Abstract
The morphological feature of tight junctions (TJs) fits well with their functions. The core of TJs is a fibril-like proteinaceous structure within the lipid bilayer, the so-called TJ strands. TJ strands in apposing plasma membranes associate with each other to eliminate the intercellular space. A network of paired TJ strands generates a continuous belt that circumscribes each cell to establish the diffusion barrier to the solutes in the paracellular pathway throughout the cellular sheet. Identification and characterization of TJ-associated proteins during the last two decades has unveiled the nature of TJ strands and how they are spatially organized. The interplay between integral membrane proteins, claudins, and cytoplasmic plaque proteins, ZO-1/ZO-2, is critical for TJ formation and function.
Structural studies reveal how claudins, ZO-1/2, and other proteins in tight junctions are assembled to produce an effective paracellular diffusion barrier.
Tight junctions (TJs) are fascinating structures in terms of their function and morphology. In 1963, using ultrathin-section electron microscopy, Farquhar and Palade described the fine structure of TJs together with adherens junctions (AJs) and desmosomes at the most luminal side of the lateral membrane (Farquhar and Palade 1963). In addition, they demonstrated insightfully that TJs function as permeability seals for mass tracers. Indeed, the structure of TJs observed in electron microscopy indicates that TJs could physically restrict the leak of solutes through the intercellular space. However, physiological studies at the same time revealed that solute transport occurred via the intercellular space in a variety of epithelial cells. A resolution of these different views of TJ function comprises the current concept that the TJ regulates the diffusion of solutes with size and charge selectivity and that it is functionally different in physiologically diverse epithelial cell types (Powell 1981; Anderson and Cereijido 2001). To understand the molecular mechanism controlling TJ structure and function, it is important to determine their molecular composition and organization.
Although purification of TJs is difficult, Stevenson and Goodenough developed an isolation method for a TJ-enriched plasma membrane fraction from rodent liver. They discovered the first TJ-associated protein, ZO-1, in 1986 by generating monoclonal antibodies against this fraction (Stevenson et al. 1986). Since then, many molecular components of TJs have been identified using immunological approaches or searches for binding proteins with known molecules, which have enabled detailed molecular cell biological analyses of TJs. Among the TJ-associated proteins, the claudin family of membrane proteins identified in 1998 by the Tsukita group are key molecules in the architecture and barrier function of TJs (Furuse et al. 1998a). Functional analyses of claudins have allowed remarkable progress in the development of a comprehensive understanding of the molecular basis of the ultrastructure and physiological characteristics of TJs (Van Itallie and Anderson 2006; Furuse and Tsukita 2006; Angelow et al. 2008). In addition, the cytoplasmic plaque proteins associated with TJs are important in regulating TJ architecture (Guillemot et al. 2008).
In this article, we present the molecular basis for the core structure of TJs based on recent progress in functional analyses of TJ-associated proteins. The current molecular basis of TJ physiology is covered in detail in Anderson and Van Itallie (2009).
MORPHOLOGY OF TJS
Electron Microscopic Observations of TJs
TJs occur in epithelial cell types, including epithelial and endothelial cells, in vertebrate species and tunicates (Lane 2001). TJs are located at the most apical part of junctional complexes, next to AJs in epithelial cells. In contrast, TJs and AJs are spatially intermingled in endothelial cells (Schulze and Firth 1993). TJs are also located in myelin sheaths formed by oligodendrocytes (Dermietzel 1974) and Schwann cells (Tetzlaff 1978).
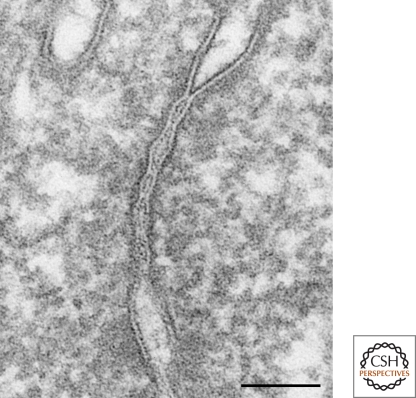
In ultrathin-section electron microscopy, TJs are visualized as a zone in which adjacent plasma membranes are closely apposed, that circumscribes the cell as a belt together with AJs. At high magnification, TJs are characterized as focal attachments of adjacent cell membranes that exclude the intercellular gap (Fig. 1). This appearance of TJs suggests their function as rate-limiting barriers to the diffusion of solutes, and this idea has been supported by electron microscopic observations using electron-opaque tracers (Farquhar and Palade 1963; Brightman and Reese 1969; Goodenough and Revel 1970).
Figure 1.
Thin section of TJs of mouse epididymis epithelial cells. TJs are defined as close contacts between plasma membranes of adjacent cells. Bar, 100 nm.
TJs have been intensely analyzed by freeze-fracture electron microscopy, in which the morphology of the central hydrophobic plane of the lipid bilayer, including the particles and pits, can be observed. In general, TJs in glutaraldehyde-fixed epithelial cells appear as anastomosing linear fibrils (TJ strands) on the cytoplasmic leaflet (P-face) and grooves on the exoplasmic leaflet (E-face) surrounding the cell (Fig. 2) (Staehelin 1973, 1974). In some cell types, however, TJs exhibit discontinuous particle strands on the P-face and grooves containing many particles on the E-face (Simionescu et al. 1975; van Deurs and Koehler 1979). Furthermore, specimens that are not fixed with glutaraldehyde before freezing also reveal particle strands on the E-face (Staehelin 1973). Detailed analyses of double replicas have revealed that these strands and grooves are complementary between the P- and E-face (Chalcroft and Bullivant 1970; van Deurs and Koehler 1979). It is widely accepted that TJ strands correspond to the focal attachments of adjacent plasma membranes observed in ultrathin sections. Freeze-fracture electron microscopy has also revealed that the number and network pattern of TJ strands varies between cell types (Friend and Gilula 1972; Claude and Goodenough 1973). By combining morphological observations and physiological measurements in various epithelial cell types, the correlation between the structure and barrier function of TJs has been discussed (Claude 1978). In several cases, however, the correlation was not perfect. At that time, there was no information about the molecular nature of TJs, and they hypothesized that each TJ strand in various cell types was equal. Currently, we know that the combination of caudin types is an important factor to determine the barrier property of TJs.
Figure 2.
Freeze-fracture replica of TJs of mouse intestinal epithelial cells. TJs appear as anastomosing fibrils, namely, TJ strands, on P-face (arrow) or complementary grooves on E-face (arrowhead) in glutaraldehyde-fixed specimens. Bar, 200 nm.
Two models have been proposed for the structural nature of the TJ strand based on morphological analyses using electron microscopy. The most plausible model is the protein model, in which proteinaceous attachment elements, i.e., adhesion molecules, hold the apposing plasma membranes together (Staehelin 1973). Detailed characterization of TJ-associated integral membrane proteins, including claudins as described in the following section, strongly support this model. The other model, proposed in the 1980s, is the lipid model in which TJ strands are cylindrical inverted lipid micelles formed by the fusion of apposed plasma membranes, resulting in the continuity of outer leaflets of plasma membranes between adjacent cells (Pinto da Silva and Kachar 1982). The observations that a fluorescent-labeled lipid that was introduced in the outer leaflet of the apical membrane of an epithelial cell never moved to the next cell (van Meer et al. 1986) and TJ strands are detergent resistant (Stevenson and Goodenough 1984) do not conform to this model, although specific lipids may contribute to the formation of TJs.
Tricellular TJs: Specialized TJs at Tricellular Corners
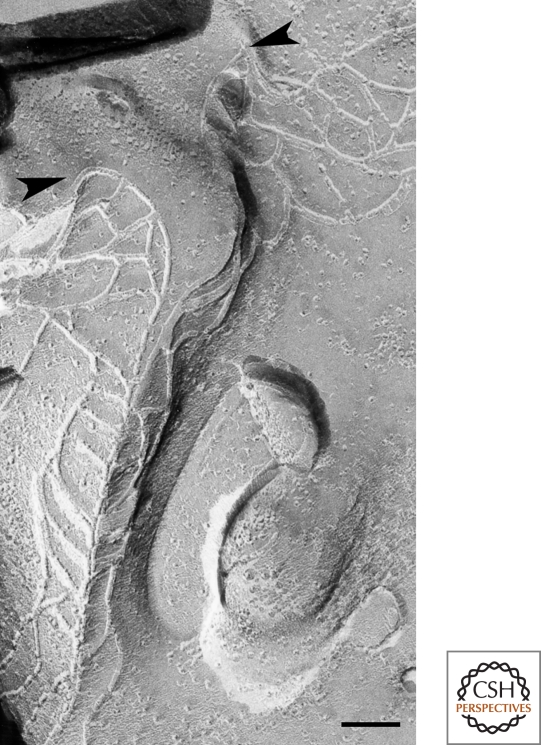
Considering that TJs function as continuous barriers throughout the cellular sheet, it is interesting to understand how tricellular contacts formed by the joining of three cells are sealed. Because TJs are thought to be zipper-like structures formed between two adjacent cells, it is not simple to see how they could exclude the extracellular space between three plasma membranes. Specialized structures, namely tricellular TJs (tTJs), are observed in these regions by freeze-fracture electron microscopy (Staehelin et al. 1969; Friend and Gilula 1972; Staehelin 1973; Wade and Karnovsky 1974). At tTJs, the most apical elements of the horizontal TJ strands from both sides attach and turn together to extend vertically in the basal direction (Fig. 3). These vertical TJ strands link to several short horizontal TJ strands that connect with the main portion of the TJ strands. As a result, three sets of parallel and closely spaced TJ strands extend vertically to form a narrow tube at the extracellular space of the tricellular contact region. This tube is thought to reduce to a negligible amount the free diffusion of solutes through the intercellular space (Staehelin 1973).
Figure 3.
Freeze-fracture replica of tTJs in MDCK cells. At tricellular corners, the most apical elements of the horizontal TJ strands from both sides (arrowheads) attach to form the central seal and extend vertically in the basal direction. Short, horizontal strands connect with the central seal. Bar, 100 nm.
MOLECULAR ORGANIZATION OF THE CORE STRUCTURE OF TJS
Overview of TJ-associated Proteins
Many molecular components of TJs have been identified and characterized. Like other intercellular junctions, TJs are protein complexes composed of integral membrane proteins, cytoplasmic plaque proteins, and cytoskeletal proteins. Among these, the claudin family membrane proteins are key components for the structure and function of TJs (Van Itallie and Anderson 2006; Furuse and Tsukita 2006; Angelow et al. 2008). Claudin molecules interact with each other between cells and probably polymerize within the plasma membrane as fibrils to generate TJ strands. Occludin and tricellulin, comprising other types of four transmembrane domain-containing proteins with related amino acid sequences, are incorporated into or localized very close to claudin-based TJ strands (Furuse et al. 1993; Fujimoto 1995; Ikenouchi et al. 2005). In addition, immunoglobulin superfamily membrane proteins with two extracellular Ig-like domains, including JAM-A, JAM4, coxsackie adenovirus receptor (CAR), and endothelial cell-selective adhesion molecule (ESAM), are also localized at TJs (Martin-Padura et al. 1998; Hirabayashi et al. 2003; Cohen et al. 2001; Nasdala et al. 2002).
On the cytoplasmic side of TJ strands, the membrane-associated guanylate kinase (MAGUK) family proteins ZO-1, ZO-2, and ZO-3 (Stevenson et al. 1986; Gumbiner et al. 1991; Haskins et al. 1998) bind to the C-terminal cytoplasmic domain of claudins, occludin, tricellulin, and JAM-A. In addition, MAGI-1, MAGI-3, MUPP1, and PATJ are known to be PDZ (PSD-95, discs-large, ZO-1) domain-containing proteins that directly bind to claudins or other TJ-associated membrane proteins (Ide et al. 1999; Laura et al. 2002; Hamazaki et al. 2002; Lemmers et al. 2002; Roh et al. 2002; Jeansonne et al. 2003). Other cytoplasmic proteins with coiled-coil domains, such as cingulin and JACOP/paracingulin, are concentrated relatively far from the plasma membrane of TJs (Citi et al. 1988; Ohnishi et al. 2004). Of these proteins, at least the ZO family of proteins and cingulin interact with F-actin (Itoh et al. 1997, 1999b; Fanning et al. 1998, 2002; Wittchen et al. 1999; D'Atri and Citi. 2001), which is present on the cytoplasmic side of TJs (Madara 1987). Signaling molecules, including the aPKC-Par3-Par6 complex, which is involved in epithelial polarity formation, are also localized on the cytoplasmic side of TJs and regulate mature TJ formation (Izumi et al. 1998; Suzuki and Ohno 2006; Ebnet et al. 2008). Furthermore, ZONAB, a transcription factor involved in cell growth, has been reported to be localized at TJs (Balda et al. 2003).
Among these TJ-associated proteins, claudins and ZO proteins are directly involved in the formation of TJ strands in epithelial cells. Two other membrane proteins with four transmembrane domains, occludin and tricellulin, appear to have intimate relationships with claudins and modulate TJ strand formation. These proteins are highlighted in this section as constituents of the core structure of TJs.
Claudins
Claudin Family
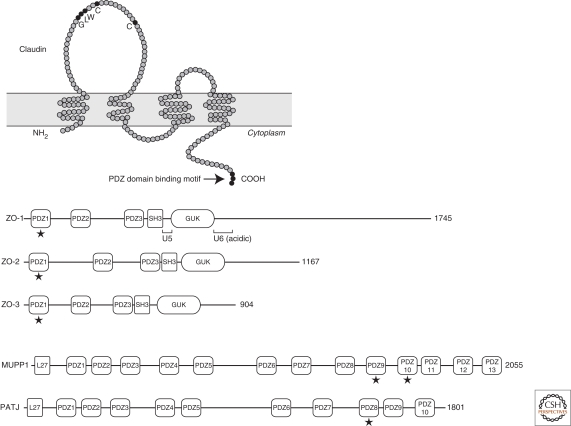
Claudins were originally identified as membrane proteins that cofractionated with occludin, the first identified integral membrane protein of TJs, by sucrose density gradient centrifugation of a sonicated TJ/AJ-enriched membrane fraction isolated from chick liver (Furuse et al. 1998a). Claudins comprise a multigene family with approximately 24 members in humans and mice (Van Itallie and Anderson 2006; Furuse and Tsukita 2006; Angelow et al. 2008). Claudins are 21–28-kD proteins and consist of four transmembrane domains, two extracellular loops, amino- and carboxy-terminal cytoplasmic domains, and a short cytoplasmic turn. Most claudins have the conserved motif GLWxxC(8-10 aa)C in the first extracellular loop, and a PDZ domain-binding motif at the carboxy-terminal, which binds to the TJ-associated PDZ domain-containing plaque proteins ZO-1, ZO-2, and ZO-3 (Fig. 4) (Itoh et al. 1999a). Claudins are key components for the structure and function of TJs. To investigate the roles of claudins in the barrier/channel property of TJs, overexpression and RNAi-mediated suppression of particular claudins in epithelial cell lines followed by physiological measurements have been carried out (see Anderson and Van Itallie 2009). In addition, many claudin knockout mice have been generated and analyzed to clarify the roles of the individual claudins in vivo (Furuse 2009).
Figure 4.
The membrane-spanning model of claudins and the domain organization of claudin-binding plaque proteins. Claudins contain four transmembrane domains. The conserved amino acid residues in the first extracellular loop are indicated. The PDZ domain binding motif is located at the carboxyl terminus, which is known to bind to PDZ-domains in ZO-1, ZO-2, ZO-3, MUPP1, and PATJ (stars). (GUK) guanylate kinase domain; (L27) LIN2, 7 homology domain; (U5) unique 5 region; (U6) unique 6 region. U6 contains the acidic region.
TJ Strand Formation by Claudins
The most obvious function of claudins is to generate TJ strands, the structural and functional core of TJs within the plasma membrane. When claudins are overexpressed in mouse L fibroblasts lacking endogenous TJs, the exogenous claudins concentrate into cell–cell contacts between adjacent cells and generate TJ strands with the induction of cell adhesive activity (Furuse et al. 1998b; Kubota et al. 1999). Immunoelectron and immunofluorescence microscopy analyses revealed that claudins themselves generate TJ strands as structural components (Furuse et al. 1998b; Sasaki et al. 2003). Importantly, addition to the culture medium of the carboxy-terminal of Clostridium perfringens enterotoxin, which binds to the second extracellular loop of some claudin types and functions as an inhibitor of these claudins, results in the removal of some claudin types from TJs accompanied by a reduction in the number of TJ strands and loss-of-barrier function (Sonoda et al. 1999). Furthermore, in claudin-11-deficient mice, TJ strands disappear in Sertoli cells, oligodendrocytes (Gow et al. 1999), and marginal cells of the inner ear (Gow et al. 2004; Kitajiri et al. 2004), where claudin-11 is the only known claudin type to be expressed. These observations from gain-of-function and loss-of-function studies confirm that claudins are involved in the structure of TJ strands. Deletion of the carboxy-terminal cytoplasmic region of claudin-1 did not abolish TJ strand reconstitution in L cells (Furuse et al. 1998b), also indicating that the ability to form TJ strands is mediated by claudins. In epithelial cells, however, the TJ-associated plaque proteins ZO-1 or ZO-2 are required for TJ strand formation by claudins (Umeda et al. 2006) (see Section ZO-1, ZO-2, and ZO-3).
TJ Strand as a Mosaic of Multiple Claudin Types
Most cell types coexpress multiple claudin subtypes in vivo in combinations and proportions that vary among different cell types. This complex expression pattern of claudins creates diversity in the barrier/channel property of TJs, depending on each epithelium (see Anderson and Van Itallie 2009 for details). The manner in which multiple claudins generate TJs between cells has been analyzed using L cells overexpressing various claudin types (Furuse et al. 1999). Coculture of L cells expressing claudin-1, -2, or -3 revealed that the combinations of claudin-1/-3 and claudin-2/-3, but not claudin-1/-2, could form heterotypic TJ strands. Furthermore, coexpression of two of these three claudins in L cells revealed that these claudins could copolymerize into single TJ strands in a heteromeric manner in individual cells. In other analyses using HeLa cells, claudin-3 and claudin-4 were unable to assemble heterotypically between cells, but their heteromeric assembly is possible (Daugherty et al. 2007). These experiments suggest that TJ strands are generally comprised of mosaics of different claudins that form homotypic and heterotypic interactions between cells and homomeric and heteromeric complexes in individual cells, although their compatibility is dependent on the right claudin combination. It is important to remember this complex architecture of claudin-based TJ strands when the barrier property of TJs is considered in terms of claudin content. Note, however, that claudin types may also be separated within TJs in the same cell surface in vivo. For example, TJ strands can be divided into two subdomains between outer hair cells and supporting Deiter cells in the organ of Corti in the inner ear; claudin-14 is concentrated in the apical TJ strands, whereas claudin-6 and claudin-9 are localized at lateral TJ strands (Nunes et al. 2006).
Structural Units of TJ Strands
Freeze-fracture replica electron microscopy studies with glutaraldehyde-fixed samples revealed that the E-face/P-face distributions of the intramembrane particles of TJ strands vary among different cell types. This variation appears to depend on the claudin types forming the TJ strands. For instance, TJ strands generated in L cells with claudin-1 and fixed with glutaraldehyde exhibit continuous rod-shaped strands on the P-face, whereas those in L cells with claudin-5 show thin and shallow grooves on the P-face and many particles on the E-face (Furuse et al. 1998b; Morita et al. 1999). An attractive hypothesis is that the P-face distribution of TJ strands is because of the interaction of the intramembrane components of TJs with cytoplasmic components (Balda and Anderson 1993). This does not appear to be the case because carboxy-terminal-deleted claudin-1, which cannot associate with cytoplasmic plaque proteins, still exhibits a P-face distribution (Furuse et al. 1999).
Because quick-freeze samples show chains of E-face-associated particles, it is reasonable to think that this is the unit particle structure of TJ strands. However, it remains unclear how this unit particle is constituted from claudin molecules. Interestingly, an intramembrane particle of TJs is similar to the diameter of a connexon, the basic unit of gap junctions consisting of the hexamer of connexins (Tsukita and Furuse 1999; Kumar and Gilula 1996). Both claudins and connexins have four transmembrane domains. Therefore, each E-face-associated particle of TJs may be composed of a claudin oligomer, similar to the hexamer of connexins, rather than claudin monomer. So far, there is no direct evidence to demonstrate this idea, although a crosslinking experiment indirectly implied the possibility of claudin oligomer formation in claudin-4-overexpressing insect cells (Mitic et al. 2003).
Modification of Claudins
Claudins are posttranslationally modified by palmitoylation and phosphorylation. All claudins have membrane proximal CxxC motifs near the ends of the second and fourth transmembrane domains. Both of these regions in claudin-14 are palmitoylated (Van Itallie et al. 2005). Site-directed mutagenesis revealed that this modification of claudin-14 is required for its efficient localization at TJs, but not the stability of claudin-14 protein or the assembly of TJ strands.
Ser or Thr residues in the carboxy-terminal cytoplasmic regions of several claudins, including claudin-3, -4, -5, and -16, are phosphorylated (D'souza et al. 2005; Aono and Hirai. 2008; Ishizaki et al. 2003; Ikari et al. 2006). This modification seems to regulate the localization of claudins and the barrier property of TJs, but the molecular mechanisms remain unknown.
Occludin
Occludin, the first-reported integral membrane protein of TJs, was identified by screening with monoclonal antibodies raised against an AJ/TJ-enriched plasma membrane fraction isolated from chick liver (Furuse et al. 1993). Occludin is an approximately 65-kD protein with four transmembrane domains, two extracellular loops, an intracellular turn, and carboxy- and amino-terminal cytoplasmic domains, but does not exhibit any sequence similarities with claudins. The first extracellular loop has a characteristic amino acid content (∼60% of the amino acids are glycine and tyrosine residues), whose physiological significance remains elusive. The carboxy-terminal cytoplasmic region of occludin binds to ZO-1, ZO-2, and ZO-3, comprising the MAGUK family plaque proteins of TJs (Furuse et al. 1994; Fanning et al. 1998; Itoh et al. 1999a; Haskins et al. 1998). Multiple Ser and Thr residues in this region are phosphorylated, and highly-phoshorylated occludin is selectively concentrated at TJs (Sakakibara et al. 1997).
Overexpression studies in cultured cells revealed some activities of occludin in TJ formation and cell adhesion, although these activities are much weaker than those of claudins. When overexpressed in mouse L fibroblasts, which lack TJs as well as cadherin-mediated cell–cell adhesion, exogenous mouse occludin assembles into cell–cell contacts formed by adjacent cells as dots observed by immunofluorescence microscopy (Furuse et al. 1998b). Short TJ strandlike structures containing occludin are observed in these regions by freeze-fracture electron microscopy. When human occludin is expressed in fibroblasts, the cells acquire weak cell–cell adhesive activity (Van Itallie and Anderson 1997). In these cells, addition of synthetic peptides corresponding to the first extracellular loop inhibits cell–cell adhesion. Overexpression of chicken occludin in MDCK cells increased the number of TJ strands (McCarthy et al. 1996), whereas overexpression of human occludin induced side-to-side aggregation of TJ strands (Medina et al. 2000). Furthermore, overexpression of occludin affected the transepithelial electric resistance as well as the paracellular flux in various epithelial cell lines (McCarthy et al. 1996; Balda et al. 1996; Bamforth et al. 1999). These observations suggest that occludin has some accessory functions in TJ formation. However, occludin-deficient epithelial cells exhibit no significant abnormalities in the morphology and physiology of their TJs, and occludin-deficient mice are viable (Saitou et al. 1998; Saitou et al. 2000). Occludin-related proteins, such as tricellulin, may have redundant functions, thereby obscuring the function of occludin.
Tricellulin
Tricellulin is the only known protein that is highly concentrated at tTJs, the vertically oriented TJ strands at tricellular contacts. Tricellulin was originally identified as an epithelium-specific transcript in a screening for putative targets of the Snail transcription repressor, which determines epithelial–mesenchymal transition (Ikenouchi et al. 2005). Tricellulin gene was also identified as a causative gene for recessive, nonsyndromic deafness DFNB49 (Riazuddin et al. 2006). Tricellulin contains four transmembrane domains and has sequence similarity with occludin in the carboxy-terminal cytoplasmic domain of approximately 130 amino acids. This region of occludin is responsible for its association with ZO-1, and tricellulin also binds ZO-1 (Riazuddin et al. 2006). RNAi-mediated suppression of tricellulin in epithelial cells results in abnormal organization of tTJs and impairment of epithelial barrier function (Ikenouchi et al. 2005). Furthermore, tricellulin knockdown also appears to affect bicellular TJ formation, as evaluated by immunostaining of occludin. Interestingly, occludin knockdown in cultured epithelial cells causes mislocalization of tricellulin to bicellular TJs, indicating a role of occludin in the concentration of tricellulin at tTJs (Ikenouchi et al. 2008).
When tricellulin is expressed in claudin-1-overexpressing L cells, it is colocalized with claudin-1-based reconstituted TJ strands and induces their crosslinking (Ikenouchi et al. 2008), suggesting that tricellulin influences the morphology of TJ strands. Although tricellulin is highly concentrated at tTJs, it is also detected at bicellular TJs and the extent of the bicellular distribution of tricellulin depends on the epithelial cell type (Ikenouchi et al. 2005). Therefore, it will be of interest to analyze the relationship between the amount of tricellulin within bicellular TJs and the morphology of TJ strand branching in different cell-types. Despite the critical role of tricellulin in tTJ formation, the function of tricellulin in the organization of tTJs together with claudins remains largely unknown.
ZO-1, ZO-2, and ZO-3
TJ MAGUKs
Just beneath the plasma membrane of TJs, PDZ domain-containing cytoplasmic proteins directly interact with TJ-associated integral membrane proteins and form cytoplasmic plaques, which function as scaffolds to recruit other structural proteins, signaling proteins, and the actin cytoskeleton to the underlying surface of TJs. Three structurally related proteins, ZO-1, ZO-2, and ZO-3, are included in these cytoplasmic plaques. ZO-1 was the first reported TJ-associated molecule. It was identified by generating monoclonal antibodies against a TJ-enriched membrane fraction isolated from mouse liver (Stevenson et al. 1986). ZO-2 and ZO-3 were later identified as coprecipitated proteins with ZO-1 from extracts of cultured epithelial cells (Gumbiner et al. 1991; Haskins et al. 1998). All of these proteins belong to the MAGUK family and consist of three PDZ domains, a SH3 domain, and a guanylate kinase (GUK) domain from their amino-termini (Guillemot et al. 2008). ZO-1 and ZO-2 have acidic domains next to their GUK domains (Fig. 4). Multiple domains of ZO-1, ZO-2, and ZO-3 are involved in complex interactions with other TJ-associated molecules. The first PDZ domains of ZO-1, ZO-2, and ZO-3 directly bind to the carboxy-termini of claudins in vitro (Itoh et al. 1999b). ZO-1 also binds to the TJ-associated integral membrane proteins occludin and JAM-A (Furuse et al. 1994; Fanning et al. 1998; Bazzoni et al. 2000; Itoh et al. 2001; Li et al. 2005), and the cytoplasmic proteins cingulin and ZONAB (Corndenonsi et al. 1999; Balda et al. 2003). ZO-1, ZO-2, and ZO3 can interact with actin filaments (Itoh et al. 1997, 1999b; Fanning et al. 1998, 2002; Wittchen et al. 1999). ZO-1 and ZO-2 form heterodimers through their second PDZ domains (Utepbergenov et al. 2006). ZO-1 and ZO-3 also form heterodimers, but trimer formation between ZO-1, ZO-2, and ZO-3 has not been detected (Wittchen et al. 1999).
Roles of ZO-1 and ZO-2 in TJ Formation
Among the three TJ MAGUK proteins, ZO-1 and ZO-2 have been shown to be indispensable for TJ formation in epithelial cells. Targeted disruption of the ZO-1 gene and RNAi-mediated depletion of ZO-2 protein in mouse epithelial cells without ZO-3 expression results in deficient TJ formation (Umeda et al. 2006). Using these cells, the role of TJ MAGUKs in TJ strand formation was further characterized. Re-expression of either ZO-1 or ZO-2 recovered normal TJ formation in these cells, whereas the introduction of ZO-3 did not. A deletion construct of ZO-1 containing only three PDZ domains did not rescue the phenotype, but a longer construct containing the three PDZ domains, SH3 domain, GUK domain, and the following acidic domain induced normal TJ formation. However, a slightly shorter construct without the acidic domain induced aberrant TJ strand formation on lateral membranes.
These observations indicate that the SH3+GUK+acidic domains of ZO-1 play important roles in the correct localization of ZO-1 as well as the induction of claudin polymerization into TJ strands. Indeed, this region containing the SH3+unique-5 (U5)+GUK+unique-6 (U6) domains is highlighted as the entire SH3-GUK module in terms of the function in TJ formation (Fanning et al. 2007). U5 is the domain that links the SH3 domain and the GUK domain, whereas the U6 domain is immediately adjacent to the GUK domain. The GUK+U6 domains can bind to the SH3 domain, suggesting that the SH3-GUK module forms intramolecular interactions. A ZO-1 mutant lacking the U5 domain expressed in cultured epithelial cells was unable to localize to TJs and was distributed in the cytoplasm. On the other hand, a ZO-1 mutant lacking the U6 domain induced ectopically extended TJ strands to the lateral plasma membrane (Fanning et al. 2007).
Despite critical roles of the SH3-GUK module containing U5 and U6 domains, it remains unclear how ZO-1 is recruited to the correct region of the lateral membrane to induce TJ strand formation. The interaction of the SH3-GUK module of ZO-1 with AJ-associated molecules such as α-catenin and afadin might be implicated in this recruitement (Itoh et al. 1997; Yokoyama et al. 2001). Interestingly, in ZO-1/ZO-2/ZO-3-depleted epithelial cells, artificial anchoring of the three PDZ domains of ZO-1 to the plasma membrane by myristylation at the amino terminus did not induce TJ formation, whereas dimerization of this construct induced TJ formation throughout the lateral membrane (Umeda et al. 2006). Therefore, dimerization of ZO-1 (or ZO-2) just beneath the plasma membrane may trigger TJ formation, for instance by inducing claudin polymerization. The SH3-GUK module of ZO-1 may be involved in this dimerization, as proposed for the other MAGUK proteins hDlg and hCASK (Nix et al. 2000).
The roles of ZO-1 and ZO-2 are not limited to TJ formation. In calcium switch assays, ZO-1/ZO-2/ZO-3-depleted epithelial cells exhibit a delay in the transition from spotlike AJs to beltlike AJs with linear actin cables. Depletion of ZO-1 in epithelial cells causes a delay in TJ formation (Umeda et al. 2004; McNeil et al. 2006), but this phenotype may also be because of a delay in AJ formation. ZO-1 and ZO-2 are required for Rac1 activation and myosin II recruitment during linear AJ formation (Ikenouchi et al. 2007; Yamazaki et al. 2008). Therefore, ZO-1 and ZO-2 appear to be involved in sequential steps in the assembly of AJ and TJ as well as their physical segregation in the membrane.
CONCLUDING REMARKS
The major molecular components of the core structure of TJs have been identified and characterized. Claudins are the main constituent of TJ strands, and cytoplasmic plaque proteins ZO-1 and ZO-2 directly regulate TJ strand formation and location in epithelial cells. However, many issues concerning the detailed structure of TJ strands remain unsolved.
A more complete understanding of the fine structure of TJ strands will require analysis by structural biological approaches, which may also provide important information about the size and charge selectivity of TJs (see review by Anderson and Van Itallie 2009). Furthermore, little is known about the mechanism of the ZO-1/ZO-2-mediated control of claudin polymerization within membranes. Recent studies indicate that dimerization of the claudin-interacting domains of ZO-1 and ZO-2 triggers claudin polymerization. On the other hand, it is unlikely that all claudins in the TJ bind to ZO-1 and ZO-2. Further research into the SH3-GUK module (the regulatory domains for TJ MAGUKs) of ZO family members will be key to understanding this problem. Interestingly, claudins lacking the carboxy-terminal cytoplasmic region, which interacts with ZO-1 and ZO-2, reconstitute TJ strands in mouse L fibroblasts, whereas TJs are not formed in epithelial cells lacking ZO-1 and ZO-2. Clarification of the molecular mechanisms behind this discrepancy in claudin-based TJ formation between epithelial cells and fibroblasts will provide important information about the polymerization of claudins within the plasma membrane.
A new research area of TJs is how tTJs are formed. Although tricellulin is required for the formation of normal tTJs and the epithelial barrier function, it is unknown how tricellulin functions in tTJs and how it is recruited there. Further functional analyses of tricellulin as well as the identification of novel components associated with tTJs will lead to better understanding of the molecular mechanism of tTJ formation and biological significance for tTJ sealing.
Finally, the function of occludin, the first identified TJ-associated integral membrane protein, is still not well understood although it was identified more than 15 years ago. It must have an important function because occludin-deficient mice cannot produce offspring (Saitou et al. 2000). Because occludin exhibits structural similarity with tricellulin, these two proteins may share redundant functions, although it is still unknown whether they are incorporated into or localized very closed to claudin-based TJ strands. Further investigation of the relationship between occludin and tricellulin may clarify the common role of these molecules in TJ formation and regulation.
ACKNOWLEDGMENTS
I would like to thank Drs. Hiroyuki Sasaki (The Jikei University School of Medicine) and Toshio Imai (KAN Research Institute, Inc.) for providing electron micrographs. M.F. is supported by a Grant-in-Aid for Cancer Research and the National Project on Targeted Protein Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Editors: W. James Nelson and Elaine Fuchs
Additional Perspectives on Cell Junctions available at www.cshperspectives.org
REFERENCES
- Anderson JM, Cereijido M 2001. Evolution of ideas on the tight junction. In Tight junctions, 2nd ed. (ed. Cereijido M, Anderson J), pp. 1–18 CRC Press, Boca Raton [Google Scholar]
- Anderson JM, Van Itallie CM 2009. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 1:002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelow S, Ahlstrom R, Yu AS 2008. Biology of claudins. Am J Physiol Renal Physiol 295:867–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono S, Hirai Y 2008. Phosphorylation of claudin-4 is required for tight junction formation in a human keratinocyte cell line. Exp Cell Res 314:3326–3339 [DOI] [PubMed] [Google Scholar]
- Balda MS, Anderson JM 1993. Two classes of tight junctions are revealed by ZO-1 isoforms. Am J Physiol 264:918–924 [DOI] [PubMed] [Google Scholar]
- Balda MS, Garrett MD, Matter K 2003. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol 160:423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Whitney JA, Flores C, Gonzalez S, Cereijido M, Matter K 1996. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J Cell Biol 134:1031–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamforth SD, Kniesel U, Wolburg H, Engelhardt B, Risau W 1999. A dominant mutant of occludin disrupts tight junction structure and function. J Cell Sci 112:1879–1888 [DOI] [PubMed] [Google Scholar]
- Bazzoni G, Martinez-Estrada OM, Orsenigo F, Cordenonsi M, Citi S, Dejana E 2000. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J Biol Chem 275:20520–20526 [DOI] [PubMed] [Google Scholar]
- Brightman MW, Reese TS 1969. Junctions between intimately apposed cell membranes in the vertebrate brain. J Cell Biol 40:648–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citi S, Sabanay H, Jakes R, Geiger B, Kendrick-Jones J 1988. Cingulin, a new peripheral component of tight junctions. Nature 333:272–276 [DOI] [PubMed] [Google Scholar]
- Chalcroft JP, Bullivant S 1970. An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture. J Cell Biol 47:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude P 1978. Morphological factors influencing transepithelial permeability: A model for the resistance of the zonula occludens. J Membr Biol 39:219–232 [DOI] [PubMed] [Google Scholar]
- Claude P, Goodenough DA 1973. Fracture faces of zonulae occludentes from “tight” and “leaky” epithelia. J Cell Biol 58:390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Shieh JT, Pickles RJ, Okegawa T, Hsieh JT, Bergelson JM 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc Natl Acad Sci 98:15191–15196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, D'Atri F, Hammar E, Parry DA, Kendrick-Jones J, Shore D, Citi S 1999. Cingulin contains globular and coiled-coil domains and interacts with ZO-1, ZO-2, ZO-3, and myosin. J Cell Biol 147:1569–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Atri F, Citi S 2001. Cingulin interacts with F-actin in vitro. FEBS Lett 507:21–24 [DOI] [PubMed] [Google Scholar]
- D'souza T, Agarwal R, Morin PJ 2005. Phosphorylation of claudin-3 at threonine 192 by cAMP-dependent protein kinase regulates tight junction barrier function in ovarian cancer cells. J Biol Chem 280:26233–26240 [DOI] [PubMed] [Google Scholar]
- da Silva PP, Kacher B 1982. On tight-junction structure. Cell 28:441–450 [DOI] [PubMed] [Google Scholar]
- Daugherty BL, Ward C, Smith T, Ritzenthaler JD, Koval M 2007. Regulation of heterotypic claudin compatibility. J Biol Chem 282:30005–30013 [DOI] [PubMed] [Google Scholar]
- Dermietzel R 1974. Junctions in the central nervous system of the cat. I. Membrane fusion in central myelin. Cell Tissue Res 148:565–576 [DOI] [PubMed] [Google Scholar]
- Ebnet K, Iden S, Gerke V, Suzuki A 2008. Regulation of epithelial and endothelial junctions by PAR proteins. Front Biosci 13:6520–6536 [DOI] [PubMed] [Google Scholar]
- Fanning AS, Ma TY, Anderson JM 2002. Isolation and functional characterization of the actin binding region in the tight junction protein ZO-1. Faseb J 16:1835–1837 [DOI] [PubMed] [Google Scholar]
- Fanning AS, Jameson BJ, Jesaitis LA, Anderson JM 1998. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem 273:29745–29753 [DOI] [PubMed] [Google Scholar]
- Fanning AS, Little BP, Rahner C, Utepbergenov D, Walther Z, Anderson JM 2007. The unique-5 and -6 motifs of ZO-1 regulate tight junction strand localization and scaffolding properties. Mol Biol Cell 18:721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE 1963. Junctional complexes in various epithelia. J Cell Biol 17:375–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend DS, Gilula NB 1972. Variations in tight and gap junctions in mammalian tissues. J Cell Biol 53:758–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K 1995. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J Cell Sci 108:3443–3449 [DOI] [PubMed] [Google Scholar]
- Furuse M 2009. Knockout animals and natural mutations as experimental and diagnostic tool for studying tight junction functions in vivo. Biochim Biophys Acta 1788:813–819 [DOI] [PubMed] [Google Scholar]
- Furuse M, Tsukita S 2006. Claudins in occluding junctions of humans and flies. Trends Cell Biol 16:181–188 [DOI] [PubMed] [Google Scholar]
- Furuse M, Sasaki H, Tsukita S 1999. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol 147:891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Sasaki H, Fujimoto K, Tsukita S 1998b. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol 143:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S 1998a. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol 141:1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S 1993. Occludin: A novel integral membrane protein localizing at tight junctions. J Cell Biol 123:1777–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S 1994. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol 127:1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough DA, Revel JP 1970. A fine structural analysis of intercellular junctions in the mouse liver. J Cell Biol 45:272–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Davies C, Southwood CM, Frolenkov G, Chrustowski M, Ng L, Yamauchi D, Marcus DC, Kachar B 2004. Deafness in Claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J Neurosci 24:7051–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA 1999. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 99:649–659 [DOI] [PubMed] [Google Scholar]
- Guillemot L, Paschoud S, Pulimeno P, Foglia A, Citi S 2008. The cytoplasmic plaque of tight junctions: A scaffolding and signalling center. Biochim Biophys Acta 1778:601–613 [DOI] [PubMed] [Google Scholar]
- Gumbiner B, Lowenkopf T, Apatira D 1991. Identification of a 160-kDa polypeptide that binds to the tight junction protein ZO-1. Proc Natl Acad Sci 88:3460–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S 2002. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J Biol Chem 277:455–461 [DOI] [PubMed] [Google Scholar]
- Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR 1998. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol 141:199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S, Tajima M, Yao I, Nishimura W, Mori H, Hata Y 2003. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol Cell Biol 23:4267–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide N, Hata Y, Nishioka H, Hirao K, Yao I, Deguchi M, Mizoguchi A, Nishimori H, Tokino T, Nakamura Y, Takai Y 1999. Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene 18:7810–7815 [DOI] [PubMed] [Google Scholar]
- Ikari A, Matsumoto S, Harada H, Takagi K, Hayashi H, Suzuki Y, Degawa M, Miwa M 2006. Phosphorylation of paracellin-1 at Ser217 by protein kinase A is essential for localization in tight junctions. J Cell Sci 119:1781–1789 [DOI] [PubMed] [Google Scholar]
- Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S 2005. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 171:939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J, Sasaki H, Tsukita S, Furuse M, Tsukita S 2008. Loss of occludin affects tricellular localization of tricellulin. Mol Biol Cell 19:4687–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J, Umeda K, Tsukita S, Furuse M, Tsukita S 2007. Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J Cell Biol 176:779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T, Chiba H, Kojima T, Fujibe M, Soma T, Miyajima H, Nagasawa K, Wada I, Sawada N 2003. Cyclic AMP induces phosphorylation of claudin-5 immunoprecipitates and expression of claudin-5 gene in blood-brain-barrier endothelial cells via protein kinase A-dependent and -independent pathways. Exp Cell Res 290:275–288 [DOI] [PubMed] [Google Scholar]
- Itoh M, Morita K, Tsukita S 1999b. Characterization of ZO-2 as a MAGUK family member associated with tight as well as adherens junctions with a binding affinity to occludin and α catenin. J Biol Chem 274:5981–5986 [DOI] [PubMed] [Google Scholar]
- Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S 1999a. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol 147:1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Nagafuchi A, Moroi S, Tsukita S 1997. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α catenin and actin filaments. J Cell Biol 138:181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, Tsukita S 2001. Junctional adhesion molecule (JAM) binds to PAR-3: A possible mechanism for the recruitment of PAR-3 to tight junctions. J Cell Biol 154:491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, Fujimoto T, Tabuse Y, Kemphues KJ, Ohno S 1998. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol 143:95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeansonne B, Lu Q, Goodenough DA, Chen YH 2003. Claudin-8 interacts with multi-PDZ domain protein 1 (MUPP1) and reduces paracellular conductance in epithelial cells. Cell Mol Biol 49:13–21 [PubMed] [Google Scholar]
- Kitajiri S, Miyamoto T, Mineharu A, Sonoda N, Furuse K, Hata M, Sasaki H, Mori Y, Kubota T, Ito J, et al. 2004. Compartmentalization established by claudin-11-based tight junctions in stria vascularis is required for hearing through generation of endocochlear potential. J Cell Sci 117:5087–5096 [DOI] [PubMed] [Google Scholar]
- Kubota K, Furuse M, Sasaki H, Sonoda N, Fujita K, Nagafuchi A, Tsukita S 1999. Ca(2+)-independent cell-adhesion activity of claudins, a family of integral membrane proteins localized at tight junctions. Curr Biol 9:1035–1038 [DOI] [PubMed] [Google Scholar]
- Kumar NM, Gilula NB 1996. The gap junction communication channel. Cell 84:381–388 [DOI] [PubMed] [Google Scholar]
- Lane JN 2001. Tight junctions in invertebrates. In Tight junctions, 2nd ed. (ed. Cereijido M and Anderson J), pp. 39–59, CRC Press, Boca Raton [Google Scholar]
- Laura RP, Ross S, Koeppen H, Lasky LA 2002. MAGI-1: A widely expressed, alternatively spliced tight junction protein. Exp Cell Res 275:155–170 [DOI] [PubMed] [Google Scholar]
- Lemmers C, Medina E, Delgrossi MH, Michel D, Arsanto JP, Le Bivic A 2002. hINADl/PATJ, a homolog of discs lost, interacts with crumbs and localizes to tight junctions in human epithelial cells. J Biol Chem 277:25408–25415 [DOI] [PubMed] [Google Scholar]
- Li Y, Fanning AS, Anderson JM, Lavie A 2005. Structure of the conserved cytoplasmic C-terminal domain of occludin: Identification of the ZO-1 binding surface. J Mol Biol 352:151–164 [DOI] [PubMed] [Google Scholar]
- Madara JL 1987. Intestinal absorptive cell tight junctions are linked to cytoskeleton. Am J Physiol 253:171–175 [DOI] [PubMed] [Google Scholar]
- Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A, Ruco L, Villa A, et al. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J Cell Biol 142:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KM, Skare IB, Stankewich MC, Furuse M, Tsukita S, Rogers RA, Lynch RD, Schneeberger EE 1996. Occludin is a functional component of the tight junction. J Cell Sci 109:2287–2298 [DOI] [PubMed] [Google Scholar]
- McNeil E, Capaldo CT, Macara IG 2006. Zonula occludens-1 function in the assembly of tight junctions in Madin-Darby canine kidney epithelial cells. Mol Biol Cell 17:1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Rahner C, Mitic LL, Anderson JM, Van Itallie CM 2000. Occludin localization at the tight junction requires the second extracellular loop. J Membr Biol 178:235–247 [DOI] [PubMed] [Google Scholar]
- Mitic LL, Unger VM, Anderson JM 2003. Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci 12:218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Sasaki H, Furuse M, Tsukita S 1999. Endothelial claudin: Claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol 147:185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasdala I, Wolburg-Buchholz K, Wolburg H, Kuhn A, Ebnet K, Brachtendorf G, Samulowitz U, Kuster B, Engelhardt B, Vestweber D, et al. 2002. A transmembrane tight junction protein selectively expressed on endothelial cells and platelets. J Biol Chem 277:16294–16303 [DOI] [PubMed] [Google Scholar]
- Nix SL, Chishti AH, Anderson JM, Walther Z 2000. hCASK and hDlg associate in epithelia, and their src homology 3 and guanylate kinase domains participate in both intramolecular and intermolecular interactions. J Biol Chem 275:41192–41200 [DOI] [PubMed] [Google Scholar]
- Nunes FD, Lopez LN, Lin HW, Davies C, Azevedo RB, Gow A, Kachar B 2006. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J Cell Sci 119:4819–4827 [DOI] [PubMed] [Google Scholar]
- Ohnishi H, Nakahara T, Furuse K, Sasaki H, Tsukita S, Furuse M 2004. JACOP, a novel plaque protein localizing at the apical junctional complex with sequence similarity to cingulin. J Biol Chem 279:46014–46022 [DOI] [PubMed] [Google Scholar]
- Powell DW 1981. Barrier function of epithelia. Am J Physiol 241:275–288 [DOI] [PubMed] [Google Scholar]
- Riazuddin S, Ahmed ZM, Fanning AS, Lagziel A, Kitajiri S, Ramzan K, Khan SN, Chattaraj P, Friedman PL, Anderson JM, et al. 2006. Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet 79:1040–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh MH, Liu CJ, Laurinec S, Margolis B 2002. The carboxyl terminus of zona occludens-3 binds and recruits a mammalian homologue of discs lost to tight junctions. J Biol Chem 277:27501–27509 [DOI] [PubMed] [Google Scholar]
- Saitou M, Fujimoto K, Doi Y, Itoh M, Fujimoto T, Furuse M, Takano H, Noda T, Tsukita S 1998. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J Cell Biol 141:397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Furuse M, Sasaki H, Schulzke JD, Fromm M, Takano H, Noda T, Tsukita S 2000. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell 11:4131–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara A, Furuse M, Saitou M, Ando-Akatsuka Y, Tsukita S 1997. Possible involvement of phosphorylation of occludin in tight junction formation. J Cell Biol 137:1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui C, Furuse K, Mimori-Kiyosue Y, Furuse M, Tsukita S 2003. Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc Natl Acad Sci 100:3971–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze C, Firth JA 1993. Immunohistochemical localization of adherens junction components in blood-brain barrier microvessels of the rat. J Cell Sci 104:773–782 [DOI] [PubMed] [Google Scholar]
- Simionescu M, Simionescu N, Palade GE 1975. Segmental differentiations of cell junctions in the vascular endothelium. The microvasculature. J Cell Biol 67:863–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S 1999. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J Cell Biol 147:195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA 1973. Further observations on the fine structure of freeze-cleaved tight junctions. J Cell Sci 13:763–786 [DOI] [PubMed] [Google Scholar]
- Staehelin LA 1974. Structure and function of intercellular junctions. Int Rev Cytol 39:191–283 [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Mukherjee TM, Williams AW 1969. Freeze-etch appearance of the tight junctions in the epithelium of small and large intestine of mice. Protoplasma 67:165–184 [DOI] [PubMed] [Google Scholar]
- Stevenson BR, Goodenough DA 1984. Zonulae occludentes in junctional complex-enriched fractions from mouse liver: Preliminary morphological and biochemical characterization. J Cell Biol 98:1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA 1986. Identification of ZO-1: A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol 103:755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Ohno S 2006. The PAR-aPKC system: Lessons in polarity. J Cell Sci 119:979–987 [DOI] [PubMed] [Google Scholar]
- Tetzlaff W 1978. The development of a zonula occludens in peripheral myelin of the chick embryo. A freeze-fracture study. Cell Tissue Res 189:187–201 [DOI] [PubMed] [Google Scholar]
- Tsukita S, Furuse M 1999. Occludin and claudins in tight-junction strands: Leading or supporting players? Trend Cell Biol 9:268–273 [DOI] [PubMed] [Google Scholar]
- Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M, Tsukita S 2006. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126:741–754 [DOI] [PubMed] [Google Scholar]
- Umeda K, Matsui T, Nakayama M, Furuse K, Sasaki H, Furuse M, Tsukita S 2004. Establishment and characterization of cultured epithelial cells lacking expression of ZO-1. J Biol Chem 279:44785–44794 [DOI] [PubMed] [Google Scholar]
- Utepbergenov DI, Fanning AS, Anderson JM 2006. Dimerization of the scaffolding protein ZO-1 through the second PDZ domain. J Biol Chem 281:24671–24677 [DOI] [PubMed] [Google Scholar]
- van Deurs B, Koehler JK 1979. Tight junctions in the choroid plexus epithelium. A freeze-fracture study including complementary replicas. J Cell Biol 80:662–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM 1997. Occludin confers adhesiveness when expressed in fibroblasts. J Cell Sci 110:1113–1121 [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM 2006. Claudins and epithelial paracellular transport. Annu Rev Physiol 68:403–429 [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Gambling TM, Carson JL, Anderson JM 2005. Palmitoylation of claudins is required for efficient tight-junction localization. J Cell Sci 118:1427–1436 [DOI] [PubMed] [Google Scholar]
- van Meer G, Gunbiner B, Simons K 1986. The tight junction does not allow lipid molecules to diffuse from one epithelial cell to the next. Nature 322:639–641 [DOI] [PubMed] [Google Scholar]
- Wade JB, Karnovsky MJ 1974. The structure of the zonula occludens. A single fibril model based on freeze-fracture. J Cell Biol 60:168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittchen ES, Haskins J, Stevenson BR 1999. Protein interactions at the tight junction. Actin has multiple binding partners, and ZO-1 forms independent complexes with ZO-2 and ZO-3. J Biol Chem 274:35179–35185 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Umeda K, Wada M, Nada S, Okada M, Tsukita S, Tsukita S 2008. ZO-1- and ZO-2-dependent integration of myosin-2 to epithelial zonula adherens. Mol Biol Cell 19:3801–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Tachibana K, Nakanishi H, Yamamoto Y, Irie K, Mandai K, Nagafuchi A, Monden M, Takai Y 2001. α-Catenin-independent recruitment of ZO-1 to nectin-based cell-cell adhesion sites through afadin. Mol Biol Cell 12:1595–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]