 2008). Interestingly, several of these small pharmacological compounds appear to share a common chemical activity as Michael acceptors and can modify thiols in p53, suggesting that they affect p53 folding through redox regulation (Lambert et al. 2009). Peptides have also been generated that interact specifically with several p53 mutants and block their nonspecific transactivation capacities, inhibiting GOF properties. These approaches complement other approaches such as gene therapy that target tumors that do not express p53 mutants (Senzer et al. 2009).
CONCLUDING REMARKS
The TP53 gene has been the most extensively sequenced tumor gene before the era of large scale sequencing, generating a large amount of data to identify links between carcinogens, mutation fingerprints, and tumorigenesis. Recent data produced by genome-wide sequencing have confirmed that overall mutation patterns in cancer genomes are quite similar to those in TP53 itself. The existence of several in vitro and in vivo mutagenesis assays such as the HUF assay provides tools to experimentally corroborate observations on mutation patterns in human cancers. Thus, studies on TP53 mutations will further contribute to the understanding of gene-environment interactions in cancer, in particular when comparing variations in TP53 mutation patterns in relation to different cohorts of patients.
In terms of clinical applications, TP53 mutations have proven to be extremely complex biomarkers. Despite impressive progress in mechanistic understanding of p53 structure and function, p53 research has not yet generated applications of wide impact on cancer management and therapy. Somehow, the complexity of the p53 field may have acted as a deterrent for clinical applications. The fact that mutations in TP53 are diverse in their biological effects and that they may occur at many different stages before or during tumor development makes it impossible to derive simple messages uniformly applicable to all clinical contexts. Translating TP53 mutation into the clinics will require large, structured clinical trials in which patients with defined p53 status are recruited on the basis of specific inclusion criteria, randomized for treatment according to determined regimens, and followed up for long-term therapeutic and clinical end points. Databases such as the current TP53 mutation databases will have a critical role in collecting, structuring, and annotating these data, allowing for the interpretation of TP53 mutation and their educated use in standard molecular pathology practice.
Footnotes
Editors: Arnold J. Levine and David Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- Aas T, Borresen AL, Geisler S, Smith-Sorensen B, Johnsen H, Varhaug JE, Akslen LA, Lonning PE 1996. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med 2:811–814 [DOI] [PubMed] [Google Scholar]
- Ambs S, Hussain SP, Marrogi AJ, Harris CC 1999. Cancer-prone oxyradical overload disease. IARC Sci Publ 150:295–302 [PubMed] [Google Scholar]
- Arlt VM, Stiborova M, vom BJ, Simoes ML, Lord GM, Nortier JL, Hollstein M, Phillips DH, Schmeiser HH 2007. Aristolochic acid mutagenesis: Molecular clues to the aetiology of Balkan endemic nephropathy-associated urothelial cancer. Carcinogenesis 28:2253–2261 [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457:608–611 [DOI] [PubMed] [Google Scholar]
- Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, et al. 2003. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell 3:387–402 [DOI] [PubMed] [Google Scholar]
- Bergh J, Norberg T, Sjogren S, Lindgren A, Holmberg L 1995. Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat Med 1:1029–1034 [DOI] [PubMed] [Google Scholar]
- Bertheau P, Espie M, Turpin E, Lehmann J, Plassa LF, Varna M, Janin A, de TH 2008. TP53 status and response to chemotherapy in breast cancer. Pathobiology 75:132–139 [DOI] [PubMed] [Google Scholar]
- Birch JM, Alston RD, McNally RJ, Evans DG, Kelsey AM, Harris M, Eden OB, Varley JM 2001. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene 20:4621–4628 [DOI] [PubMed] [Google Scholar]
- Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR 2008. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci 105:10360–10365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonafe M, Ceccarelli C, Farabegoli F, Santini D, Taffurelli M, Barbi C, Marzi E, Trapassi C, Storci G, Olivieri F, et al. 2003. Retention of the p53 codon 72 arginine allele is associated with a reduction of disease-free and overall survival in arginine/proline heterozygous breast cancer patients. Clin Cancer Res 9:4860–4864 [PubMed] [Google Scholar]
- Bougeard G, Baert-Desurmont S, Tournier I, Vasseur S, Martin C, Brugieres L, Chompret A, Bressac-de Paillerets B, Stoppa-Lyonnet D, Bonaiti-Pellie C, et al. 2006. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J Med Genet 43:531–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougeard G, Brugieres L, Chompret A, Gesta P, Charbonnier F, Valent A, Martin C, Raux G, Feunteun J, Bressac-de Paillerets B, Frebourg T 2003. Screening for TP53 rearrangements in families with the Li-Fraumeni syndrome reveals a complete deletion of the TP53 gene. Oncogene 22:840–846 [DOI] [PubMed] [Google Scholar]
- Brachmann RK, Vidal M, Boeke JD 1996. Dominant-negative p53 mutations selected in yeast hit cancer hot spots. Proc Natl Acad Sci 93:4091–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressac B, Kew M, Wands J, Ozturk M 1991. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature 350:429–431 [DOI] [PubMed] [Google Scholar]
- Bullock AN, Henckel J, Fersht AR 2000. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: Definition of mutant states for rescue in cancer therapy. Oncogene 19:1245–1256 [DOI] [PubMed] [Google Scholar]
- Bykov VJ, Issaeva N, Zache N, Shilov A, Hultcrantz M, Bergman J, Selivanova G, Wiman KG 2005. Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J Biol Chem 280:30384–30391 [DOI] [PubMed] [Google Scholar]
- Caron de Fromentel C, Gruel N, Venot C, Debussche L, Conseiller E, Dureuil C, Teillaud JL, Tocque B, Bracco L 1999. Restoration of transcriptional activity of p53 mutants in human tumour cells by intracellular expression of anti-p53 single chain Fv fragments. Oncogene 18:551–557 [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP 1994. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science 265:346–355 [DOI] [PubMed] [Google Scholar]
- Denissenko MF, Pao A, Tang M, Pfeifer GP 1996. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274:430–432 [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455:1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix BR, Robbins PD, Spagnolo DV, Padovan GL, House AK, Iacopetta BJ 1995. Clonal analysis of colorectal tumors using K-ras and p53 gene mutations as markers. Diagn Mol Pathol 4:261–265 [DOI] [PubMed] [Google Scholar]
- Dudgeon C, Kek C, Demidov ON, Saito S, Fernandes K, Diot A, Bourdon JC, Lane DP, Appella E, Fornace AJ Jr, et al. 2006. Tumor susceptibility and apoptosis defect in a mouse strain expressing a human p53 transgene. Cancer Res 66:2928–2936 [DOI] [PubMed] [Google Scholar]
- Eeles RA 1995. Germline mutations in the TP53 gene. Cancer Surv 25:101–124 [PubMed] [Google Scholar]
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B 1992. Definition of a consensus binding site for p53. Nat Genet 1:45–49 [DOI] [PubMed] [Google Scholar]
- Felley-Bosco E, Weston A, Cawley HM, Bennett WP, Harris CC 1993. Functional studies of a germ-line polymorphism at codon 47 within the p53 gene. Am J Hum Genet 53:752–759 [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hollstein M, Xu Y 2006. Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle 5:2812–2819 [DOI] [PubMed] [Google Scholar]
- Franklin WA, Gazdar AF, Haney J, Wistuba II, La Rosa FG, Kennedy T, Ritchey DM, Miller YE 1997. Widely dispersed p53 mutation in respiratory epithelium. A novel mechanism for field carcinogenesis. J Clin Invest 100:2133–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frebourg T, Barbier N, Kassel J, Ng YS, Romero P, Friend SH 1992. A functional screen for germ line p53 mutations based on transcriptional activation. Cancer Res 52:6976–6978 [PubMed] [Google Scholar]
- Gemignani F, Moreno V, Landi S, Moullan N, Chabrier A, Gutierrez-Enriquez S, Hall J, Guino E, Peinado MA, Capella G, et al. 2004. A TP53 polymorphism is associated with increased risk of colorectal cancer and with reduced levels of TP53 mRNA. Oncogene 23:1954–1956 [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G, Sarasin A 2003. TP53 mutations in human skin cancers. Hum Mutat 21:217–228 [DOI] [PubMed] [Google Scholar]
- Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, Han JH, Lowstuter K, Longmate J, Sommer SS, et al. 2009. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol 27:1250–1256 [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446:153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochola LF, Zeron-Medina J, Mériaux S, Bond GL 2010. Single nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol 2:a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, Suzuki N, Fernandes A, Rosenquist T, Medverec Z, et al. 2007. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci 104:12129–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Weinberg RA 2002. Modelling the molecular circuitry of cancer. Nat Rev Cancer 2:331–341 [DOI] [PubMed] [Google Scholar]
- Hainaut P, Hollstein M 2000. p53 and human cancer: The first ten thousand mutations. Adv Cancer Res 77:81–137 [DOI] [PubMed] [Google Scholar]
- Hergenhahn M, Luo JL, Hollstein M 2004. p53 designer genes for the modern mouse. Cell Cycle 3:738–741 [PubMed] [Google Scholar]
- Hinds PW, Finlay CA, Quartin RS, Baker SJ, Fearon ER, Vogelstein B, Levine AJ 1990. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: A comparison of the “hot spot” mutant phenotypes. Cell Growth Differ 1:571–580 [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC 1991. p53 mutations in human cancers. Science 253:49–53 [DOI] [PubMed] [Google Scholar]
- Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC 1991. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature 350:427–428 [DOI] [PubMed] [Google Scholar]
- Hussain SP, Schwank J, Staib F, Wang XW, Harris CC 2007. TP53 mutations and hepatocellular carcinoma: Insights into the etiology and pathogenesis of liver cancer. Oncogene 26:2166–2176 [DOI] [PubMed] [Google Scholar]
- Inga A, Storici F, Darden TA, Resnick MA 2002. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol Cell Biol 22:8612–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishioka C, Frebourg T, Yan YX, Vidal M, Friend SH, Schmidt S, Iggo R 1993. Screening patients for heterozygous p53 mutations using a functional assay in yeast. Nat Genet 5:124–129 [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G 2004. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med 10:1321–1328 [DOI] [PubMed] [Google Scholar]
- Jia L, Liu Y, Yi X, Miron A, Crum CP, Kong B, Zheng W 2008. Endometrial glandular dysplasia with frequent p53 gene mutation: A genetic evidence supporting its precancer nature for endometrial serous carcinoma. Clin Cancer Res 14:2263–2269 [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR 2007. Structure-function-rescue: The diverse nature of common p53 cancer mutants. Oncogene 26:2226–2242 [DOI] [PubMed] [Google Scholar]
- Joerger AC, Ang HC, Fersht AR 2006. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs 6. Proc Natl Acad Sci 103:15056–15061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR 2005. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations 8. J Biol Chem 280:16030–16037 [DOI] [PubMed] [Google Scholar]
- Jones PA, Rideout WM, Shen JC, Spruck CH, Tsai YC 1992. Methylation, mutation and cancer. Bioessays 14:33–36 [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321:1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Feng Z, Sun Y, Atwal G, Murphy ME, Rebbeck TR, Rosenwaks Z, Levine AJ, Hu W 2009. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci 106:9761–9766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C 2003. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci 100:8424–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk GD, Camus-Randon AM, Mendy M, Goedert JJ, Merle P, Trepo C, Brechot C, Hainaut P, Montesano R 2000. Ser-249 p53 mutations in plasma DNA of patients with hepatocellular carcinoma from The Gambia. J Natl Cancer Inst 92:148–153 [DOI] [PubMed] [Google Scholar]
- Klug SJ, Wilmotte R, Santos C, Almonte M, Herrero R, Guerrero I, Caceres E, Peixoto-Guimaraes D, Lenoir G, Hainaut P, et al. 2001. TP53 polymorphism, HPV infection, and risk of cervical cancer. 10:1009–1012 [PubMed] [Google Scholar]
- Lalloo F, Varley J, Moran A, Ellis D, O'dair L, Pharoah P, Antoniou A, Hartley R, Shenton A, Seal S, et al. 2006. BRCA1, BRCA2 and TP53 mutations in very early-onset breast cancer with associated risks to relatives 1. Eur J Cancer 42:1143–1150 [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119:861–872 [DOI] [PubMed] [Google Scholar]
- Langerod A, Bukholm IR, Bregard A, Lonning PE, Andersen TI, Rognum TO, Meling GI, Lothe RA, Borresen-Dale AL 2002. The TP53 codon 72 polymorphism may affect the function of TP53 mutations in breast carcinomas but not in colorectal carcinomas. Cancer Epidemiol Biomarkers Prev 11:1684–1688 [PubMed] [Google Scholar]
- Langerod A, Zhao H, Borgan O, Nesland JM, Bukholm IR, Ikdahl T, Karesen R, Borresen-Dale AL, Jeffrey SS 2007. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res 9:R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323–331 [DOI] [PubMed] [Google Scholar]
- Li FP, Fraumeni JFJ, Mulvihill JJ, Blattner WA, Dreyfus MG, Tucker MA, Miller RW 1988. A cancer family syndrome in twenty-four kindreds. Cancer Res 48:5358–5362 [PubMed] [Google Scholar]
- Lindahl T 1993. Instability and decay of the primary structure of DNA. Nature 362:709–715 [DOI] [PubMed] [Google Scholar]
- Liu Z, Hergenhahn M, Schmeiser HH, Wogan GN, Hong A, Hollstein M 2004. Human tumor p53 mutations are selected for in mouse embryonic fibroblasts harboring a humanized p53 gene. Proc Natl Acad Sci 101:2963–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M 2001. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: A new biomedical research tool 9. Oncogene 20:320–328 [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JFJ, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA 1990. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250:1233–1238 [DOI] [PubMed] [Google Scholar]
- Marcel V, Palmero EI, Falagan-Lotsch P, Martel-Planche G, Ashton-Prolla P, Olivier M, Brentani RR, Hainaut P, Achatz MI 2009. TP53PIN3 and MDM2 SNP309 polymorphisms as genetic modifiers in the Li-Fraumeni syndrome: Impact on age at first diagnosis. J Med Genet (in press). [DOI] [PubMed] [Google Scholar]
- Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, et al. 2000. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 25:47–54 [DOI] [PubMed] [Google Scholar]
- Matakidou A, Eisen T, Houlston RS 2003. TP53 polymorphisms and lung cancer risk: A systematic review and meta-analysis. Mutagenesis 18:377–385 [DOI] [PubMed] [Google Scholar]
- Montesano R, Hainaut P 1998. Molecular precursor lesions in oesophageal cancer. Cancer Surv 32:53–68 [PubMed] [Google Scholar]
- Nedelko T, Arlt VM, Phillips DH, Hollstein M 2009. TP53 mutation signature supports involvement of aristolochic acid in the aetiology of endemic nephropathy-associated tumours. Int J Cancer 124:987–990 [DOI] [PubMed] [Google Scholar]
- Nelson HH, Wilkojmen M, Marsit CJ, Kelsey KT 2005. TP53 mutation, allelism and survival in non-small cell lung cancer 1. Carcinogenesis 26:1770–1773 [DOI] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849–862 [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119:847–860 [DOI] [PubMed] [Google Scholar]
- Olivier M, Goldgar DE, Sodha N, Ohgaki H, Kleihues P, Hainaut P, Eeles RA 2003. Li-Fraumeni and related syndromes: Correlation between tumor type, family structure, and TP53 genotype. Cancer Res 63:6643–6650 [PubMed] [Google Scholar]
- Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC 2004. TP53 mutation spectra and load: A tool for generating hypotheses on the etiology of cancer. IARC Sci Publ 157:247–270 [PubMed] [Google Scholar]
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, et al. 2006. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12:1157–1167 [DOI] [PubMed] [Google Scholar]
- Oren M, Rotter V 2010. Mutant p53 gain of function in cancer. Cold Spring Harb Perspect Biol 2:a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M 2007a. TP53 mutations in human cancers: Functional selection and impact on cancer prognosis and outcomes. Oncogene 26:2157–2165 [DOI] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M 2007b. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat 28:622–629 [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Besaratinia A 2009. Mutational spectra of human cancer. Hum Genet 125:493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Hainaut P 2003. On the origin of G –> T transversions in lung cancer. Mutat Res 526:39–43 [DOI] [PubMed] [Google Scholar]
- Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P 2002. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking- associated cancers. Oncogene 21:7435–7451 [DOI] [PubMed] [Google Scholar]
- Pistritto G, Puca R, Nardinocchi L, Sacchi A, D'Orazi G 2007. HIPK2-induced p53Ser46 phosphorylation activates the KILLER/DR5-mediated caspase-8 extrinsic apoptotic pathway. Cell Death Differ 14:1837–1839 [DOI] [PubMed] [Google Scholar]
- Ponten F, Berg C, Ahmadian A, Ren ZP, Nister M, Lundeberg J, Uhlen M, Ponten J 1997. Molecular pathology in basal cell cancer with p53 as a genetic marker. Oncogene 15:1059–1067 [DOI] [PubMed] [Google Scholar]
- Reinbold M, Luo JL, Nedelko T, Jerchow B, Murphy ME, Whibley C, Wei Q, Hollstein M 2008. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene 27:2788–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9:402–412 [DOI] [PubMed] [Google Scholar]
- Schmidt MK, Reincke S, Broeks A, Braaf LM, Hogervorst FB, Tollenaar RA, Johnson N, Fletcher O, Peto J, Tommiska J, et al. 2007. Do MDM2 SNP309 and TP53 R72P interact in breast cancer susceptibility? A large pooled series from the breast cancer association consortium. Cancer Res 67:9584–9590 [DOI] [PubMed] [Google Scholar]
- Schmutte C, Yang AS, Nguyen TT, Beart RW, Jones PA 1996. Mechanisms for the involvement of DNA methylation in colon carcinogenesis. Cancer Res 56:2375–2381 [PubMed] [Google Scholar]
- Senzer N, Nemunaitis J 2009. A review of contusugene ladenovec (Advexin) p53 therapy. Curr Opin Mol Ther 11:54–61 [PubMed] [Google Scholar]
- Shi H, Tan SJ, Zhong H, Hu W, Levine A, Xiao CJ, Peng Y, Qi XB, Shou WH, Ma RL, et al. 2009. Winter temperature and UV are tightly linked to genetic changes in the p53 tumor suppressor pathway in Eastern Asia. Am J Hum Genet 84:534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314:268–274 [DOI] [PubMed] [Google Scholar]
- Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M 2008. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res 36:3639–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, et al. 2005. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet 37:590–592 [DOI] [PubMed] [Google Scholar]
- Song H, Hollstein M, Xu Y 2007. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 9:573–580 [DOI] [PubMed] [Google Scholar]
- Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L 1998. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature 393:229–234 [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA 2009. The cancer genome. Nature 458:719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer 6:909–923 [DOI] [PubMed] [Google Scholar]
- Tommasino M, Accardi R, Caldeira S, Dong W, Malanchi I, Smet A, Zehbe I 2003. The role of TP53 in Cervical carcinogenesis. Hum Mutat 21:307–312 [DOI] [PubMed] [Google Scholar]
- Vaninetti NM, Geldenhuys L, Porter GA, Risch H, Hainaut P, Guernsey DL, Casson AG 2008. Inducible nitric oxide synthase, nitrotyrosine and p53 mutations in the molecular pathogenesis of Barrett's esophagus and esophageal adenocarcinoma. Mol Carcinog 47:275–285 [DOI] [PubMed] [Google Scholar]
- Vikhanskaya F, Siddique MM, Kei LM, Broggini M, Sabapathy K 2005. Evaluation of the combined effect of p53 codon 72 polymorphism and hotspot mutations in response to anticancer drugs. Clin Cancer Res 11:4348–4356 [DOI] [PubMed] [Google Scholar]
- vom Brocke J, Schmeiser HH, Reinbold M, Hollstein M 2006. MEF immortalization to investigate the ins and outs of mutagenesis. Carcinogenesis 27:2141–2147 [DOI] [PubMed] [Google Scholar]
- Wang Y, Helland A, Holm R, Skomedal H, Abeler VM, Danielsen HE, Trope CG, Borresen-Dale AL, Kristensen GB 2004a. TP53 mutations in early-stage ovarian carcinoma, relation to long-term survival. Br J Cancer 90:678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kringen P, Kristensen GB, Holm R, Baekelandt MM, Olivier M, Skomedal H, Hainaut P, Trope CG, Abeler VM, et al. 2004b. Effect of the codon 72 polymorphism (c.215G>C, p.Arg72Pro) in combination with somatic sequence variants in the TP53 gene on survival in patients with advanced ovarian carcinoma. Hum Mutat 24:21–34 [DOI] [PubMed] [Google Scholar]
- Wang-Gohrke S, Rebbeck TR, Besenfelder W, Kreienberg R, Runnebaum IB 1998. p53 germline polymorphisms are associated with an increased risk for breast cancer in German women. Anticancer Res 18:2095–2099 [PubMed] [Google Scholar]
- Wang-Gohrke S, Weikel W, Risch H, Vesprini D, Abrahamson J, Lerman C, Godwin A, Moslehi R, Olipade O, Brunet JS, et al. 1999. Intron variants of the p53 gene are associated with increased risk for ovarian cancer but not in carriers of BRCA1 or BRCA2 germline mutations. Br J Cancer 81:179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M 2009. p53 polymorphisms: cancer implications. Nat Rev Cancer 9:95–107 [DOI] [PubMed] [Google Scholar]
- Zawlik I, Kita D, Vaccarella S, Mittelbronn M, Franceschi S, Ohgaki H 2009. Common polymorphisms in the MDM2 and TP53 genes and the relationship between TP53 mutations and patient outcomes in glioblastomas. Brain Pathol 19:188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski B, Liu Z, Hollstein M, Hergenhahn M, Luo JL 2002. Mouse models for generating P53 gene mutation spectra. Toxicol Lett 134:31–37 [DOI] [PubMed] [Google Scholar]