Abstract
The Darwinian concept of biological evolution assumes that life on Earth shares a common ancestor. The diversification of this common ancestor through speciation events and vertical transmission of genetic material implies that the classification of life can be illustrated in a tree-like manner, commonly referred to as the Tree of Life. This article describes features of the Tree of Life, such as how the tree has been both pruned and become bushier throughout the past century as our knowledge of biology has expanded. We present current views that the classification of life may be best illustrated as a ring or even a coral with tree-like characteristics. This article also discusses how the organization of the Tree of Life offers clues about ancient life on Earth. In particular, we focus on the environmental conditions and temperature history of Precambrian life and show how chemical, biological, and geological data can converge to better understand this history.
“You know, a tree is a tree. How many more do you need to look at?”
–Ronald Reagan (Governor of California), quoted in the Sacramento Bee, opposing expansion of Redwood National Park, March 3, 1966
Environmental temperature significantly influenced the evolution of early life forms. Reconstruction of ancestral DNA sequences is helping to reveal Earth’s temperature history.
The following article addresses a period in life most removed from life’s origins compared with other articles in this collection. The article discusses an advanced form of life that seems to have lived on the order of 3.5–4.0 billion years ago, around the time when life as we know it began to diversify in a Darwinian sense. The life from this geological period is located deep within an illustrated taxonomic tree of life. The hope is that by understanding how early life evolved, we can better understand how life originated. In this sense, the article attempts to travel backwards in time, starting from modern organisms, to understand life’s origin.
The Darwinian concept of evolution suggests that all modern life shares a single common ancestor, often referred to as the last universal common ancestor (LUCA). Throughout evolutionary history, this ancestor has for the most part generated descendants as successive bifurcations in a tree-like manner. This so called Tree of Life, and phylogenetics in general provides much of the framework for the field of molecular evolution. Taxonomic trees allow us to better understand relationships and commonalities shared by life. For instance, a tree may tell us whether a trait or phenotype shared between two organisms is the result of shared-common ancestry (termed homologous traits) or whether the trait has evolved multiple times independent of ancestry (analogous traits such as wings).
Taxonomic trees can be built using diverse sources of information. These can include morphological and phenotypic data at the macro-level down to DNA and protein sequence data at the micro-level. Ideally, trees built from multiple sources of input have identical taxonomic relationships and branching patterns, and such trees are said to be congruent. In practice, however, trees built from morphological data (say, presence or absence of wings) are often different than a tree built from molecular data (DNA or protein sequences). This requires the biologist to determine which of the two data sets is misleading and/or which taxonomic tree-building algorithm is most appropriate to use for a particular data set. Such an artform is common in the field of molecular evolution because rarely are trees congruent when built from two sources of input data.
In light of this fact, we have provided the quote at the beginning of this article as a reflection about the field of molecular evolution and its interpretations of taxonomic trees. Although Reagan was not speaking about taxonomic trees in his quote, the same sort of disconnect exists between evolutionary biologists and molecular biologists (Woese and Goldenfeld 2009), as it did between conservationists and Ronald Reagan. A molecular biologist may be inclined to say that once you have seen one phylogenetic tree, you have seen them all. And in fairness, there is some validity to such a notion because historically a phylogenetic tree could not help a molecular biologist to better describe their system. An evolutionary biologist, however, will argue that individual trees have nuances that can dramatically alter our interpretation of evolutionary processes.
We intend to show in this article that not all (taxonomic) trees look similar and describe identical evolutionary scenarios. We will discuss how our concept of the Tree of Life has changed over the past couple of decades, how trees can be interpreted, and what a tree can tell us about early life. In particular, the article will focus on the temperature conditions of early life because this topic has received much attention over the past few years as a direct result of improved DNA sequencing technology and a better understanding of molecular evolutionary processes. We will also describe how trees can be used to guide laboratory experiments in our attempt to understand ancient life. Lastly, we will discuss how phylogenetic trees will serve as the foundation for an “evolutionary synthetic biology” that should allow us to better understand the evolution of cellular pathways, macromolecular machines such as the ribosome, and other emergent properties of early life.
BACKGROUND
Prokaryotes and Eukaryotes
All natural and physical scientists have been taught that biological classification is the manner in which organisms are categorized according to common or shared traits. Two organisms will be located in close proximity within a classification system if those two organisms have similar characteristics. The greater the number of shared characteristics, the closer the two organisms will be grouped within the classification system.
The ability to classify organisms is probably a reflection of the notion that all living organisms share a common ancestor. In essence then, Darwinian evolution has already created a classification scheme, and it is our job to illustrate this scheme in a taxonomic context. Our ability to recapitulate life’s phylogeny depends of course on our ability to identify all life forms and describe these life forms at a sufficiently detailed level, allowing us to identify shared characteristics resulting from common ancestry.
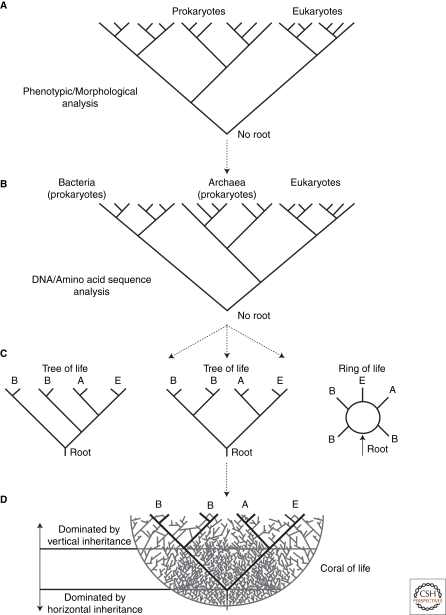
Biologists have made tremendous progress in their classification scheme during the past couple of centuries. For instance, Edouard Chatton outlined his classification scheme that divided life into two categories in the 1920s—he divided life into prokaryotes and eukaryotes (Fig. 1A) (Chatton 1925). Eukaryotic cells had a nucleus that encapsulates their genomic DNA, whereas prokaryotic cells had no nuclear organelle. From an evolutionary perspective, and by definition, prokaryotes (meaning before nucleus) were the progenitors to eukaryotes (dawn of the nucleus). Prokaryotic organisms were microscopic and morphologically similar but metabolically very diverse in their ability to inhabit “extreme” environments. Conversely, eukaryotic organisms were both microscopic and macroscopic and metabolically similar but morphologically they are very diverse because of their multicellularity. The prokaryotic/eukaryotic perspective seemed reasonable at the time because prokaryotes were for the most part morphologically “simple” single-celled organisms, whereas eukaryotes were for the most part “complex” multicellular organisms. The use of simple and complex were of course anthropomorphic. A flatworm would be considered complex because it morphologically looks more similar to humans than say to a simple bacterium, even if that bacterium can live in an anoxic, lightless, and boiling hot environment. The notion of similarity and complexity, however, would be uprooted nearly 50 years after Chatton’s classification scheme as a result of a revolution in biochemistry and molecular biology.
Figure 1.
Tree of Life and its evolution over 100 years. (A) Taxonomy of life based on morphological characteristics (Chatton 1925). (B) Phylogenetic tree of life based on DNA sequence analysis (Woese and Fox 1977). (C) Competing views for the rooting of the phylogenetic tree of life. Initials A, B, and E represent Archaea, Bacteria, and Eukarotes, respectively. The tree on the left is based on membrane architecture and insertion/deletion events in gene (Cavalier-Smith 2002), the tree in the center is based on ancient gene duplication events (Gogarten et al. 1989; Iwabe et al. 1989; Brown and Doolittle 1995; Brown et al. 1997; Gribaldo and Cammarano 1998), and the tree on the right is based on phylogenetic analysis of hundreds of genes (Rivera and Lake 2004). (D) Most recent view about the tree of life in light of vertical and horizontal gene transmission (Fournier et al. 2009).
Three Domains of Life
Although the field of biology is fundamentally concerned with the classification of living and extinct organisms, the field is highly dependent on technological advances that allow the biologist to gather ever more information that can in turn be used to classify organisms (e.g., the microscopic). One such technological advance took place in the 1970s, when chemists developed efficient methods to sequence DNA. The ability to extract information from the heritable material of life and use this information to classify life would revolutionize the classification system.
Carl Woese and George Fox would turn out to be the leaders of the revolution (Woese and Fox 1977). These microbiologists sequenced the DNA that encodes the RNA components of the ribosome from a diverse set of organisms. The DNA sequence information was then used to construct taxonomic trees (or so-called phylogenetic trees when they are generated from sequence data). Using the basic principles of taxonomy, a phylogenetic analysis attempts to group organisms, or gene sequences, based on similarity. The rRNA gene was an ideal gene to study because certain portions of the gene accumulate mutations very slowly so these portions could be used to elucidate ancient evolutionary relationships “deep” in the Tree of Life. The phylogenetic analysis of rRNA sequences by Woese and Fox would show that prokaryotes are divided into two groups, and that one of these groups shared a common ancestor with eukaryotes to the exclusion of the other prokaryotic group (Fig. 1B). This meant that prokaryotes should no longer be considered monophyletic (that all prokaryotes share a common ancestor to the exclusion of eukaryotes) and it meant that our Lemarckian notion of eukaryotes evolving from prokaryotes needed to be abandoned.
The use of DNA sequence information not only changed our view about the classification of life, but it also changed our confidence in taxonomy. Comparing sequences provides a discrete observation on a level at which development proceeds from and evolution acts on. Biologists identified the level at which natural selection and Darwinian evolution enabled life to diversify, thus comparing organisms at this level would allow biologists to accurately illustrate the natural classification scheme of life.
Root of the “Tree of Life”
The Darwinian notion of the Tree of Life implies that a trunk exists from which all branches extend if life does indeed share a common ancestor. The point where the all branches collapse and connect to the trunk is called the root in taxonomy and phylogenetics. Rooting (curiously not termed trunking) trees is theoretically possible if life shared a common ancestor and if a gene made a duplicate copy of itself (paralog) before the three domains of life diverged and both copies have since been retained in all three domains. A phylogenetic analysis of such anciently duplicated paralogs could then generate a tree consisting of two subtrees. Each subtree would be topologically identical to the Tree of Life and the point or node where the two subtrees connected to one another would then represent the root of the phylogeny. Rooting a tree in this manner requires explicit models of sequence evolution because the analysis is attempting to extract a very ancient signal from the DNA/amino acid sequences.
The accumulation of DNA sequence data has allowed biologists to identify multiple paralogous gene families that appear to have undergone duplications before the three domains of life emerged and yet these genes have been evolving slowly enough to identify them clearly as paralogs. Some examples of gene families include ATPases, elongation factors, tRNA synthetases, signal recognition particles, and inter alia (Gogarten et al. 1989; Iwabe et al. 1989; Brown and Doolittle 1995; Brown et al. 1997; Gribaldo and Cammarano 1998). Initial phylogenetic analyses of these families place the root of the tree on the branch that separates bacteria from archaea/eukaryotes (Fig. 1C). This implies that the oldest separation or bifurcation on the Tree of Life was when LUCA split to give rise to the branch that would evolve into bacteria on one side of the tree and a branch that would later serve as the common ancestor of archaea and eukaryotes on the other side of the tree.
This is currently the prevailing view for the root of the Tree of Life. We must note, however, that other studies have criticized details of the approach discussed previously and reach different conclusions. In one alternative view, a ring has replaced the tree properties for illustrating the taxonomy of life. The so-called Ring of Life developed by Jim Lake suggests that genomes have been created by multiple fusion events during the evolution of early life and that these obviate a bifurcating branching pattern in the tree (Fig. 1C) (Rivera and Lake 2004). One advantage of this view is that it considers the likely widespread horizontal or lateral transfer of DNA during early life. Such transfer violates assumptions of the phylogenetic models used to analyze sequence data.
Another approach to root the Tree of Life is to use morphological or phenotypic observations in an attempt to define a clear splitting or bifurcation in the tree. For instance, membrane architecture and insertion/deletion events (indels) in gene sequences have been used to argue that the root of the Tree of Life exists within the bacterial domain, not on the branch that separates bacteria from archaea/eukaryotes (Fig. 1C) (Cavalier-Smith 2002; Cavalier-Smith 2006). Whereas a phylogenetic analysis of paralogous genes is heavily dependent on explicit models of sequence evolution (which can potentially and grossly mislead or bias results), the use of membrane architecture and indel events are conversely independent of models (but which cannot account for parallel or convergent evolution).
Biologists' ability to model evolution has improved substantially over the past decade. Biologists concede that the models are not perfect and that it will be a long road before the models accurately capture all evolutionary processes. Despite this hurdle, they are energized by the prospect of delineating evolution at the sequence level as opposed to being paralyzed by the challenges.
Lateral Gene Transfer: The “Coral of Life”
Biology’s confidence in illustrating the relationships among living organisms has been a rollercoaster ride during the past 20 years. One of the most recent challenges has been the realization that DNA is not solely transmitted in a vertical manner to descendents. Multiple studies have shown that horizontal gene transfer (HGT) has played a major role in the flow of genetic information between organisms—especially deep in life’s phylogeny. This obviously blurs the phylogenetic picture for life because a basic assumption of the Tree of Life is that information only flows in a one-way vertical direction (from parent to offspring in its broadest sense), not a horizontal direction (from one species to another species). This would be the equivalent of violating the linearity of time in the Universe because you cannot be in two places at a single time. HGT essentially allows two identical pieces of DNA to exist at the same time in complete disregard to evolutionary relatedness.
Does this require that we abandon the concept of vertical transmission and the Tree of Life? Yes and no. As mentioned previously, one alternative view is the Ring of Life. This model assumes that life intermixed so much genetic information shortly after LUCA diverged that there are no dominate traces of vertical inheritance until well after the three of domains of life emerged.
Another alternative not yet mentioned is the Coral of Life (Fig. 1D). This concept is being developed by Peter Gogarten, and like the Ring of Life, allows for genetic information to flow in a horizontal manner from species to species (Fournier et al. 2009). Unlike the Ring of Life, however, the Coral of Life permits a dominant path of vertical inheritance to have occurred for ancient life deep in a phylogeny. This path is thought to be present in the tree by the observation that some genes appear to be resistant to HGT. Some genes and their protein products are so entrenched in biochemical and cellular pathways, and protein–protein interactions that have evolved covariantly, that there is no selective advantage, and in all likelihood there would probably be a disadvantage, for a species to acquire a foreign copy of the gene.
These observations have resuscitated the notion of the Tree of Life (or whatever life-like creature it illustrates) and shown that biologists need not be paralyzed by HGT when attempting to understand early life. Now that we have hopefully convinced the reader that some phylogeny of life exists, we now discuss how researchers exploit this phylogeny in attempts to understand early life.
Early Life and Its Temperature History
The temperature history of life is a topic that has interested scientists for at least two centuries since Darwin’s famous statement regarding a warm little pond and the origins of life. Although this article does not deal with the origins of life per se, there has been an equal interest in the temperature history for early life, in particular the close descendents of LUCA.
All of modern life is categorized into one of four temperature ranges. Heat-loving organisms come in the form of thermophiles (grow optimally ∼45° to 80°C) and hyperthermophiles (grow optimally ≥80°C). Cold-loving organisms are called psychrophiles (grow optimally ≤15°C), and middle-loving organisms are called mesophiles (grow optimally ∼15°C to 45°C).
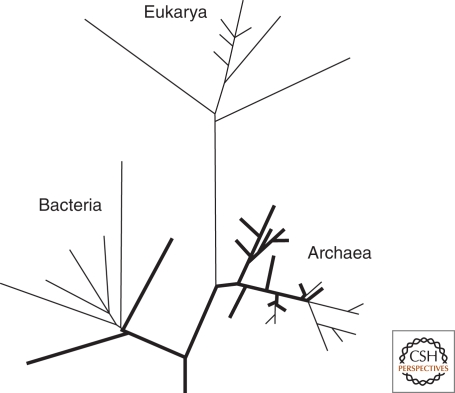
Before the mid 1990s, most conclusions about the temperature history of life were based on chemical considerations and the physical behaviors of biomolecules. This changed with the accumulation of DNA sequence information from a broad range of species and the topology of the inferred phylogenies built from this sequence information. For instance, the first comprehensive discussion about thermostability and ancient life was based on the distribution of hyperthermophilic archaea and bacteria in the Tree of Life (Stetter 1996). The grouping of hyperthermophilic species on short branches near the bases of both the bacterial and archaeal domains of the tree parsimoniously suggested that LUCA was a hyperthermophile (Fig. 2).
Figure 2.
Distribution of modern hyperthermophilic organisms (Stetter 1996). Thick terminal branches lead to hyperthermophiles and thin terminal branches lead to nonhyperthermophiles. Thick internal branches are inferred based on the distribution and relatedness of modern hyperthermophiles.
This conclusion, however, was disputed shortly after it was presented. Some argued for a long-branch attraction artifact in the phylogenetic approach that caused hyperthermophiles to be randomly attracted/grouped instead of grouped because of common ancestry. Others argued that species sampling was sparse and the distribution of hyperthermophiles was coincidental. Still others argued that all hyperthermophiles require a particular protein to survive (reverse gyrase) and this protein evolved through a fusion of two other nonrelated proteins (Forterre 2002). So, if reverse gyrase is required for hyperthermophiles, and if the reverse gyrase cannot be “spontaneously” evolved from a random sequence, then hyperthermophiles must have evolved from a species that lived at a lower temperature.
A genomic-wide approach to understanding the temperature history of early life exploited the observation that the genomic G+C content of modern organisms correlates to the optimal growth temperature of the host organism itself (Galtier et al. 1999). Higher G+C content equates to a higher growth temperature because Gs and Cs form an extra hydrogen bond between base pairs (bps) compared with an A:T bp. The accumulation of extra hydrogen bonds throughout the genome would therefore make it more stable and resistant to heat denaturation. These researchers used models of molecular sequence evolution to infer the G+C content for gene families believed to have been present in LUCA and inferred to have traversed a mostly vertical descent through the Tree of Life with minimal horizontal gene transfer. The researchers concluded that LUCA did not have a genomic G+C content consistent with a hyperthermophilic life style.
To show how sensitive inferences can be to the use of evolutionary models as input for phylogenetic analysis, Di Giulio analyzed the same genomic dataset as previously discussed but used a different phylogenetic algorithm to infer the G+C content of LUCA (Di Giulio 2000). This analysis resulted in an ancient G+C content for LUCA that is consistent with thermophilic and hyperthermophilic life styles.
The previous contention represents one of multiple examples in which analyses have led to competing conclusions. Table 1 presents a condensed chronological list of studies that have attempted to determine the temperature history of early life. The majority of these studies are strictly computational and were not verified in any experimental manner. The next sections show how phylogenetic analysis can be used to guide laboratory experiments to address the temperature history of early life.
Table 1.
Temperature History of Life
| Citation | Taxonomic unit? | Study design and observations | Conclusion |
|---|---|---|---|
| (Stetter 1996) | LUCA | A review of hyperthermophilic archaea and bacteria. In the 16S rRNA-based universal phylogenetic tree, hyperthermophiles are represented in the deepest and shortest lineages. | Hyperthermophile |
| (Forterre 1996) | LUCA | Reverse gyrase is a hyperthermophile-specific protein formed by the association of a putative topoisomerase and helicase. If reverse gyrase is a prerequisite to life at high temperatures, it suggests that hyperthermophiles descended from less thermophilic organisms that possessed these putative enzymes. | Mesophile or thermophile |
| (Galtier et al. 1999) | LUCA | A model of sequence evolution, assuming varying G+C content among lineages and unequal substitution rates among sites, was applied to estimate ancestral base compositions of rRNA sequences. The inferred G+C content of the LUCA is incompatible with survival at a high temperature. | Mesophile |
| (Di Giulio 2000) | LUCA | Reanalysis of the alignment used by Galtier (Science 1999) by maximum parsimony implies that the LUCA may have been a thermophile or hyperthermophile. | Thermophile or hyperthermophile |
| (Brochier and Philippe 2002) | LUB | Applied the heterotacy method on the rRNA bacterial phylogeny and found that the Planctomycetales are the first branching bacterial group; therefore, concluding the most recent common ancestor of bacteria was not hyperthermophilic. | Mesophile or thermophile |
| (Gaucher et al. 2003) | LUB | The most probabilistic ancestral sequences of elongation factor Tu (EF-Tu) were reconstructed at nodes in the bacterial evolutionary tree. These resurrected proteins were assayed and their temperature optima of 55°–65°C corresponds to ancient bacteria living as thermophiles. | Thermophile |
| (Brooks et al. 2004) | LUCA | Inferred amino acid composition of 65 proteins dating to the LUCA by maximum-likelihood using expectation-maximization. The inferred protein sequences were more similar to those found in modern-day thermophilic organisms than mesophilic ones. | Thermophile |
| (Knauth and Lowe 2003; Knauth 2005) | Ocean | Low oxygen isotopes in diagenetic cherts (3.5–3.2 Ga) in South Africa indicate extremely high ocean temperatures of 55°–85°C. Early thermophilic microbes could have been global and not huddled around hydrothermal vents. | Thermophile |
| (Iwabata et al. 2005) | LUCA | Studied the thermostabilty of ancestral isocitrate dehydrogenase (ICDH) mutants. The incorporation of ancestral residues into a modern ICDH led to an increase in thermostability. | Hyperthermophile |
| (Robert and Chaussidon 2006) | Ocean | Study of oxygen isotope ratios of cherts (siliceous sediments) as a measure of the Earth’s climate in the Precambrian. The observed silicon isotope variations imply seawater temperature changes from 70°C 3.5 billion years ago to 20°C about 800 million years ago. | Thermophile |
| (Becerra et al. 2007) | LUCA | A study on the evolution of protein disulfide oxidoreductases (PDO) and its implications to then thermostabilty of the LUCA. The results imply that the LUCA lacked PDO-encoding sequences, and may not have been a thermophile. | Mesophile |
| (Shimizu et al. 2007) | LUCA | Ancestral glycyl-tRNA synthetases (GlyRS) were deduced and residues were introduced in Thermus thermophilus GlyRS. The thermostabilty of these mutants were studied and several were found with higher thermostabilty and activity than wild-type Thermus. These results suggest a highly thermophilic protein translation system in the LUCA. | Hyperthermophile |
| (Gaucher et al. 2008) | LUB | Extensions of earlier work with more than 25 phylogenetically dispersed ancestral EF-Tu’s. The resurrected proteins at basal nodes are compatible with thermophilic environments. | Thermophile |
| (Boussau et al. 2008) | LUCA | A computational analysis of both rRNAs and protein sequences whose results imply that the LUCA was a mesophile. This implies that the two lineages descending from LUCA and leading to the ancestors of Bacteria and Archaea-Eukaryota convergently adapted to high temperatures. | Mesophile |
| (Glansdorff et al. 2008) | LUCA | Archaea have a uniform membrane lipid composition that is suited to life at extreme conditions (heat and pH); in contrast, bacterial membranes show a high variability in composition. The authors suggest that Archaea emerged from a nonthermophilic LUCA under strong selective pressure for adaptation to high temperature; whereas, bacteria were initially nonthermophilic and adapted by convergent evolution to high temperatures. | Mesophile or thermophile |
(LUCA) last universal common ancestor of life; (LUB) last common ancestor of bacteria.
Ancestral Sequence Reconstruction
The recent accumulation of DNA sequence data, combined with advances in evolutionary theory and computational power, have paved the way for innovative approaches to understanding the origins, evolution, and distribution of life and its constituent biomolecules (Pauling and Zuckerkandl 1963; Benner et al. 2002; Gaucher et al. 2004). One approach to understanding ancestral states follows a present-day-backwards strategy, whereby genomic sequences from extant (modern) organisms are incorporated into evolutionary models that estimate the extinct (ancient) character states of genes no longer present on Earth (Fitch 1971; Shih et al. 1993; Benner 1995; Koshi and Goldstein 1996; Schultz et al. 1996; Cunningham 1999; Omland 1999; Pagel 1999; Schultz and Churchill 1999; Chang and Donoghue 2000; Thornton 2004; Hall 2006; Liberles 2007). These inferred ancestral gene sequences act as hypotheses that can be tested in the laboratory through the resurrection of the ancestral proteins themselves. Results from functional assays of the protein products from these ancient genes permit us to accept/reject hypotheses about the sequence themselves, or about their interactions/binding specificities/environments, etc.
Ancestral sequence reconstruction uses standard statistical theory to generate posterior probabilities of different reconstructions given the data at a site from aligned sequences. For each site of the inferred sequence at a phylogenetic node, posterior values for all 20 amino acids are calculated and represent the probability of a particular amino acid occupying a specific site in the protein during its evolutionary history. This posterior probability distribution is calculated from patterns of amino acids in modern sequences as described by a phylogeny, a matrix of amino acid replacement probabilities, amino acid equilibrium (stationary) frequencies, phylogenetic branch lengths, and site-specific replacement rates. The most-probabilistic ancestral sequence (M-PAS) uses the amino acid with the highest posterior probability at each site within the distribution.
RECENT RESULTS
Elongation factor Tu (Bacteria)/1A (Archaea and Eukarya) is an ideal protein family to computationally reconstruct and then resurrect in the laboratory in our attempts to better understand the temperature history of life. There is no evidence that EF genes have been laterally transferred between bacterial lineages, and the thermal stabilities of EFs correlate with the growth temperature of their host organisms. Thus, EFs are optimally stable at temperatures of 15°–45°C, 45°–80°C, and >80°C when isolated from mesophiles, thermophiles, and hyperthermophiles, respectively. This relationship is consistent with a correlation coefficient of 0.91 between melting temperatures of proteins and environmental temperatures of their host organisms (Gromiha et al. 1999).
Reconstruction of ancestral EF sequences were computed across two bacterial phylogenies selected from the literature (Battistuzzi et al. 2004; Ciccarelli et al. 2006). Both phylogenies were constructed from the concatenation of numerous gene families and are thus less susceptible to systematic error compared with phylogenies based on single genes. The two phylogenies capture the main competing views for bacterial relationships. One scenario posits that hyperthermophilic lineages occupy basal branches of the bacterial tree, whereas the other places these lineages in a more derived portion of the tree. To accommodate the latter scenario, a phylogeny was selected in which the Firmicute lineage (void of hyperthermophiles) is located at the base of the bacterial tree, although other topologies have been suggested (Brochier and Philippe 2002).
Thermostability of modern and ancestral EF proteins was monitored using circular dichroism spectroscopy. Melting temperatures (Tm) of two modern EFs were determined. The Tm values for EFs from Escherichia coli and T. thermophilus (HB8) are 42.8°C and 76.7°C. These values highlight the relationship between EF stability and the optimal growth temperature of their respective hosts, ∼40°C and ∼74°C (Williams and da Costa 1992).
Tm values for ancestral EF proteins were determined across the two phylogenies. The thermostability profiles of the ancestral proteins display the same general trend despite the fact that the two phylogenies represent competing hypotheses. Ancestral EF proteins resurrected at basal nodes are compatible with thermophilic environments, whereas ancestral proteins from more derived nodes are compatible with cooler environments. Consistent with this temperature trend is the observation that the node representing the presumed last common ancestor of bacteria (and thus oldest) had the most thermostable protein within each phylogeny (64.8°C and 73.3°C). The similarity in thermostability (<9°C) between these two ancestral proteins is noteworthy because the sequences were identical across only 78% of the amino acid sites.
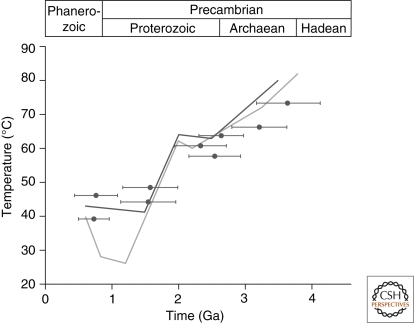
The environmental temperature of ancient bacteria inferred from resurrected EF proteins can be connected to divergence times of major bacterial lineages to gain a more detailed understanding of temperature trends for Precambrian life (Battistuzzi et al. 2004). Divergence estimates from Battistuzzi et al. (2004) were applied to nodes in the current study. Figure 3 highlights the progressive cooling trend of ancient EF proteins from approximately 3.5 billion to 500 million years ago. This temperature trend is strikingly similar to the temperature trend of the ancient ocean inferred from deposition of oxygen and silicon isotopes (Knauth and Lowe 1978; Knauth and Lowe 2003; Robert and Chaussidon 2006).
Figure 3.
Plot of ancestral EF melting temperatures versus geologic time in billions of years (Ga) (Gaucher et al. 2008). Molecular clock estimates and their confidence intervals (horizontal bars) from Battistuzzi et al., using a 2.3 Ga minimum constraint for the great oxidation event (Battistuzzi et al. 2004). Solid lines are temperature curves of the ancient ocean inferred from maximum δ18O (light gray [Knauth and Lowe 1978; Knauth and Lowe 2003], dark gray [Robert and Chaussidon 2006]). Although not shown, an analogous trend is seen with δ30Si isotopes (Robert and Chaussidon 2006).
Reconstruction of ancestral EF proteins throughout the bacterial domain of life suggests that the organisms that hosted these extinct biomolecules lived in environments that have progressively cooled for approximately 3 billion years. This evidence is predicated on multiple assumptions. For instance, it assumes that ancestral sequence reconstruction recapitulates ancient phenotypes and that phylogenies and divergence dates capture the evolutionary relationships and timing of bacterial divergences.
The inability (short of time travel) to know the true relationships of bacterial lineages and their divergence times should not preclude attempts to understand Precambrian life. Rather, a coherent description of ancient life can be generated when empirical evidence from diverse studies converge on analogous conclusions. For instance, the same paleotemperature trend was observed for ancestral EF proteins regardless of the phylogeny. And for the phylogeny with divergence dates, this trend was substantiated when aligned to the inferred paleotemperature curve of the ancient ocean.
These descriptions are particularly useful when they have predictive value. For instance, the last common ancestor of the mitochondrial bacterium is estimated to have lived 1.66–1.88 Ga based on the Tm's for ancestral EF proteins from the node representing the origins of mitochondria (51.0°C–53.0°C). This is consistent with the origins of mitochondria estimated at 1.8 Ga based on a molecular clock (Hedges et al. 2001), despite the controversial nature of the clock (Graur and Martin 2004) and assuming the last common mitochondrial bacterium lived at a time close to the endosymbiotic event between α-proteobacteria and eukaryotic cells.
Our results suggest early life lived at an environmental temperature similar to today’s hot springs. Particular geologic theory and evidence suggests the ancient ocean also had temperatures similar to hot springs (Hoyle 1972; Knauth and Lowe 1978; Knauth and Lowe 2003). As the ocean cooled from 3.5 to 0.5 billion years ago, life may have responded by adapting its range of growth temperatures to correspond to its surrounding environment. This connection assumes early life lived in the ancient ocean, which seems practical based on geologic and biologic constraints such as ocean depth/circulation, land mass exposed to the atmosphere, susceptibility to desiccation, and ultraviolet radiation, among others. Alternatively, it is possible that the inferred paleotemperature trend reflects an ecological trajectory as ancient bacteria transitioned from hot springs/thermal vents to the open ocean.
We note that correlating isotope ratios (d18O and d30Si) to ancient ocean temperatures is controversial (Kasting et al. 2006; Jaffres et al. 2007). In particular, the correlation could be invalid if isotope ratios were caused by variation in seawater composition alone. This would translate into a more temperate ancient ocean and be consistent with ancient glaciation events. The similarity, however, in paleotemperature trends inferred from d18O, d30Si and ancient EF proteins is striking. Further, the overall trend is compatible with biological evolution. For instance, the thermostability of ancient EFs suggest the origins of cyanobacteria occurred at an environmental temperature approximating 63.7°C. This is consistent with an upper temperature limit of typical cyanobacterial mats in hot springs (∼65°C) (Ward et al. 1998).
Overall, the results show that ancient EF thermostability profiles (phenotypes) are robust to uncertainties and potential biases associated with inferring ancestral character states (genotypes). The results also show how ancestral sequence reconstruction can connect physical and natural sciences in our attempts to understand the environmental conditions that hosted early life.
CHALLENGES
Statistical Models of Molecular Sequence Evolution
Despite insightful studies, the field of ancestral sequence reconstruction is encumbered by its inability to know whether inferred sequences truly recapitulate ancestral forms (Williams et al. 2006). Practitioners in the field acknowledge a certain degree of inaccuracy associated with reconstructing ancestral sequences. The concern is not necessarily whether the resurrected form has the exact composition (genotype) of the true ancestral form, but rather that the resurrected form displays the exact behavior (phenotype). A reconstructed sequence can be considered a consensus of a gene distributed throughout a population before species diverge, or before gene duplication. Inaccuracies in a reconstructed sequence can result from sequence variation of the gene itself within an ancient population. Assuming the variants of a homologous gene within a population had the same phenotype at a specific geologic time, it does not necessarily matter which individual genotype is reconstructed.
This assumption is invalid if recombination of individual genotypes generate new phenotypes and if the reconstructed ancestral gene itself represents a consensus of those genotypes. Additional concerns arise if the reconstruction process generates inaccurate sequences because of (1) bias in the evolutionary models used to infer ancestral states or (2) phylogenetic conditions such as long branches and incorrect branching patterns (Felsenstein 1978; Williams et al. 2006; Kelchner and Thomas 2007).
All methods of phylogenetic inference make assumptions about the underlying evolutionary process of their characters and it is these assumptions that determine their relative successes and failures in the estimation of the true phylogeny for a group (Hillis et al. 1992). Much like the manner in which phylogenetic tree building algorithms were developed, tested, and critiqued during the 1990s, we anticipate that ancestral sequence reconstruction algorithms and methods will go through a similar process in the next couple of years now that the reconstruction field is burgeoning. In particular, we anticipate that the development and use of mixture models will play an important role in the development of the field (Gaucher et al. 2002a; Pagel and Meade 2004; Gaucher and Miyamoto 2005).
Experimental Phylogenetics as a Way to Benchmark Ancestral Sequence Reconstruction
Computer simulations of reconstructed ancestral sequences have unequivocally shown the superior performance of the “maximum likelihood” (ML) sequence in terms of accuracy in recovering a true ancestral sequence when it is inferred from tip/leaf/extinct/modern sequences (Huelsenbeck 1995; Yang et al. 1995; Zhang and Nei 1997; Cai et al. 2004; Krishnan et al. 2004; Williams et al. 2006). Although computer simulations of ancestral genotypes and phenotypes are an intriguing approximation for reality, a true benchmark of method performance requires an evaluation of biological sequences and phenotypes measured in the laboratory. As such, it would be useful to use members of the green fluorescent protein family to generate an “experimental phylogeny” (Hillis et al. 1992; Bull et al. 1993). Green fluorescent proteins (GFP) and their varying-colored homologs are widely used as in vivo fluorescent markers and have also been used in experimental paleogenetic studies (Matz et al. 1999; Matz et al. 2002; Ugalde et al. 2004).
Research in our lab is currently generating leaf/tip sequences from an evolved experimental GFP phylogeny that will in turn be used to estimate ancestral genotypes and phenotypes. Because the leaf/tip sequences will be sequentially evolved from nodes on the experimental phylogeny in the laboratory, we will know the true ancestral genotypes and phenotypes. This presents us with the unique opportunity to compare/contrast different approaches attempting to reconstruct ancestral sequences from biologically relevant conditions. Our work represents the first time evolved sequences will be used to benchmark ancestral sequence reconstruction approaches to address issues of ambiguity and bias associated with both reconstructed genotypes and phenotypes.
Sequences at the tips (leaves) of the evolved phylogeny will then be used to computationally reconstruct the inferred ancestral fluorescent sequences at all nodes of the experimental-derived tree. DNA-, codon-, and amino acid-based approaches will be exploited (Yang et al. 1995; Chang et al. 2002; Thornton 2004; Thomson et al. 2005). For each type of data input, we will test different models of sequence evolution and their potential effects on ancestral sequence reconstruction (e.g., transition/transversion ratios, codon tables, amino acid matrices, rate heterogeneity, and others) (Gaucher et al. 2001; Gaucher et al. 2002b; Gaucher and Miyamoto 2005).
RESEARCH DIRECTIONS
We anticipate that our understanding of the temperature history of early life will continue to improve in the coming years. This improvement will not be driven by any single advancement. Rather, a combination of advances in multiple scientific disciplines will enhance our understanding. This is due in large part to the multidisciplinary nature of studying the temperature history of life. For instance, our understanding of taxonomic and evolutionary relationships of bacteria and archaea will greatly enhance our understanding of deep phylogeny, and this in turn will improve our understanding of the environmental conditions that supported these ancient life forms.
More sophisticated models of molecular sequence evolution will help us to better understand ancient life. Such models will improve our ability to accurately construct phylogenetic trees as well as add rigor to ancestral sequence reconstruction methods. The biologists and computational scientists will not be making improvements alone. We anticipate that advances in chemical and geological techniques will also help us define properties of early life.
We are further energized by the prospect of joining evolutionary biology and synthetic biology in our attempts to dissect early life. The next logical extension of molecular reconstruction beyond natural history is to synthetic biology. Synthetic biology means different things to different scientific disciplines (Benner and Sismour 2005; Endy 2005). Surprisingly, however, biologists seem to have taken a backseat to chemists and engineers in the development of this field. It seems apparent that synthetic biology would stand to benefit if “molecular reconstructionists” contributed to its progress. In this way, an evolutionary synthetic biology is formed. A couple of examples come to mind: cellular machines and recombinant genomes.
Cellular machines have a broad range of potentials, from simple expression of heterologous genes for laboratory analysis to the synthesis of minimal artificial cells (Deamer et al. 2002; Martin et al. 2003; Noireaux and Libchaber 2004; Chen et al. 2005). We anticipate that ancestral reconstructed sequences will provide some of the foundation of genetic information for these machines in the future. As a first step, we have shown that ancestral EF proteins can participate in a reconstituted in vitro translation system designed to incorporate unnatural amino acids (unpubl. data). Further, experimental evolution studies of these ancestral genes introduced into laboratory organisms will enhance our biological understanding of adaptive and sequence landscapes, shed light on the transition to protein synthesis by early life, and help elucidate the evolution and adaptation of biochemical pathways. This work will have obvious extensions to natural history and the origins of (early) life.
We also anticipate that the synthesis of recombinant, minimal, and/or ancestral genomes will have a profound effect on our understanding of early life. The Venter Institute, for instance, is in the process of constructing a minimal synthetic Mycoplasma genome (Glass et al. 2006; Lartigue et al. 2007; Gibson et al. 2008). As molecular reconstructionists, we would ask why not construct a complete ancestral biochemical pathway (e.g., operon), or even a complete ancestral genome? The ancestral reconstruction field would no longer be confined to single gene analysis. It is also quite possible that our understanding of what constitutes a sustaining minimal genome required to support life will be altered through ancestral reconstructions. In this way, homologous genes performing two different, but related, functions may share a single common ancestor that performed both of these functions, albeit with less efficiency or specificity.
We anticipate that our understanding of the origins of life and its early evolution will be greatly enhanced by advances in molecular evolution techniques in the coming years. Phylogenetic methods and ancestral sequence reconstruction will continue to be combined in innovative ways to contribute to the Origins of Life field.
Footnotes
Editors: David Deamer and Jack Szostak
Additional Perspectives on The Origin of Life available at www.cshperspectives.org
REFERENCES
- Battistuzzi FU, Feijao A, Hedges SB 2004. A genomic timescale of prokaryote evolution: Insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evolutionary Biol 4:44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra A, Delaye L, Lazcano A, Orgel LE 2007. Protein disulfide oxidoreductases and the evolution of thermophily: Was the last common ancestor a heat-loving microbe? J Mol Evol 65:296–303 [DOI] [PubMed] [Google Scholar]
- Benner SA 1995. Reconstructing Ancient Forms Of Life. J Cell Biochem: 200–200 [Google Scholar]
- Benner SA, Sismour AM 2005. Synthetic biology. Nat Rev Genet 6:533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner SA, Caraco MD, Thomson JM, Gaucher EA 2002. Planetary biology–paleontological, geological, and molecular histories of life. Science 296:864–868 [DOI] [PubMed] [Google Scholar]
- Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M 2008. Parallel adaptations to high temperatures in the Archaean eon. Nature 456:942–945 [DOI] [PubMed] [Google Scholar]
- Brochier C, Philippe H 2002. Phylogeny: A non-hyperthermophilic ancestor for bacteria. Nature 417:244. [DOI] [PubMed] [Google Scholar]
- Brooks DJ, Fresco JR, Singh M 2004. A novel method for estimating ancestral amino acid composition and its application to proteins of the Last Universal Ancestor. Bioinformatics 20:2251–2257 [DOI] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF 1995. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci 92:2441–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Robb FT, Weiss R, Doolittle WF 1997. Evidence for the early divergence of tryptophanyl- and tyrosyl-tRNA synthetases. J Mol Evol 45:9–16 [DOI] [PubMed] [Google Scholar]
- Bull JJ, Cunningham CW, Molineux IJ, Badgett MR, Hillis DM 1993. Experimental molecular evolution of bacteriophage-T7. Evolution 47:993–1007 [DOI] [PubMed] [Google Scholar]
- Cai W, Pei J, Grishin NV 2004. Reconstruction of ancestral protein sequences and its applications. Bmc Evol Biol 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T 2002. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol 52:7–76 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 2006. Rooting the tree of life by transition analyses. Biol Direct 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BSW, Donoghue MJ 2000. Recreating ancestral proteins. Trends In Ecology & Evolution 15:109–114 [DOI] [PubMed] [Google Scholar]
- Chang BS, Jonsson K, Kazmi MA, Donoghue MJ, Sakmar TP 2002. Recreating a functional ancestral archosaur visual pigment. Mol Biol Evol 19:1483–1489 [DOI] [PubMed] [Google Scholar]
- Chatton E 1925. Pansporella perplexa. Réflexions sur la biologie et la phylogénie des protozoaires. Ann Sci Nat Zool (Ser 10) 8:5–84 [Google Scholar]
- Chen IA, Salehi-Ashtiani K, Szostak JW 2005. RNA catalysis in model protocell vesicles. J Am Chem Soc 127:13213–13219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287 [DOI] [PubMed] [Google Scholar]
- Cunningham CW 1999. Some limitations of ancestral character-state reconstruction when testing evolutionary hypotheses. Systematic Biology 48:665–674 [Google Scholar]
- Deamer D, Dworkin JP, Sandford SA, Bernstein MP, Allamandola LJ 2002. The first cell membranes. Astrobiology 2:371–381 [DOI] [PubMed] [Google Scholar]
- Di Giulio M 2000. The universal ancestor lived in a thermophilic or hyperthermophilic environment. J Theor Biol 203:203–213 [DOI] [PubMed] [Google Scholar]
- Endy D 2005. Foundations for engineering biology. Nature 438:449–453 [DOI] [PubMed] [Google Scholar]
- Felsenstein J 1978. Cases in Which Parsimony or Compatibility Methods Will Be Positively Misleading. Systematic Zoology 27:401–410 [Google Scholar]
- Fitch W 1971. Towards defining the course of evolution. Minimum change for a specific tree topology. Syst Zoology 20:406–416 [Google Scholar]
- Forterre P 1996. A hot topic: The origin of hyperthermophiles. Cell 85:789–792 [DOI] [PubMed] [Google Scholar]
- Forterre P 2002. A hot story from comparative genomics: Reverse gyrase is the only hyperthermophile-specific protein. Trends Genet 18:236–237 [DOI] [PubMed] [Google Scholar]
- Fournier GP, Huang J, Gogarten JP 2009. Horizontal gene transfer from extinct and extant lineages: Biological innovation and the coral of life. Philos Trans R Soc Lond B Biol Sci 364:2229–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Tourasse N, Gouy M 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283:220–221 [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Miyamoto MM 2005. A call for likelihood phylogenetics even when the process of sequence evolution is heterogeneous. Mol Phylogenet Evol 37:928–931 [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Das UK, Miyamoto MM, Benner SA 2002a. The crystal structure of eEF1A refines the functional predictions of an evolutionary analysis of rate changes among elongation factors. Mol Biol Evolution 19:569–573 [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Govindarajan S, Ganesh OK 2008. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature 451:704–707 [DOI] [PubMed] [Google Scholar]
- Gaucher E, Graddy L, Li T, Simmen R, Simmen F, Schreiber D, Liberles D, Janis C, Benner S 2004. The planetary biology of cytochrome P450 aromatases. BMC Biology 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Gu X, Miyamoto MM, Benner SA 2002b. Predicting functional divergence in protein evolution by site-specific rate shifts. Trends Biochem Sci 27:315–321 [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Miyamoto MM, Benner SA 2001. Function-structure analysis of proteins using covarion-based evolutionary approaches: Elongation factors. Proc Natl Acad Sci 98:548–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Thomson JM, Burgan MF, Benner SA 2003. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425:285–288 [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitaliumgenome. Science 319:1215–1220 [DOI] [PubMed] [Google Scholar]
- Glansdorff N, Xu Y, Labedan B 2008. The last universal common ancestor: Emergence, constitution and genetic legacy of an elusive forerunner. Biol Direct 3:29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA 3rd, Smith HO, Venter JC 2006. Essential genes of a minimal bacterium. Proc Natl Acad Sci 103:425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten JP, Kibak H, Dittrich P, Taiz L, Bowman EJ, Bowman BJ, Manolson MF, Poole RJ, Date T, Oshima T, et al. 1989. Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. Proc Natl Acad Sci 86:6661–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Martin W 2004. Reading the entrails of chickens: Molecular timescales of evolution and the illusion of precision. Trends Genet 20:80–86 [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Cammarano P 1998. The root of the universal tree of life inferred from anciently duplicated genes encoding components of the protein-targeting machinery. J Mol Evol 47:508–516 [DOI] [PubMed] [Google Scholar]
- Gromiha MM, Oobatake M, Sarai A 1999. Important amino acid properties for enhanced thermostability from mesophilic to thermophilic proteins. Biophys Chem 82:51–67 [DOI] [PubMed] [Google Scholar]
- Hall BG 2006. Simple and accurate estimation of ancestral protein sequences. Proc. Natl Acad Sci 103:5431–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Chen H, Kumar S, Wang DY, Thompson AS, Watanabe H 2001. A genomic timescale for the origin of eukaryotes. BMC Evol Biol 1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ, White ME, Badgett MR, Molineux IJ 1992. Experimental phylogenetics—generation of a known phylogeny. Science 255:589–592 [DOI] [PubMed] [Google Scholar]
- Hoyle F 1972. History of Earth. Q J Roy Astron Soc 13:328–345 [Google Scholar]
- Huelsenbeck JP 1995. Performance of phylogenetic methods in simulation. Systematic Biology 44:17–48 [Google Scholar]
- Iwabata H, Watanabe K, Ohkuri T, Yokobori S, Yamagishi A 2005. Thermostability of ancestral mutants of Caldococcus noboribetus isocitrate dehydrogenase. FEMS Microbiol Lett 243:393–398 [DOI] [PubMed] [Google Scholar]
- Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T 1989. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci 86:9355–9359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffres JBD, Shields GA, Wallmann K 2007. The oxygen isotope evolution of seawater: A critical review of a long-standing controversy and an improved geological water cycle model for the past 3.4 billion years. Earth-Science Rev 83:83–122 [Google Scholar]
- Kasting JF, Howard MT, Wallmann K, Veizer J, Shields G, Jaffres J 2006. Paleoclimates, ocean depth, and the oxygen isotopic composition of seawater. Earth and Planetary Science Letts 252:82–93 [Google Scholar]
- Kelchner SA, Thomas MA 2007. Model use in phylogenetics: Nine key questions. Trends in Ecology & Evolution 22:87–94 [DOI] [PubMed] [Google Scholar]
- Knauth LP 2005. Temperature and salinity history of the Precambrian ocean: Implications for the course of microbial evolution. Palaeogeogr Palaeocl 219:53–69 [Google Scholar]
- Knauth LP, Lowe DR 1978. Oxygen isotope geochemistry of cherts from onverwacht group (3.4 billion years), Transvaal, South-Africa, with implications for secular variations in isotopic composition of cherts. Earth and Planetary Science Lett 41:209–222 [Google Scholar]
- Knauth LP, Lowe DR 2003. High Archean climatic temperature inferred from oxygen isotope geochemistry of cherts in the 3.5 Ga Swaziland Supergroup, South Africa. Geol Soc Am Bull 115:566–580 [Google Scholar]
- Koshi JM, Goldstein RA 1996. Probabilistic reconstruction of ancestral protein sequences. J Mol Evolution 42:313–320 [DOI] [PubMed] [Google Scholar]
- Krishnan NM, Seligmann H, Stewart CB, de Koning APJ, Pollock DD 2004. Ancestral sequence reconstruction in primate mitochondrial DNA: Compositional bias and effect on functional inference. Mol Biol Evolution 21:1871–1883 [DOI] [PubMed] [Google Scholar]
- Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA 3rd, Smith HO, Venter JC 2007. Genome transplantation in bacteria: Changing one species to another. Science 317:632–638 [DOI] [PubMed] [Google Scholar]
- Liberles DA 2007. Ancestral Sequence Reconstruction Oxford University Press, Oxford [Google Scholar]
- Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21:796–802 [DOI] [PubMed] [Google Scholar]
- Matz MV, Lukyanov KA, Lukyanov SA 2002. Family of the green fluorescent protein: Journey to the end of the rainbow. Bioessays 24:953–959 [DOI] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol 17:969–973 [DOI] [PubMed] [Google Scholar]
- Noireaux V, Libchaber A 2004. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci 101:17669–17674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omland KE 1999. The assumptions and challenges of ancestral state reconstructions. Systematic Biol 48:604–611 [Google Scholar]
- Pagel M 1999. Inferring the historical patterns of biological evolution. Nature 401:877–884 [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A 2004. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Syst Biol 53:571–581 [DOI] [PubMed] [Google Scholar]
- Pauling L, Zuckerkandl E 1963. Chemical paleogenetics molecular restoration studies of extinct forms of life. Acta Chem Scand 17:89 [Google Scholar]
- Rivera MC, Lake JA 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431:152–155 [DOI] [PubMed] [Google Scholar]
- Robert F, Chaussidon M 2006. A palaeotemperature curve for the Precambrian oceans based on silicon isotopes in cherts. Nature 443:969–972 [DOI] [PubMed] [Google Scholar]
- Schultz TR, Churchill GA 1999. The role of subjectivity in reconstructing ancestral character states: A Bayesian approach to unknown rates, states, and transformation asymmetries. Systematic Biol 48:651–664 [Google Scholar]
- Schultz TR, Cocroft RB, Churchill GA 1996. The reconstruction of ancestral character states. Evolution 50:504–511 [DOI] [PubMed] [Google Scholar]
- Shih P, Malcolm BA, Rosenberg S, Kirch JF, Wilson AC 1993. Reconstruction and testing of ancestral proteins. in molecular evolution: Producing The Biochemical Data, pp. 576–590 [DOI] [PubMed] [Google Scholar]
- Shimizu H, Yokobori S, Ohkuri T, Yokogawa T, Nishikawa K, Yamagishi A 2007. Extremely thermophilic translation system in the common ancestor commonote: ancestral mutants of Glycyl-tRNA synthetase from the extreme thermophile Thermus thermophilus. J Mol Biol 369:1060–1069 [DOI] [PubMed] [Google Scholar]
- Stetter KO 1996. Hyperthermophilic procaryotes. Fems Microbiology Reviews 18:149–158 [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA 2005. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet 37:630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW 2004. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet 5:366–375 [DOI] [PubMed] [Google Scholar]
- Ugalde JA, Chang BS, Matz MV 2004. Evolution of coral pigments recreated. Science 305:1433. [DOI] [PubMed] [Google Scholar]
- Ward DM, Ferris MJ, Nold SC, Bateson MM 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev 62:1353–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RAD, da Costa MS 1992. The genus Thermus and related microorganisms. in The prokaryotes (ed. Balows A., Truper H.G., Dworkin M., Harder W., Schleifer K-H.), pp. 3745–3753Springer-Verlag, New York [Google Scholar]
- Williams PD, Pollock DD, Blackburne BP, Goldstein RA 2006. Assessing the accuracy of ancestral protein reconstruction methods. PLoS Comput Biol 2: pe69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci 74:5088–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Goldenfeld N 2009. How the microbial world saved evolution from the scylla of molecular biology and the charybdis of the modern synthesis. Microbiol Mol Biol Rev 73:14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Nei M 1997. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evolution 44:139–146 [DOI] [PubMed] [Google Scholar]