Abstract
Exquisite control of the activity of p53 is necessary for mammalian survival. Too much p53 is lethal, whereas too little permits tumorigenesis. MDM2 and MDM4 are structurally related proteins critical for the control of p53 activity during development, homeostasis, and the response to stress. These two essential proteins regulate both the activation of p53 in response to stress and the recovery of cells following resolution of the damage, yet both are oncogenic when overexpressed. Thus, multiple regulatory circuits ensure that their activities are fine-tuned to promote tumor-free survival. Numerous diverse stressors activate p53, and much research has gone into trying to find commonalities between them that would explain the mechanism by which p53 becomes active. It is now clear that although these diverse stressors activate p53 by different biochemical pathways, one common feature is the effort they direct, through a variety of means, toward disrupting the functions of both MDM2 and MDM4. This article provides an overview of the relationship between MDM2 and MDM4, features the various biochemical mechanisms by which p53 is activated through inhibition of their functions, and proposes some emerging areas for investigation of the p53-mediated stress response.
MDM2 and MDM4 proteins inhibit the p53 tumor suppressor in unstressed cells. Stress signals such as radiation relieve this inhibition by blocking MDM2/4 and/or promoting their degradation.
Regulation of the p53-mediated stress response by the essential inhibitory proteins MDM2 and MDM4 is critical for survival. In response to stressors such as ionizing radiation, p53 induces a number of potentially lethal but tumor-suppressive processes, including cell cycle arrest, senescence, and apoptosis (reviewed by Horn and Vousden 2007). Both MDM2 and MDM4 are critical to surviving the p53-mediated stress response to whole body ionizing irradiation as mice with reduced levels of either protein undergo p53-dependent death after exposure to doses of radiation that are sublethal to wild-type mice (Mendrysa et al. 2003; Terzian et al. 2007). MDM2 and MDM4 are also required to control p53 function during development, as shown by the early embryonic death of mice lacking either MDM2 or MDM4, unless they also lack p53 (Jones et al. 1995; Montes de Oca Luna et al. 1995; Parant et al. 2001; Migliorini et al. 2002).
Although both MDM2 and MDM4 are essential for development, they are detrimental to long-term survival when in excess, because both are oncogenic. Both MDM2 and MDM4 confer the tumorigenic phenotype on cultured cells when experimentally overexpressed (Fakharzadeh et al. 1991; Danovi et al. 2004). In addition, targeted expression of MDM2 in the mammary gland results in tumorigenesis (Lundgren et al. 1997). In people, single nucleotide polymorphisms that reduce expression of either of the orthologs of MDM2 or MDM4 (also referred to as Hdm2 and Hdm4) correlate with increased risk for breast cancer (Bond et al. 2004; Atwal et al. 2009). Approximately 10% of human tumors have been found to overexpress either MDM2 or MDM4 and many of these express wild-type p53 (reviewed in Toledo and Wahl 2006). Because the majority of human cancers express mutant forms of p53, overexpression of MDM2 and MDM4 in the subset of tumors expressing wild-type p53 supports the notion that excessive MDM2 and MDM4 promote tumorigenesis, at least in part, by blocking p53 function. Thus, limiting the activities of MDM2 and MDM4 is important to prevent cancer.
NATURE OF THE RELATIONSHIP BETWEEN MDM2 AND MDM4
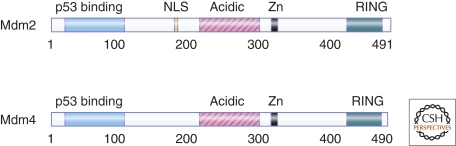
MDM2 and MDM4 are structurally related proteins that each bind to p53 and inhibit its activity (Fig. 1). MDM2 was discovered through its ability to confer the tumorigenic phenotype on murine cells (Fakharzadeh et al. 1991) and later found to bind p53 (Momand et al. 1992). The esoteric name “MDM2” reflects its discovery as a murine gene amplified on double minute chromosomes in a spontaneously transformed cell line (Fakharzadeh et al. 1991). Two genes coamplified with MDM2, MDM1, and MDM3, encode proteins that are neither oncogenic nor related structurally to MDM2 (Fakharzadeh et al. 1991). A few years after MDM2 was discovered, MDM4, also called MDMX, was identified through a screen for proteins that interact with p53 and found to be highly homologous to MDM2 (Shvarts et al. 1996). The human orthologs of both MDM2 and MDM4 are 491- and 490-amino acid phosphoproteins, respectively, that bind to p53 and inhibit its ability to transactivate gene expression. In addition to the amino-terminal domain that binds to p53, they share a number of structural domains including a central acidic domain and a carboxy-terminal RING finger through which they form MDM2/MDM4 heterodimers (Sharp et al. 1999).
Figure 1.
Domain structure of human homologs of MDM2 and MDM4. The amino-termini of both proteins bind p53. Only MDM2 has a nuclear localization signal (NLS). The central acidic region of MDM2, but not MDM4, binds ribosomal proteins. The RING finger domains are required for heterodimerization between MDM2 and MDM4.
Despite their many similarities, MDM2 and MDM4 differ functionally, as revealed by experiments with genetically engineered mice (reviewed by Marine et al. 2006). Although mouse embryos undergo p53-mediated death in utero when both copies of either MDM2 or MDM4 are deleted from the germline, they fail to develop because of distinct mechanisms. Embryos deficient in MDM2 undergo massive apoptosis before implantation and die, whereas those without MDM2 die postimplantation from either growth arrest or apoptosis (Parant et al. 2001; Migliorini et al. 2002; Chavez-Reyes et al. 2003). The embryonic death of MDM2-deficient mice can be rescued by deletion of the proapoptotic BAX protein, whereas MDM4 null mice can be partially rescued by deletion of the cell cycle arrest protein p21, indicating that different p53-mediated pathways can result in embryonic death (Chavez-Reyes et al. 2003; Steinman et al. 2004). Although all mice homozygous null for either protein die during embryogenesis, 70% of mice heterozygous for both MDM2 and MDM4 are born live (Terzian et al. 2007), demonstrating that the two proteins synergize to restrain p53. In some cell types, the consequences of MDM2 deletion differ from those of MDM4 deletion, suggesting that these proteins also have some nonredundant functions. Tissue-specific deletion of MDM2 in either progenitor neuronal cells or cardiomyocytes results in embryonic lethality, whereas deletion of MDM4 in the same cell population results in milder tissue defects and live births (Grier et al. 2006; Francoz et al. 2006). On a biochemical level, MDM2 deletion results in accumulation of p53 and a concomitant increase in p53-dependent transactivation, whereas MDM4 deletion results in increased p53-dependent transactivation without p53 stabilization (Francoz et al. 2006; Toledo et al. 2006). This difference in outcomes can be ascribed to the ubiquitin ligase (E3) function of MDM2, which is not shared by MDM4 (reviewed in Marine and Lozano 2009). Thus, whereas both MDM2 and MDM4 can block the transcriptional activation function of p53, only MDM2 stimulates p53 degradation through ubiquitylation.
The RING-finger domains of MDM2 and MDM4 are major determinants of their functions toward p53 and each other. The MDM2 RING differs from that of MDM4 by facilitating ubiquitylation of several proteins, including MDM4 (Pan and Chen 2003). MDM2-mediated ubiquitylation of MDM4 facilitates accumulation of MDM4 in the nucleus, where it is degraded (Pereg et al. 2006). MDM4 can influence the ubiquitin ligase function of MDM2 through its own RING-finger domain, which heterodimerizes with that of MDM2 (Gu et al. 2002; Kawai et al. 2007). Under different circumstances, MDM4 can either stimulate or inhibit the E3 ligase function of MDM2 toward p53 (Jackson et al. 2001; Kawai et al. 2007; Barboza et al. 2008). This is important because ubiquitylation of p53 by MDM2 is critical for embryogenesis (Itahana et al. 2007), and is modulated during the stress response, as discussed below. MDM2-mediated ubiquitylation of p53 regulates both p53 turnover and cellular localization. Monoubiquitylation of p53 results in export of p53 to the cytoplasm and polyubiquitylation stimulates proteasome degradation (recently reviewed by Kruse and Gu 2009). MDM2 can ubiquitylate itself, and this activity increases in response to stress, as does its ability to ubiquitylate MDM4.
MDM2 and MDM4 also differ at the genetic level. The MDM2 gene contains a p53-response element and its expression is induced by p53 during the stress response (Chen et al. 1994), whereas MDM4 gene expression is not regulated by p53 (Shvarts et al. 1996). The spectrum of tumors overexpressing MDM2 differs from that overexpressing MDM4 (Toledo and Wahl 2006). For example, MDM4 is overexpressed in some colon cancers, whereas MDM2 is not. In addition, the MDM4 gene was found to be amplified in retinoblastoma, whereas MDM2 was not (Laurie et al. 2006). These differences in gene regulation appear to reflect different roles for MDM2 and MDM4 in both the stress response and tumorigenesis.
DIVERSE OUTCOMES FROM ACTIVATION OF P53 BY DIFFERENT STRESSORS
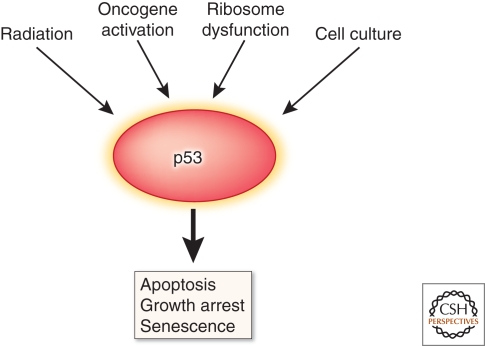
In homeostatic tissues, most if not all functions of p53 are undetectable and become evident only following a stimulus (Mendrysa et al. 2003; Horn and Vousden 2007). When activated, p53 can arrest the cell cycle, induce apoptosis, or promote senescence, exerting a protective effect on the species, the organism, or the cell (Fig. 2). Although any one stressor can stimulate multiple p53-mediated outcomes, a particular response to a stressor may help ensure survival. For example, exposure to ultraviolet radiation and many other DNA-damaging agents causes stabilization and activation of p53, resulting in apoptosis, which eliminates cells with severe DNA damage. Prolonged, excessive, or mistimed oncogene function leads to p53 activation and senescence, limiting the oncogenic potential of preneoplastic cells. Deficient ribosome function promotes p53 activation, promoting cell cycle arrest until translation is restored. Although it is not entirely clear how stressors manifest different outcomes, all of them activate p53 by altering the functions of MDM2 and MDM4, and there are several mechanisms for doing so.
Figure 2.
Multiple stressors activate p53 with several possible outcomes. Apoptosis, senescence, and growth arrest have each been shown to be tumor suppressive. However, if unrestrained, they can be lethal.
MDM2 and MDM4 are implicated as factors that influence whether p53 induces cell cycle arrest, senescence, or apoptosis. Both MDM2 and MDM4 can be detected bound to p53 on its target DNA, where they appear to repress transcription of p53-responsive genes (Tang et al. 2008). Acetylation of p53 in response to some stressors abrogates the binding of MDM2 and MDM4 to p53 at most but not all p53-responsive promoters. By directly repressing only those promoters on which they are bound to p53, MDM2 and MDM4 could influence whether cell cycle arrest genes such as p21 are induced in favor of proapoptotic genes such as BAX (recently reviewed by Kruse and Gu 2009). The ubiquitin ligase function of MDM2 also appears to contribute to the decision between growth arrest and apoptosis by stimulating the degradation of both upstream and downstream effectors of p53. When pharmacologically released from p53, MDM2 stimulates the turnover of hnRNP K, a transcriptional co-factor that assists p53 in transactivating the p21 gene (Enge et al. 2009). Under these circumstances, MDM2 also stimulates the degradation of the p21 protein, such that the resulting low levels of p21 are insufficient to establish growth arrest and fail to protect the cell from apoptosis. The kinase HIPK2, which phosphorylates p53 on serine 46 in response to some stressors, is an upstream regulator of p53 that is ubiquitylated by MDM2 (Rinaldo et al. 2007). Phosphorylation of p53 on serine 46 promotes apoptosis, and MDM2-mediated degradation of HIPK2 reduces the amount of p53 phosphorylated at serine 46, inhibiting apoptosis. A proteosome-independent function of the ubiquitin ligase function of MDM2 is also involved in the decision between cell cycle arrest and apoptosis. In stressed cell types prone to undergo apoptosis, p53 is transported to the mitochondria, where it stimulates apoptosis by blocking the function of the antiapoptotic Bcl2 protein. MDM2 has been shown to facilitate this translocation through monoubiquitylation of p53 (Marchenko et al. 2007). In sum, multiple, sometimes nonredundant and sometimes overlapping pathways converge to regulate the p53-mediated response to stress with consequences that can be either dire or life-saving.
THE ROLES OF MDM2 AND MDM4 IN THE RESPONSE TO IONIZING RADIATION
Ionizing radiation is an important stressor that activates both the growth suppressive and apoptotic functions of p53 (reviewed by Kastan 2008). p53 is a major determinant of organismal survival following exposure to ionizing radiation because its activation in response to this stress results in depletion of hematopoietic cells (Westphal et al. 1998). Whereas wild-type mice die within 10–20 days after exposure to a dose of 10 Gray, p53 null mice seem unperturbed. On a histological level, bone marrow and spleen from irradiated p53-null mice do not show the radiation damage seen in these tissues from irradiated wild-type mice (Westphal et al. 1998). The importance of MDM2 and MDM4 in regulating p53 activity in the response to ionizing radiation is shown by the observation that mice with reduced levels of either protein are radiosensitive, dying following exposure to doses of radiation that are sublethal to wild-type mice (Mendrysa et al. 2003; Terzian et al. 2007). Moreover, unirradiated mice engineered to have low (30% of wild type) levels of MDM2 appear as though they have been irradiated (Mendrysa et al. 2003), suggesting that p53 is poised to act in the absence of stress and that MDM2 prevents it from doing so.
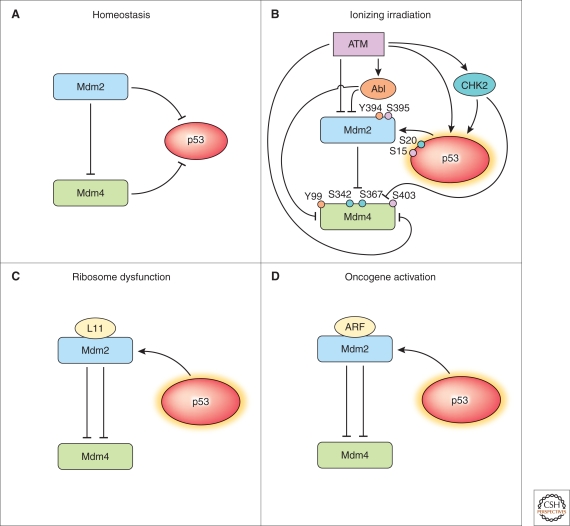
The major signal for the response to ionizing radiation is transmitted by a kinase mutated in the human condition, ataxia telangiectasia, ATM (for ataxia telangiectasia mutated) (reviewed by Kastan 2008). ATM phosphorylates the amino terminus of human p53 on serine 15 such that its affinity for MDM2 and MDM4 is reduced (Fig. 3). This modification alone would be expected to result in p53 accumulation. However, ATM guarantees an unambiguous p53 response by simultaneously stimulating the rapid degradation of both MDM2 and MDM4 by phosphorylating the c-termini of both proteins (Stommel and Wahl 2005; Chen et al. 2005). ATM also activates a second kinase, c-Abl, which phosphorylates MDM2 at tyrosine 394 (Goldberg et al. 2002) and MDM4 at tyrosine 99 (Zuckerman et al. 2009), reducing their ability to inhibit p53. Another ATM-activated kinase, CHK2, phosphorylates additional sites within MDM4, facilitating its association with 14-3-3 proteins and accumulation in the nucleus where it is ubiquitylated by MDM2 (Okamoto et al. 2005; Pereg et al. 2006). Additionally, CHK2-mediated phosphorylation of MDM4 disrupts its interaction with a deubiquitinating enzyme, “herpesvirus associated ubiquitin-specific protease” (HAUSP), thereby allowing unopposed ubiquitylation and proteasomal degradation (Meulmeester et al. 2005; Pereg et al. 2006). The importance of phosphorylation of MDM4 in allowing p53 to mount a stress response to ionizing radiation was recently shown by the radioresistance of mice in which the serines phosphorylated by ATM and CHK2 had been mutated to alanine (Wang et al. 2009). Compared with wild-type mice, the mutant mice mounted a diminished p53-mediated transcriptional response, had fewer apoptotic lymphocytes, and withstood higher doses of ionizing radiation. Thus, ATM uses a multipronged approach to maximally activate p53 during the response to radiation.
Figure 3.
Pathways for regulation of p53 by MDM2 and MDM4 under different conditions. (A) In homeostatic conditions (e.g., unstressed, adult tissues), MDM2 and MDM4 inhibit p53. (B) Following exposure to ionizing radiation, ATM phosphorylates several substrates to block the abilities of MDM2 and MDM4 to inhibit p53. (C) Ribosomal dysfunction leads to direct binding of L11 to MDM2, and to a redirection of its ubiquitin ligage activity away from p53 to MDM4. (D) Oncogenic activation causes increased expression of ARF, which binds to MDM2 and inhibits it from ubiquitylating p53.
In cells that survive the stress response, the activities of p53, MDM2, and MDM4 must be temporally regulated first to allow p53 to exert its function and then to restrain p53 such that the cell can recover. However, little is known about these regulatory events. It has been hypothesized that, in the early stages of the response to ionizing radiation, the ubiquitin ligase function of MDM2 becomes redirected from p53 to MDM4 and to MDM2 itself (Okamoto et al. 2005) with the degradation of MDM2 and MDM4 augmenting the activation of p53. p53 then exerts its growth suppressive or apoptotic function, and the cell either undergoes growth arrest or dies. The expression of MDM2 is induced in response to ionizing radiation through a p53-response element in intron 2 of the MDM2 gene, upstream of the initiation codon for full-length MDM2 (Juven et al. 1993; Wu et al. 1993; Chen et al. 1994). This negative feedback loop is thought to be critical for the recovery of cells following exposure to stressors, although this has not been formally shown. Another mechanism that may be critical for the recovery phase is the dephosphorylation of MDM2 by the Wip1 phosphatase (Lu et al. 2007). Wip1 dephosphorylates MDM2 at serine 395, the site phosphorylated by ATM. In this way, Wip1 facilitates the interaction between MDM2 and p53, and suppresses the autoubiquitylation of MDM2. The delayed induction of Wip1 in the damage response makes it a compelling candidate for a major determinant of the recovery phase. Insight into the recovery phase is likely to be accelerated by widespread adoption of the approach of Lahav et al. (reviewed in Batchelor et al. 2009), who study single cells to observe oscillations in the levels and activities of p53 in the stress response that are hidden in studies of populations. This appears to be a promising approach to discover the mechanisms underlying the activation and deactivation of p53 in the stress response.
p53 clearly suppresses tumorigenesis in response to ionizing radiation (Kemp et al. 1994), yet the relationship between the acute, apoptosis-mediated, pathological response to ionizing radiation and tumor suppression is not entirely clear, as revealed by Evan and colleagues, who devised an elegant strategy to investigate the stage of the stress response at which p53 exerts its tumor suppressive effect in irradiated tissues (Christophorou et al. 2006). They took advantage of mice genetically engineered to express a p53 protein that could be switched from functional to inactive at will. They irradiated the mice when p53 was either functional or inactive and then allowed recovery with p53 in the inactive state. Although only the mice that were irradiated when p53 was in the functional state showed a pathological response to radiation (e.g., an increase in the percentage of apoptotic lymphocytes), they developed tumors at the same rate as those irradiated with inactive p53. This observation suggests that the immediate p53-mediated stress response to irradiation is not tumor suppressive, or is less critical than a later p53 action. Even more surprisingly, Christophorou et al. (2006) found that converting p53 to the functional state eight days post-irradiation protected the mice against lymphomagenesis even though the acute, pathological response to irradiation was undetectable in these circumstances. Thus, at least in this experimental system, p53 exerts a tumor suppressive function that is independent of the acute response to ionizing radiation. The mechanism behind this tumor suppression is not understood, but preliminary results show it is dependent on expression of the alternative reading frame (ARF) tumor suppressor, which inactivates MDM2 to allow p53 to function (Christophorou et al. 2006) (see section below on oncogene stress). Additional, perhaps even more ingenious experiments, may be necessary to elucidate the tumor suppressive function of p53 following irradiation.
MDM2 AND MDM4 INFLUENCE THE RESPONSE TO ULTRAVIOLET RADIATION
Ultraviolet (UV) light is another type of radiation that induces a p53-mediated stress response that appears critical for tumor suppression. The p53 gene is mutated in over 50% of human squamous cell carcinomas, a type of skin cancer caused by overexposure to UV light (Brash et al. 1991). In an elegant study, Ziegler et al. (1994) found that a majority of UV-induced, precancerous lesions (actinic keratoses) contained p53 mutations with the hallmarks of UV-induce mutagenesis (e.g., C to T and CC to TT transitions resulting from pyrimidine dimers). In the same study, experimental exposure of mice to UV light caused accumulation of p53 and apoptosis in the epidermis. In contrast, p53-minus mice lacked a proficient apoptotic response to UV light. More recently, p53 has been proposed to play a central role in inducing the tanning response, which appears important in protecting against the mutagenic effects of UV light (Cui et al. 2007). Together, these studies suggest that p53 is an important determinant of both cell survival and cancer susceptibility following UV exposure.
UV and ionizing radiation activate p53 through different but similar signaling cascades. Although ionizing radiation causes mainly DNA double strand breaks, UV light in the UVB wavelengths causes covalent adducts and single stranded breaks that block replication (reviewed in Marrot and Meunier 2007). These different types of damage are detected by different sensors which activate different kinases. Whereas ATM is the major transducer of the response to ionizing radiation, the “ATM and Rad 9-related protein” (ATR) is activated preferentially by UV light. Like ATM, ATR phophorylates p53, MDM2, and MDM4. ATR also phosphorylates and activates CHK1, a kinase similar to CHK2 (reviewed by Stracker et al. 2009), and CHK1 phosphorylates both MDM2 and MDM4, resulting in degradation of MDM2 and MDM4 and accumulation of active p53. Thus, like the response to ionizing radiation, the UV response stimulates multiple kinases to activate p53.
The dose of UV light influences the p53-mediated stress response as measured by the timing and magnitude of the increases in p53 and MDM2 (Latonen et al. 2001). At low doses, phosphorylation may be the dominant determinant of the response, whereas at higher doses, blocks to transcription and translation of MDM2 and MDM4 contribute to activate p53. UV light also results in increased transcription of alternatively spliced MDM2 and MDM4 gene products (Chandler et al. 2006), the consequences of which are not completely understood. Moreover, high doses of UV light may also activate novel functions of MDM2 or MDM4. In response to lethal but not sublethal doses of UV light, MDM4 translocates to the mitochondria, where it blocks Bcl2 function and promotes apoptosis (Mancini et al. 2009). This counterintuitive finding suggests that MDM4 has pro- or anti-apoptotic functions under certain circumstances, through which it could influence the probability of cell survival. It will be important to clarify the multiple roles of MDM2 and MDM4 that contribute to the initial stress response and to the recovery of cells following resolution of the damage.
RIBOSOMAL STRESS ACTIVATES P53 BY INHIBITING MDM2 AND MDM4
Ribosomal stress is medically relevant. Heterozygous mutations in ribosomal protein genes are associated with a predisposition to nerve sheath tumors in zebrafish (Lai et al. 2009) and to Diamond-Blackfan syndrome in people (reviewed in Lipton and Ellis 2009). Diamond-Blackfan syndrome is a genetic disorder characterized by congenital anomalies, anemia, and a predisposition to cancer. Mice with mutations in the genes encoding small ribosomal proteins 6, 19, and 20 show features of Diamond-Blackfan syndrome, including small stature, anemia, and abnormally pigmented skin. Recently, Barsh and colleagues (McGowan et al. 2008) discovered that p53 is activated in these mice, and contributes to the small stature, anemia, and dark skin phenotypes. Thus, activation of p53 in response to aberrant ribosome function is physiologically significant.
Although the molecular mechanisms by which p53 is activated in Diamond-Blackfan syndrome have not been elucidated, several ribosomal proteins (L5, L11, and L23) can activate p53 in cultured cells by binding the acidic region of MDM2 and inhibiting ubiquitylation of p53 (Gilkes et al. 2006). Although MDM4 has a similar acidic domain, it does not appear to bind these ribosomal proteins. L11, but not L5 or L23, has been shown to stimulate MDM2-mediated ubiquitylation of MDM4, thereby reducing the level of this p53 inhibitor. Unlike radiation, ribosomal stress does not stimulate phosphorylation of MDM4 serine 367, binding of MDM4 to 14-3-3 protein, nor nuclear localization of MDM4. Less effective inhibition of MDM4 in the response to ribosomal stress may indicate a specific role for MDM4 in this particular stress response. As both MDM4 and MDM2 bind to p53 on DNA, where they could influence the transcriptional program (Tang et al. 2008; Vousden and Prives 2009), these different stress responses may require specific ratios of MDM2 and MDM4 to orchestrate the relevant outcome.
MDM2 AND MDM4 ARE INHIBITED IN RESPONSE TO ONCOGENIC ACTIVATION
The p53-mediated response to oncogenic activation appears critical for tumor suppression (Christophorou et al. 2006; Halazonetis et al. 2008; Efeyan et al. 2009). The term “oncogene activation” refers to a condition in which the function of a proto-oncogene or oncogene product is active either for an aberrantly long time or at an abnormally high level. Disregulated signaling or mutations in proto-oncogenes can lead to oncogene activation. p53 becomes activated in response to oncogene activation through multiple mechanisms, one of which involves the ARF tumor suppressor protein that inhibits MDM2. Under some conditions, oncogene activation has been found to lead to DNA strand breaks (reviewed by Halazonetis et al. 2008). However, the contribution of DNA damage to the p53-mediated stress response to oncogene activation is not yet clear.
ARF is important for the p53-mediated stress response to oncogene activation in cultured cells and in mice (Christophorou et al. 2006; Efeyan et al. 2009). ARF levels rise when oncogenes are active and ARF binds to MDM2, inhibiting its ability to interact with p53 (reviewed by Sherr et al. 2005). Although ARF appears central to the response to oncogenic stress, and oncogenic stress appears critical for tumorigenesis, surprisingly little data directly implicate ARF as an important tumor suppressor in people (Halazonetis et al. 2008). It will be an important mission to delineate the contribution of ARF to human tumor suppression.
CELL CULTURE CAUSES STRESS THAT ACTIVATES P53
Cells in culture typically are under stress because of the high oxygen concentration in the tissue culture incubator (DiMicco et al. 2008). p53 becomes stabilized and induces cell cycle arrest and senescence. This p53-mediated response to culture stress limits the lifespan of primary cells, such as mouse embryo fibroblasts, which spontaneously immortalize if they lack either ARF or p53. Only a portion of the biochemical pathways induced by stress in cultured cells have been confirmed to be physiologically relevant. For example, although p53 induces MDM2 expression constitutively in cultured cells, it does not regulate basal levels of MDM2 in homeostatic tissues (Mendrysa et al. 2000). Thus, it is likely that culture stress contributes to some of the outcomes seen in studies of p53 with experimentally added stressors. For a more in-depth comparison of results from cell culture and animal models, see the review by Toledo and Wahl (2006).
CONCLUDING REMARKS
Despite the tens of thousands of papers published about p53, the p53-mediated response to stress remains a complicated phenomenon that is not completely understood. Several major questions remain unresolved:
What mechanisms regulate the timing of the activation and deactivation of p53 in response to stress?
What is the purpose of so many different ways to activate p53 in response to stress?
How important are the different p53-mediated responses to stress in suppressing tumorigenesis?
Can the lethal effects of p53 be separated pharmacologically from its tumor suppressive function?
What is the role of ARF in tumor suppression in people?
How physiologically relevant are p53-independent functions of MDM2 and MDM4 in the stress response?
Can MDM2 and MDM4 activities be manipulated safely to prevent cancer or reduce radiation sickness?
The answers to these and other questions will aid in the design of strategies to target p53, MDM2, and MDM4 for cancer prevention and therapy (Wade and Wahl 2009).
ACKNOWLEDGMENTS
The author thanks Drs. Guillermina Lozano of MD Anderson Medical Center and Peter Johnson and Allan Weissman of the National Cancer Institute, National Institutes of Health (NIH), for critical and constructive comments. This work was supported by the intramural research program of the NIH National Cancer Institute, Center for Cancer Research.
Footnotes
Editors: Arnold J. Levine and David Lane
Additional Perspectives on The p53 Family available at www.cshperspectives.org
REFERENCES
- Atwal GS, Kirchhoff T, Bond EE, Monagna M, Menin C, Bertorelle R, Scaini MC, Bartel F, Bohnke A, Pempe C, et al. 2009. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene. PNAS doi /10.1073/pnas.0901298106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza JA, Iwakuma T, Terzian T, El-Naggar AK, Lozano G 2008. MDM2 and MDM4 loss regulates distinct p53 activities. Mol Cancer Res 6:947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Loewer A, Lahav G 2009. The ups and downs of p53: Understanding protein dynamics in single cells. Nature Rev Cancer 9:371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119:591–602 [DOI] [PubMed] [Google Scholar]
- Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Pontén J 1991. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci 88:10124–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G 2006. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res 66:9502–9508 [DOI] [PubMed] [Google Scholar]
- Chavez-Reyes A, Parant JM, Amelse LL, Montes de Oca Luna R, Korsmeyer SJ, Lozano G 2003. Switching mechanisms of cell death in MDM2- and MDM4-null mice by deletion of p53 downstream targets. Cancer Res 63:8664–8669 [PubMed] [Google Scholar]
- Chen L, Gilkes DM, Pan Y, Lane WS, Chen J 2005. ATM and CHK2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 24:3411–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Oliner JD, Zhan Q, Fornace AJ Jr, Vogelstein B, Kastan MB 1994. Interactions between p53 and MDM2 in a mammalian cell cycle checkpoint pathway. Proc Natl Acad Sci 91:2684–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Ringshausen I, Ginch AJ, Brown-Swigart L, Evan GI 2006. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature 443:214–217 [DOI] [PubMed] [Google Scholar]
- Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D'Orazio J, Fung CY, Schanbacher CF, Granter SR, et al. 2007. Central role for p53 in the suntan response and pathological hyperpigmentation. Cell 128:853–864 [DOI] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al. 2004. Amplification of MDMX (or MDM4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol 24:5835–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMicco R, Cicalese A, Fumagalli M, Dobreva M, Verrecchia A, Pelicci PG, diFagagna F 2008. DNA damage response activation in mouse embryo fibroblasts undergoing replicative senescence and following spontaneous immortalization. Cell Cycle 7:3601–3606 [DOI] [PubMed] [Google Scholar]
- Efeyan A, Murga M, Martinez-Pastor B, Ortega-Molina A, Soria R, Collado M, Fernandez-Capetillo O, Serrano M 2009. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS ONE 4:doi:10.1371/journal.pone.0005475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enge M, Bao W, Hedström E, Jackson SP, Moumen A, Selivanova G 2009. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell 15:171–183 [DOI] [PubMed] [Google Scholar]
- Fakharzadeh SS, Trusko SP, George DL 1991. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 10:1565–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumount G, Bellefroid E, Marine J-C 2006. MDM4 and MDM2 cooperate to inhibit p53 activity in proliferation and quiescent cells in vivo. Proc Natl Acad Sci 103:3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chen L, Chen J 2006. MDMX regulation of p53 response to ribosomal stress. EMBO J 25:5614–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg Z, Vogt Sionov R, Berger M, Zwang Y, Perets R, Van Etten RA, Oren M, Taya Y, Haupt Y 2002. Tyrosine phosphorylation of MDM2 by c-Abl: Implications for p53 regulation. EMBO J 21:3715–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier J, Xiong S, Elizondo-Fraire AC, Parant J, Lozano G 2006. Tissue-specific differences of p53 inhibition by MDM2 and MDM4. Mol Cell Biol 26:192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, Parant J, Lozano G, Yua Z-M 2002. Mutual Dependence of MDM2 and MDMX in their functional inactivation. J Biol Chem 277:19251–19254 [DOI] [PubMed] [Google Scholar]
- Halazonetis DT, Gorgoulis VG, Bartek J 2008. An Oncogene-induced DNA damage model for cancer development. Science 319:1352–1355 [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH 2007. Coping with stress: Multiple ways to activate p53. Oncogene 26:1306–1316 [DOI] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindström MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y 2007. Targeted inactivation of MDM2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 12:355–366 [DOI] [PubMed] [Google Scholar]
- Jackson MW, Lindstrom MS, Berberich SJ 2001. MDMX binding to ARF affects MDM2 protein stability and p53 transactivation. J Biol Chem 276:25336–25341 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A 1995. Rescue of embryonic lethality in MDM2-deficent mice by absence of p53 Nature 378:206–208 [DOI] [PubMed] [Google Scholar]
- Juven T, Barak Y, Zauberman A, George DL, Oren M 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the MDM2 gene. Oncogene 8:3411–3416 [PubMed] [Google Scholar]
- Kastan MB 2008. DNA Damage Responses: Mechanisms and roles in human disease. Mol Cancer Res 6:517–524 [DOI] [PubMed] [Google Scholar]
- Kawai H, Lopex-Pajares V, Kim MM, Wiederschain D, Yuan Z-M 2007. RING domain-mediated interaction is a requirement for MDM2's E3 ligase activity. Cancer Res 67:6026–6030 [DOI] [PubMed] [Google Scholar]
- Kemp CJ, Wheldon T, Balmain A 1994. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nature Genet 8:66–69 [DOI] [PubMed] [Google Scholar]
- Kruse J-P, Gu W 2009. Modes of p53 regulation. Cell 137:609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K, Amsterdam A, Farrington S, Bronson RT, Hopkins N, Lees JA 2009. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev Dyn 238:76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latonen L, Taya Y, Laiho M 2001. UV radiation induces dose-dependent regulation of p53 response and modulates p53-HDM2 interaction in human fibroblasts. Oncogene 20:6784–6793 [DOI] [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih C-S, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. 2006. Inactivation of the p53 pathway in retinoblastoma. Nature 444:61–66 [DOI] [PubMed] [Google Scholar]
- Lipton JM, Ellis SR 2009. Diamond-Blackfan anemia: Diagnosis, treatment and molecular pathogenesis. Hematol Oncol Clin North Am 23:261–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ma O, Nguyen T-A, Jones SN, Oren M, Donehower L 2007. The Wip1 phosphatase acts as a gatekeeper in the p53-MDM2 autroregulatory loop. Cancer Cell 12:342–354 [DOI] [PubMed] [Google Scholar]
- Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, Lozano G, Rosenberg MP, Finlay CA 1997. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev 11:714–725 [DOI] [PubMed] [Google Scholar]
- Mancini F, Di Conza G, Pellegrino M, Rinaldo C, Prodosmo A, Giglio S, D'Agnano I, Florenzano F, Felicioni L, Buttitta F, et al. 2009. MDM4 (MDMX) localizes at the mitochondria and facilitates the p53-mediated intrinsic-apoptotic pathway. EMBO J 28:1926–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Wolff S, Erster S, Becker K, Moll UM 2007. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J 26:923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine J-C, Lozano G 2009. MDM2-mediated ubiquitylation: p53 and beyond. Cell Death and Differ doi: 10.1038/cdd.2009.68 [DOI] [PubMed] [Google Scholar]
- Marine J-C, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G 2006. Keeping p53 in check: Essential and synergistic functions of MDM2 and MDM4. Cell Death and Differ 13:927–934 [DOI] [PubMed] [Google Scholar]
- Marrot L, Meunier JR 2008. Skin DNA photodamage and its biological consequences. J Am Acad Dermatol 58:139–148 [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, Angelis M, Myers RM, Attardi LD, Barsh GS 2008. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nature Genet 40:963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa SM, Perry ME 2000. The p53 tumor suppressor protein does not regulate expression of its own inhibitor, MDM2, except under conditions of stress. Mol Cell Biol 20:2023–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa SM, McElwee MK, Michalowski J, O'Leary KA, Young KM, Perry ME 2003. MDM2 is critical for inhibition of p53 during lymphopoiesis and the response to ionizing radiation. Mol Cell Biol 23:462–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG 2005. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 18:565–576 [DOI] [PubMed] [Google Scholar]
- Migliorini D, Denchi EL, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine J-C 2002. MDM4 (MDMX) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 22:5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ 1992. The MDM2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237–1245 [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G 1995. Rescue of early embryonic lethality in MDM2-deficent mice by deletion of p53. Nature 378:203–206 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A, Teunisse A, Migliorini D, Kitabayashi I, Marine J-C, et al. 2005. DNA damage-induced phosphorylation of MDMX at serine 367 activates p53 by targeting MDMX for MDM2-dependent degradation. Mol Cell Biol 25:9608–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Chen J 2003. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol 23:5113–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G 2001. Rescue of embryonic lethality in MDM4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nature 29:92–95 [DOI] [PubMed] [Google Scholar]
- Pereg Y, Lam S, Teunisse A, Biton S, Meulmeester E, Mittelman L, Buscemi G, Okamoto K, Taya Y, Shiloh Y, et al. 2006. Differential roles of ATM- and CHK2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biology 26:6819–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S 2007. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 25:739–750 [DOI] [PubMed] [Google Scholar]
- Sharp DA, Kratowicz SA, Sank MJ, George DL 1999. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 274:38189–38196 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Bertwistle D, den Besten W, Kuo ML, Sugimoto M, Tago K, Williams RT, Zindy F, Roussel MF 2005. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harbor Symp Quant Biol 70:129–137 [DOI] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RCA, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG 1996. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J 15:5349–5357 [PMC free article] [PubMed] [Google Scholar]
- Steinman HA, Sluss HK, Sands AT, Pihan G, Jones SN 2004. Absence of p21 partially rescues MDM4 loss and uncovers an antiproliferative effect of MDM4 on cell growth. Oncogene 23:303–306 [DOI] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM 2005. A new twist in the feedback loop: Stress-activated MDM2 destabilization is required for p53 activation. Cell Cycle 4:411–417 [DOI] [PubMed] [Google Scholar]
- Stracker TH, Usu T, Petrini JH 2009. Taking the time to make important decisions: The checkpoint effector kinases CHK1 and CHK2 and the DNA damage response. DNA Repair doi: 10.1016.dnarep.2009.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W 2008. Acetylation is indispensible for p53 activation. Cell 133:612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T, Wang Y, Van Pelt SC, Box NF, Travis EL, Lozano G 2007. Haploinsufficiency of MDM2 and MDM4 in tumorigenesis and development. Mol Cell Biol 27:5479–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nature Rev Cancer 6:909–923 [DOI] [PubMed] [Google Scholar]
- Toledo F, Krummel KA, Lee CJ, Liu C-W, Luo-Wei R, Tang M, Wahl GM 2006. A mouse p53 mutant lacking the proline-rich domain rescues MDM4 deficiency and provides insight into the MDM2-MDM4-p53 regulatory network. Cancer Cell 9:273–285 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: The growing complexity of p53. Cell 137:413–431 [DOI] [PubMed] [Google Scholar]
- Wade M, Wahl GM 2009. Targeting MDM2 and MDMX in cancer therapy: Better living through medicinal chemistry. Mol Cancer Res 7:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl G 2009. Increased radioresistance and accelerated B cell lymphomas in mice with MDMX mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell 16:33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal CH, Hoyes KP, Canman CE, Huang X, Kastan MB, Hendry JH, Leder P 1998. Loss of atm radiosensitizes multiple p53 null tissues. Cancer Res 58:5637–5639 [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ 1993. The p53-MDM2 autroegulatory feedback loop. Genes Dev 7:1126–1132 [DOI] [PubMed] [Google Scholar]
- Ziegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, Remington L, Jacks T, Brash DE 1994. Sunburn and p53 in the onset of skin cancer. Nature 372:773–776 [DOI] [PubMed] [Google Scholar]
- Zuckerman V, Leons K, Popwicz GM, Silberman I, Grossman T, Marine J-C, Holak TA, Jochemsen AG, Haupt Y 2009. c-Abl phosphorylates Hdmx and regulates its interaction with p53. J Biol Chem 284:4031–4039 [DOI] [PubMed] [Google Scholar]