Abstract
In the past few years, the field of methylotrophy has undergone a significant transformation in terms of discovery of novel types of methylotrophs, novel modes of methylotrophy, and novel metabolic pathways. This time has also been marked by the resolution of long-standing questions regarding methylotrophy and the challenge of long-standing dogmas. This chapter is not intended to provide a comprehensive review of metabolism of methylotrophic bacteria. Instead we focus on significant recent discoveries that are both refining and transforming the current understanding of methylotrophy as a metabolic phenomenon. We also review new directions in methylotroph ecology that improve our understanding of the role of methylotrophy in global biogeochemical processes, along with an outlook for the future challenges in the field.
Keywords: methylotrophy, biochemistry, physiology, genomics, ecology
INTRODUCTION
Methylotrophic bacteria utilize reduced carbon substrates containing no carbon-carbon bonds (such as methane, methanol, and other methylated compounds) as their sole sources of carbon and energy. Methylotrophy as a phenomenon has been known since the late nineteenth century (104), and during the twentieth century, methylotrophy-directed research activities formed a small field within the field of microbiology, formalized by a dedicated conference (C1 Conference originating in 1978 and transforming into the Gordon Research Conference format in 1998). The former resulted in seven dedicated publications highlighting the up-to-date breakthroughs in methylotrophy. The now classic monograph by Prof. Christopher Anthony published in 1982 provided an excellent and comprehensive review of the state-of-the-art knowledge in the methylotrophy field (2). His monograph has been followed by a number of more specialized reviews that both reiterated the basic information on the known modes of single-carbon (C1) metabolism and reflected new findings in the field (3, 42, 68, 104, 107). We refer the reader to these excellent reviews for a comprehensive overview. The focus of this review is on recent discoveries within the field that contribute significantly to a better understanding of the molecular basis of methylotrophy as a specialized metabolic capability and that challenge some of the long-held dogmas in the field. Also not covered in this review are topics dealing with anaerobic methane oxidation by Archaea and with autotrophic methylotrophy by Clostridia, for which we refer the reader to another review in this volume (9) and to the recent review by Ragsdale and Pierce (91), respectively.
EXPANDED VIEW OF METHYLOTROPHIC CAPABILITY AMONG PHYLOGENETIC GROUPS
Most of the work in the methylotrophy field has involved a handful of fast-growing, laboratory-condition-loving organisms that have been employed as model methylotrophs. The most prominent organisms that shaped the major concepts and assumptions within the field have been Methylomonas methanica, Methylosinus trichosporium, and Methylococcus capsulatus, representing the three recognized classes (I, II and X) of typical obligate methane utilizers (known as methanotrophs); Methylobacterium extorquens and Paracoccus denitrificans, representing typical facultative and typical autotrophic methylotrophs, respectively; and a few species within the family Methylophilaceae, representing obligate or restricted facultative nonmethane-utilizing methylotrophs. The knowledge gained from studying these model organisms has been used to classify methylotrophs into both functional and phylogenetic groups, such as facultative versus obligate methylotrophs, Type I versus Type II methylotrophs, and autotrophic versus heterotrophic methylotrophs (2, 42, 68, 104). Most of these have been placed into a small number of genera within Alpha-, Beta-, and Gammaproteobacteria and Actinobacteria. Thus, the focus on easily cultivated strains resulted in a restricted diversity of recognized methylotrophs.
In contrast, more recent efforts in methylotroph cultivation, many of which employ conditions mimicking in situ conditions rather than standard laboratory conditions, have resulted in a dramatic expansion of our view of the phylogenetic distribution and environmental occurrence of methylotrophs. The notion of defined functional groups has also been changing. For example, the long-standing notion of methane utilizers as obligate methylotrophs has been reversed by the identification of bona fide facultative methanotrophs, all so far belonging to the genus Methylocella. The representatives of this genus utilize a number of multicarbon compounds in addition to methane and methanol (28) and tightly regulate expression of methane monooxygenase in response to the presence of multicarbon compounds (102). The terms themselves, obligate versus facultative, appear of questionable validity when it comes to methylotrophs, as traditionally only a standard set of substrates, many of which are sugars or amino acids, are tested when characterizing the new species. Considering environmental distribution, even for the known guilds of methylotrophs (oligotrophic marine waters, lake sediments, soils, plant roots, and phylospheres), these substrates appear to have little classifying power. The notion of methylotrophy as an attribute of specialized bacterial groups is also morphing, because with more sampling, more bacterial groups are identified as capable of methylotrophy (8). Below, we describe some of the major groups in which methylotrophy has been discovered to occur.
Methylotrophy in Verrucomicrobia
One of the most spectacular new discoveries leading to a new, expanded outlook on the distribution of methylotrophy beyond the well-characterized phylogenetic groups was the demonstration of methane-oxidizing capability in representatives of the phylum Verrucomicrobia. Although they are abundant in a variety of environments, most Verrucomicrobia remain uncultivated, and thus their involvement in specific biogeochemical processes cannot be tested (110). Three research groups have now independently isolated from geographically separate environments pure cultures of methane-oxidizing species of Verrucomicrobia that are closely related to each other phylogenetically and physiologically, and are all thermophilic acidophiles (31, 47, 86). Although these strains are unrelated to the previously characterized proteobacterial methanotrophs, these organisms appear to employ typical if evolutionarily diverged particulate methane monooxygenase to metabolize methane (31, 47, 86). However, beyond the methane oxidation step, the metabolic pathways for C1 oxidation and assimilation in these bacteria remain largely unknown and may differ significantly from the well-understood metabolic schemes.
Methylotrophy in Burkholderiales
Until recently, within Betaproteobacteria, methylotrophy has been known only in species representing a single family, Methylophilaceae, encompassing the so-called obligate or the so-called restricted facultative varieties (2, 68). However, methylotrophs have been described belonging to the order Burkholderiales. One of these, Methylibium petroleiphilum, a potent degrader of a synthetic methylated compound, methyl tert-butyl ether (MTBE), that belongs to the family Comamonadaceae, grows actively on methanol as well as on a wide variety of multicarbon compounds, including aromatic compounds (78). In addition, methylotrophic representatives of the family Rhodocyclaceae within Burkholderiales were isolated from both pristine and polluted environments and were also true facultative methylotrophs, thus challenging the dogma of the obligate or restricted facultative nature of betaproteobacterial methylotrophs (52). Biochemically, methylotrophic Burkholderiales also seem to differ from their Methylophilaceae counterparts. They appear to utilize the serine cycle instead of the ribulosemonophosphate (RuMP) cycle for formaldehyde assimilation (52, 58), and they also lack the typical methanol dehydrogenase (MDH) and instead employ an alternative enzyme (53).
Methylotrophy in Filamentous Bacteria
Methylotrophy capability has also been established recently in bacterial species that have been observed for a long time and noted for the property of forming filaments consisting of groups of sheathed cells that exhibit complex morphological differentiation: Crenothrix polyspora Cohn (99) and Clonothrix fusca Roze (106). The former organism has not yet been successfully cultivated in the laboratory. Instead, its filaments can be physically enriched and subjected to a variety of tests (99, 106). In contrast, C. fusca could be enriched in the laboratory in the presence of methanol, formaldehyde, or both, but not methane, to form an almost pure culture (106). Phylogenetically, both organisms are related to gammaproteobacterial methanotrophs. Further evidence for methanotrophy in these organisms has been obtained via microscopic observations of elaborate membrane systems resembling those of methanotrophic Gammaproteobacteria, demonstration of methane uptake and assimilation by the cells, and identification of genes for methane monooxygenase (99, 106) and MDH (106).
Beggiatoa represents another filament-forming group within the Gammaproteobacteria, characterized by an unusually large cell size. These bacteria are ubiquitous in marine and freshwater environments and are often found as parts of microbial communities involved in C1 metabolism, for example, communities associated with methane seeps (61). So far only two organisms of this group have been implicated in methylotrophy; both are freshwater species classed as Beggiatoa alba (48). These organisms use methanol, but not methane, as a sole carbon and energy source under microoxic conditions. The presence of signature methylotrophy genes was tested by PCR. Although no evidence for a typical MDH was obtained in this way, genes for key enzymes of the tetrahydromethanopterin (H4MPT)-dependent formaldehyde oxidation pathway were identified in one of the strains (48).
Methylotrophy Linked to Denitrification
Most well-studied schemes of methylotrophy involve oxidation of reduced C1 compounds to CO2 using oxygen as a terminal electron acceptor. Denitrifying methylotrophs are known, such as Paracoccus denitrificans and many Hyphomicrobium species (5, 7), but most well-studied aerobic methylotrophs are obligate aerobes (2, 68). However, the use of other electron acceptors in methylotrophy is more common than previously recognized. Methane oxidation can be linked to denitrification in the absence of oxygen. Although originally described as a process mediated by a consortium of bacteria and archaea (90), it was later demonstrated that a single species of bacteria must be responsible for this process (35). No pure cultures of these bacteria have yet been isolated, but they have been highly enriched in a reactor. The bacteria in question belong to a deeply divergent phylum without any cultivated representatives (35, 90). The mechanism of coupling methane oxidation to nitrate reduction also remains unknown.
Although methanol is traditionally employed to enhance denitrification in industrial waste treatment reactors (37, 66) and a number of known methylotrophs (belonging to Methylophilaceae, Rhodocyclaceae, Paracoccus, and Hyphomicrobium) have been detected in the sludge communities (37, 66, 79), it remained unclear in most cases whether the same organisms that perform denitrification consume methanol. Recently we demonstrated that some methylotrophs belonging to Methylophilaceae, and most prominently to the genus Methylotenera, require nitrate for growth on methanol (M.G. Kalyuhznaya, W. Martens-Habbena, T. Wang, S.M. Stolyar & M. Hackett, et al., manuscript submitted). Some of these species appeared to only denitrify in the presence of oxygen, while others appeared to favor anaerobic conditions. From the genomic analysis of Methylotenera, involvement of a traditional MDH in this process is unlikely (54).
NEW INSIGHTS INTO THE BIOCHEMISTRY AND PHYSIOLOGY OF METHYLOTROPHY
Although the biochemistry of methylotrophy had been relatively well characterized by the early 1980s, as reflected in Anthony’s monograph (2), certain gaps remained. One of the longest-standing problems in biochemistry of the serine cycle microbes lacking isocitrate lyase has been the conversion of acetyl-CoA into glyoxylate. This problem has finally been solved owing to the combined effort of a number of groups. Less pressing but academically no less interesting has been the problem of the requirement of two different folate-linked pathways for C1 group transfers by some methylotrophs. New insights have been gained into the specific roles of these pathways. Some new biochemical problems emerged from genomic studies, such as the lack of known genes for MDH in methylotrophs of the Burkholderiales, thus prompting research directed at discovery of an alternative enzyme. New insights into the problem of low-affinity methane oxidation suggest a simple solution for this mystery. Finally, the well-characterized methylamine dehydrogenase (MADH) may not be a major enzyme in methylated amine oxidation by environmental microbes. The newly identified genetic determinants of the glutamate-mediated oxidation will allow for physiological and ecological predictions.
High-Affinity Methane Monooxygenase
Methane consumption from the atmosphere by upland soils is connected to microbially mediated methane oxidation (6, 11). However, this result created a question about the strains responsible because such soils exhibit a much higher affinity for methane than any known methanotrophs (6, 11). The microbes in question were believed to be a special group of methanotrophs, but these phantom bacteria remained uncultivated (11). Recent work by Baani & Liesack (4) demonstrated that a methanotroph of the well-described genus Methylocystis (Methylocystis sp. strain SC2) is capable of atmospheric methane consumption. By specifically disrupting each of the three gene clusters encoding particulate methane monooxygenases (two encoding the almost identical conserved types of enzyme, PmoCAB1, and one encoding a divergent type of enzyme, PmoCAB2), they demonstrated that the conserved Pmo1 was responsible for low-affinity methane oxidation and Pmo2 was responsible for high-affinity methane oxidation. Because many Type II (but not Type I) methanotrophs possess Pmo2 (4), it is likely that high-affinity methane oxidation is a common trait of this group. This finding is of considerable ecological significance because it suggests a solution for a long-standing question and shifts yet again the paradigm of methanotroph ecology.
Novel Methanol Dehydrogenase
The activity of MDH first reported in M. extorquens (2, 3) has been considered an important attribute of methylotrophic growth, and this enzyme has remained a hallmark of methylotrophy for decades. The genes encoding the MDH subunits (mxaFI) and cytochrome c electron acceptor (mxaG) were originally identified in M. extorquens (3) and later detected in most of the model methylotrophs, along with the accessory genes encoding functions required for generating an active MDH, genes for biosynthesis of the coenzyme pyrroloquinoline quinone (PQQ), and regulatory genes (3, 68, 69). Because the mxaF gene is highly conserved among the well-characterized methylotrophs, it has been employed as a tool for environmental detection of methylotrophy (30).
However, some of the newly identified methylotrophs, such as methylotrophic Burkholderiales, appeared to lack the mxaFI genes, and the properties of the partially purified enzymes responsible for MDH activity were somewhat different from those exhibited by typical mxaFI-encoded enzymes. They appeared to be composed of a single type of subunit, possessed high pI values, and required high buffer molarity for activity (53). After peptide matching with a known genome, Kalyuzhnaya et al. (53) identified genes encoding these enzymes in a number of Burkholderiales. These encoded polypeptides (named Mdh2) had low similarity to MxaF polypeptides (less than 35% amino acid identity) but were highly similar (up to 80% amino acid identity) to a different class of PQQ-linked dehydrogenases, the well-characterized alcohol dehydrogenases, which have low affinity for methanol (53). These findings have implications for environmental detection of methylotrophy and indicate that this ability is widespread beyond populations possessing mxaF, the gene traditionally used as a genetic marker for environmental detection of methanol-oxidizing capability. In addition, sequence comparisons show that this newly identified group of MDH enzymes would not be detected by the current environmental probes for mxaF and would at best be misannotated as alcohol dehydrogenases in genomic databases by annotation engines. These findings also have implications for understanding the evolution of methanol oxidation, suggesting a convergence toward the enzymatic function for methanol oxidation in MxaF- and Mdh2-type proteins.
Genetics of Glutamate-Mediated Pathways for Methylamine Oxidation
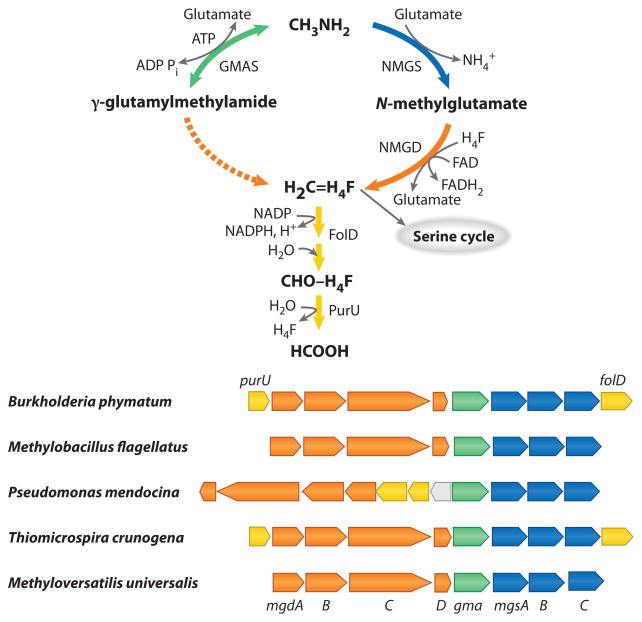
It has been recognized since the 1960s that oxidation of methylamine can be carried out either via direct oxidation to formaldehyde, by MADH (in gram-negative bacteria) or by methylamine oxidase (in gram-positive bacteria), or via indirect pathways that involve transfer of the methyl group onto an amino acid residue (glutamate or alanine), followed by oxidation of the respective methylated amino acid (68). Although MADH has been intensely studied for decades, including the identification of the genes involved and their functions (27), the genetics of indirect methylamine oxidation remained completely unknown until recently. The first support for the potential importance of γ-glutamylmethylamide as an intermediate in methylamine metabolism was obtained by the purification of γ-glutamylmethylamide synthase (GMAS) from the theanine-producing methylotroph Methylovorus mays No. 9 (113). This enzyme showed high activity toward methylamine but not ammonia and was induced in the presence of methylamine (112). The function of this gene and other genes associated with metabolism of methylamine was investigated in Methyloversatilis universalis FAM5, a methylotroph demonstrating robust growth on methylamine but lacking detectable MADH (52). Mutants in the gene encoding GMAS (gma) lacked the activity of GMAS and were affected in their ability to utilize methylamine. However, GMAS was not essential for methylamine oxidation and appeared to play a role in balancing single carbon/ammonium flow during growth on methylamine. However, mutations in genes surrounding gma, mgdABCD encoding N-methylglutamate dehydrogenase (NMGDH), and mgsABC encoding N-methylglutamate synthase (NMGS) resulted in complete arrest of growth on methylamine, suggesting their involvement in a pathway for methylamine oxidation (E. Latypova, Y.-S. Wang, T. Wang, M. Hackett, H. Schäfer & M.G. Kalyuzhnaya, manuscript submitted) (Figure 1). This work identified the genetic determinants for the N-methylglutamate (NMG) pathway and demonstrated for the first time that amino acid–linked methylamine oxidation is essential for methylotrophy in some species.
Figure 1.
The proposed roles of N-methylglutamate synthase (NMGS), γ-glutamylmethylamide synthase (GMAS), and N-methylglutamate dehydrogenase (NMGD) in the oxidation of methylamine and gene clusters encoding enzymes involved in this pathway.
Gene clusters similar to the one identified in M. universalis were identified in the genomes of other methylotrophs that utilize methylated amines: Methylobacillus flagellatus KT, Burkholderia phymatum, and Pseudomonas mendocina (Figure 1). While M. flagellatus possesses MADH that appears to be essential for growth on methylamine (36), the NMG pathway in this organism may play a role in ammonia balancing during growth on methylamine, similar to the role played by the glutamine synthetase/glutamate synthase pathway as proposed for Methylotenera mobilis (10).
Alternative Folate-Linked C1 Transfer Pathways Are Not Redundant
The pathway for formaldehyde oxidation linked to H4MPT was first discovered in M. extorquens AM1 and later shown to be widespread in methylotrophs, possibly representing one of the most ancient methylotrophy metabolic modules (17, 24). It has also become a signature pathway for detecting methylotrophy capacity in environmental samples (51, 55). However, an analogous pathway linked to tetrahydrofolate (H4F) exists in many methylotrophs, including M. extorquens. In this organism, the methylene H4F-dehydrogenase and methenyl H4F-cyclohydrolase reactions are catalyzed by two novel enzymes, MtdA and Fch (21, 88, 108). Intriguingly, MtdA is also active with H4MPT.
Mutant analysis demonstrated that both pathways are necessary for growth on C1 compounds, but it was not clear why two apparently redundant pathways would both be required (107). It was hypothesized that the pathway linked to H4F might be essential for feeding methylene H4F into the serine cycle as part of assimilatory metabolism (21), and this hypothesis was consistent with the kinetic properties of MtdA (108). This hypothesis was further supported by the discovery of formate as the product in the H4MPT-linked oxidation pathway, rather than formylmethanofuran (87), highlighting the importance of formate as an intermediate.
However, one obstacle for developing this hypothesis further was a long-held dogma that formaldehyde reacted spontaneously with H4F under in vivo conditions. Thus, an energetically costly pathway (requiring one molecule of ATP and one molecule of NADPH per molecule of formate assimilated) fulfilling the same function as a direct condensation reaction seemed unreasonable. Although labeling evidence supported significant flux through the direct condensation pathway (72), the mutant evidence did not support this conclusion, and the significance of the direct condensation route remained uncertain.
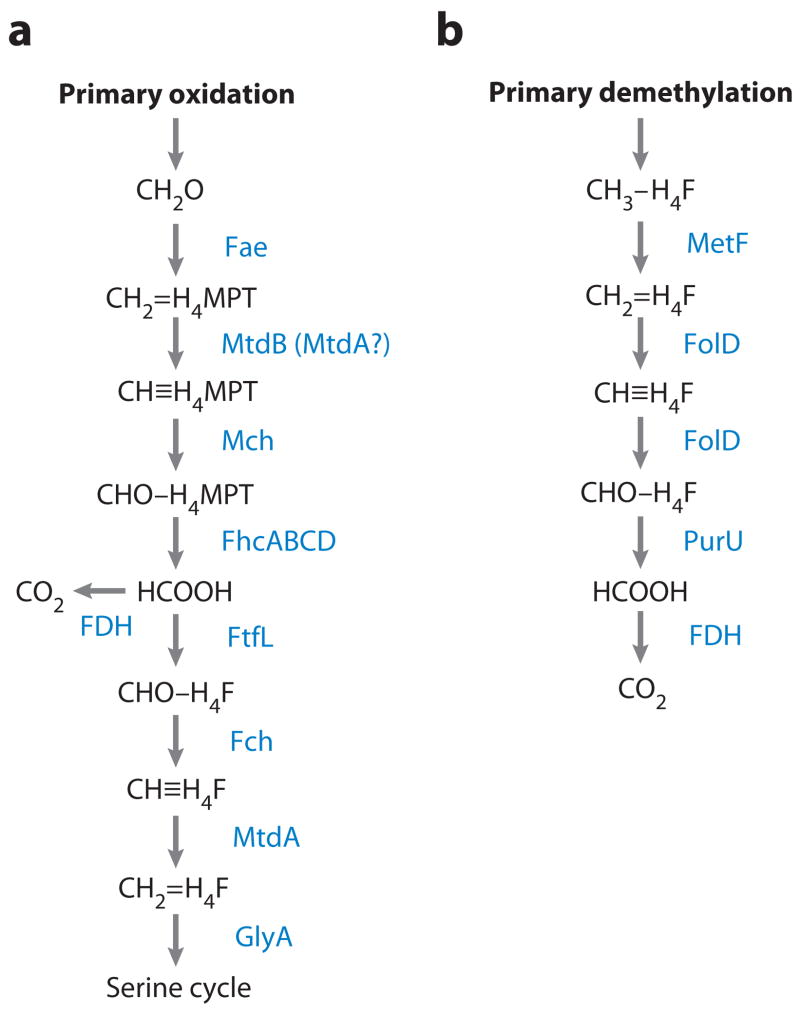
Recently, flux analysis was performed on mutants defective in the H4F-linked pathway on the premise that direct condensation is the only potential pathway for assimilation of methanol in such mutants (26). Although these mutants are unable to grow on C1 compounds, they can be grown under conditions in which C1 pathways are partially induced. In these conditions assimilation from methanol was near zero. In accordance with these results, the H4F-linked pathway appears to be the dominant pathway supplying C1 units into the serine cycle (26). This finding points to formate, not formaldehyde, as the main branch point for methylotrophic metabolism in M. extorquens AM1 and likely in other serine cycle methylotrophs, and it challenges yet another long-held dogma in methylotrophy (Figure 2a).
Figure 2.
Distinct roles of enzymes catalyzing H4F-linked C1 transfer reactions. (a) FtfL, Fch, and MtdA are involved in assimilatory metabolism in Methylobacterium species during growth on methanol or methylamine. (b) MetF, FolD, and PurU are involved in dissimilatory metabolism of methylated compounds such as chloromethane.
The significance of the H4MPT-linked activity of MtdA remains unclear, since an enzyme (MtdB) that carries out this reaction and is also required for growth on C1 compounds is present (24, 107). It could be an indication that MtdA has diverged from MtdB, having acquired affinity toward H4F but still retaining its affinity for H4MPT. Alternatively, it may be an indication of a specific, not yet understood, connection between the two analogous pathways.
Some methylotrophs possess FolD, the bifunctional methylene-H4F dehydrogenase/methenyl- H4F cyclohydrolase, in addition to the MtdA/Fch pair, whereas some only possess FolD. In Methylobacterium chloromethanicum CM4, FolD appears to be specifically involved in dissimilation of the methyl-H4F that is a product of demethylation of chloromethane (100). FolD from M. chloromethanicum, when introduced into the mtdA mutant of M. extorquens, failed to complement it for growth on methanol (70), highlighting the different roles of MtdA and FolD in Methylobacterium species (Figure 2a,b).
Pathway for Glyoxylate Regeneration Is Finally Resolved
One of the biochemical peculiarities of many methylotrophs employing the serine cycle for formaldehyde assimilation is the lack of isocitrate lyase, the key enzyme in the glyoxylate shunt, and the presence of an alternative pathway for the conversion of acetyl-CoA to glyoxylate. This unusual metabolic feature was noted soon after the discovery of the serine cycle and was intensely studied in the Quayle laboratory and later in the Anthony laboratory, using M. extorquens AM1 as a model (summarized in Reference 2). A major conclusion from this work was the involvement of this pathway in C2 metabolism as well, supported by the phenotypes of the mutants in the pathway: They were all defective in growth on both C1 and C2 compounds, and growth was rescued for both substrates by the addition of glyoxylate (2). Although a great deal of effort was expended to discover it, the nature of this pathway remained a mystery for another two decades. The breakthroughs achieved in the 1990s were enabled by the availability of DNA sequencing technologies in combination with newly developed techniques for directed mutagenesis (22). As a result, two intermediates in the pathway (originally called the glyoxylate regeneration cycle and later the ethylmalonyl-CoA pathway) were predicted, propionyl-CoA and methylmalonyl-CoA, based on the identification of one of the essential genes as the propionyl-CoA carboxylase gene ( pccB) (22). Further efforts in solving the pathway in M. extorquens were complemented by mutant analysis and metabolite detection, resulting by 2005 in the unequivocal identification of a number of reactions in the pathway and of a large number of genes involved (62–65). However, the exact role of some enzymes, such as MeaA, a homolog of methylmalonyl-CoA mutase (98), and thus some intermediates remained unknown.
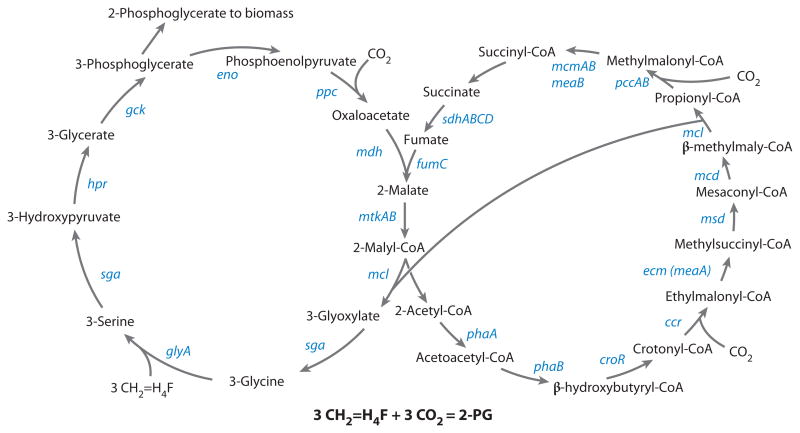
The ultimate solution of the pathway was in large part due to the recent breakthroughs in understanding the details of acetate metabolism in Rhodobacter sphaeroides, which proceeds via a pathway similar to the glyoxylate regeneration pathway in serine cycle methylotrophs, as demonstrated in the Fuchs laboratory (1, 33, 34, 74, 115) (Figure 3). The most important findings leading to the solution of the pathway for converting acetyl-CoA to glyoxylate included the following. (a) Mesaconyl-CoA dehydratase was determined to be an important enzyme in the pathway, suggesting that mesaconyl-CoA and methylmalyl-CoA must be the intermediates (115). (b) The reaction converting ethylmalonyl-CoA into methylsuccinyl-CoA was demonstrated, catalyzed by ethylmalonyl-CoA mutase, and encoded by meaA (33). (c) The function of crotonyl-CoA reductase (encoded by ccr) was revised, demonstrating that this enzyme catalyzes both reduction and carboxylation of crotonyl-CoA to produce ethylmalonyl-CoA from crotonyl-CoA (34). (d ) Malyl-CoA lyase was shown to possess dual specificity toward malyl-CoA and methylmalyl-CoA and was proposed to perform both reactions (1, 74).
Figure 3.
The refined scheme of the glyoxylate regeneration cycle, now termed ethylmalonyl-CoA pathway (right), and its connection to the serine cycle (left). Gene names are shown in blue. Each of these genes has been mutated in Methylobacterium extorquens AM1 and the mutant phenotypes analyzed. The gene for succinyl-CoA hydrolase remains unknown.
Confirmation for this new variant of the glyoxylate regeneration cycle (now called the ethylmalonyl-CoA pathway) in methylotrophs during growth on C1 compounds was obtained in elegant studies in the Vorholt laboratory, using M. extorquens AM1 as a model (60, 85). This group utilized a high-resolution mass spectrometry approach to demonstrate the presence of most of the CoA thioesters specific to this pathway. Furthermore, evidence was obtained for the operation of this pathway by short-term 13C-pulse experiments, demonstrating the sequence of reactions from the order of label incorporation into the predicted CoA derivatives. It is clear from this work that in methanol-grown cells methylsuccinyl-CoA is converted into glyoxylate and propionyl-CoA via mesaconyl-CoA and methylmalyl-CoA, solving the questions that remained for this portion of the pathway (Figure 3).
The new scheme for the conversion of acetyl-CoA to glyoxylate has major implications for C1 assimilation in terms of carbon balance. In accordance with the new scheme, two carbons are assimilated from CO2 and two molecules of glyoxylate are regenerated per each molecule of acetyl-CoA. This challenges the widely accepted serine cycle balance of two formaldehydes per one CO2 (2) and points toward one formaldehyde per one CO2 (Figure 3). Ironically, this equation balance is consistent with labeling studies conducted by the Quayle group (67) over four decades ago that showed a 1:1 relationship, as well as with the recent measurements (26).
GENOMICS OF METHYLOTROPHS
During the past decade and a half, the field of microbiology, as most of biology, has been transformed by the insights gained from whole-genome sequencing (genomics). It was only a matter of time before the first methylotroph genomic sequences became available. The draft genome sequence of M. extorquens AM1 has been publicly accessible and in active use by our lab as well as colleagues working with Methylobacterium species since 2003 (15). The first complete methylotroph sequence to be published was that of another model methylotroph, the methanotroph Methylococcus capsulatus Bath (111). These were quickly followed by the sequences of betaproteobacterial methylotroph genomes (19, 38, 58), as well as the genomes of methylotrophs not primarily studied for C1-utilizing capability, such as the genome of the emerging human pathogen Granulibacter bethesdensis (41). As a sign of the times, the first genomic sequence of a methylotrophic Verrucomicrobia appeared less than a year after the organism was first cultivated (44).
Genome of Methylococcus capsulatus
Publication of the first methylotroph genome was an important event, presenting the first complete blueprint for an obligate methane utilizer (111). The relatively small size of the genome (3.3 Mb) is consistent with the simple lifestyle of the organism. As the biochemistry of methane oxidation in this organism has been thoroughly studied, including the identification of key methylotrophy gene clusters, the genome revealed few surprises in these terms. Indeed, all the genes encoding the methane monooxygenases, the enzymes of the RuMP cycle, and the enzymes for H4MPT-linked formaldehyde oxidation were identified, as were the genes for the Calvin-Benson-Bassham (CBB) cycle and three nonhomologous formate dehydrogenases (FDHs). In addition, most of the genes for the serine cycle were found, including MtdA and Fch (111). Genome-based proteomic analysis confirmed expression of all three assimilatory pathways (59). However, no gene has been identified for one of the serine cycle enzymes, phosphoenolpyruvate carboxylase, and neither the ethylmalonyl-CoA pathway nor the glyoxylate shunt was encoded, suggesting that the serine cycle cannot operate as a major assimilatory pathway in M. capsulatus. Given these considerations, a possible role has been suggested for this pathway as an additional pathway for detoxification of formaldehyde or formate (23). Another interesting discovery was the presence of all the genes for the tricarboxylic acid cycle. In addition, all the functions that should allow the organism to grow on sugars were encoded (111). Thus, from the genomic sequence no answer has been gained for why most methanotrophs, including M. capsulatus, are incapable of growth on multicarbon compounds.
Methylobacterium extorquens Genomes
The complete genome sequences of two strains of M. extorquens, strain AM1 and the dichloromethane-degrading strain DM4, have now been fully analyzed (109). They possess genomes much larger than that of M. capsulatus (6.9 and 6.1 Mb, respectively). Strain AM1 carries a megaplasmid of 1.2 Mb and three small plasmids in addition to the chromosome, while the genome of strain DM4 has two plasmids in addition to the chromosome. Most of the methylotrophy genes are highly conserved between the two strains. The two chromosomes, unlike the plasmids, overall shared a large majority of the genes at high sequence identity, and they were highly syntenic. The main difference between the chromosomes was in the content and the number of insertion sequence elements, which were highly abundant in both genomes, suggesting a significant potential for genomic plasticity. Some of these elements are common to both strains, whereas others are strain specific.
One surprising discovery from genomic comparisons was the lack of the gene cluster encoding methylamine utilization (mau) in the genome of strain DM4. As this strain, like strain AM1, is capable of growth on methylamine, it was concluded to employ a nonhomologous system for methylamine utilization. As expected, the dichloromethane utilization (dcm) gene cluster was unique to strain DM4. Both the mau gene cluster in strain AM1 and the dcm gene cluster in strain DM4 are associated with mobile elements, suggesting that both may be subjects of lateral transfers.
Recently, complete genomic sequences of six other Methylobacterium strains have been generated by the Joint Genome Institute, representing members of M. extorquens as well as other species (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi/). These sequences are awaiting manual annotation analysis. The availability of the two characterized genomes will serve as an important platform for intra- and interspecies genomic comparisons, and for investigations of the adaptive mechanisms that allow Methylobacterium to thrive in different ecological niches.
While most methylotrophy genes in M. extorquens AM1 have been identified and characterized in the pregenomic era, the availability of the genome sequence allowed the study of functions not previously assessed via traditional genetic techniques, such as the multiple FDH functions. Genes encoding four different FDH enzymes (FDH1, 2, 3, and 4) were recognized in both genomes, and these were investigated in strain AM1 (16, 20). When each of the enzymes was disrupted in a wild-type background, mutants in only one of them, the novel type of FDH (FDH4), revealed a phenotype: These mutants accumulated formate in the late stationary phase of growth (16). However, when FDH4 was intact, all three other enzymes had to be removed to observe a phenotype: These mutants transiently accumulated formate in early exponential growth phase (20). So far, the limited information on the expression and regulation of the four FDH enzymes suggests complex regulatory patterns involving metal and cofactor availability and, in the case of FDH4, a connection to acid stress. In accordance with the new paradigm of formate as the main branchpoint of methylotrophic metabolism in Methylobacterium, such a complex genetic background is not entirely surprising, as it potentially provides a robust mechanism for balancing C1 fluxes between energy generation and assimilation.
Genomes of Betaproteobacterial Methylotrophs
Analysis of the first genome of a betaproteobacterial methylotroph was an interesting exercise in discovering the noncanonical ways of methylotrophy, conducted by a group outside the field of methylotrophy, as the main focus in studying this organism was pollutant (MTBE) degradation (58). As mentioned above, Methylibium petroleiphilum is a facultative methylotroph with a broad substrate range, defying the notion of the obligate or restricted facultative nature of this group. Accordingly, the organism possesses a reasonably large genome (4 Mb chromosome plus 0.6 Mb megaplasmid) and encodes all major pathways for metabolizing sugars and organic acids. In terms of methylotrophy, the genome revealed the lack of the key genes for the RuMP cycle but the presence of all the genes for the serine cycle, including mtdA and fch. In fact, the clustering of the serine cycle genes in M. petroleiphilum is similar to that in M. extorquens, suggesting these genes may have been transferred laterally between Alpha- and Betaproteobacteria. Unlike M. extorquens, M. petroleiphilum contains genes encoding the glyoxylate shunt and lacks key genes for the ethylmalonyl-CoA pathway. At the time of genome analysis, the lack of the mxa gene cluster was surprising, as the organism grows robustly on methanol (78). However, the recent discovery of Mdh2 in this organism has solved this problem (53). The same enzyme is utilized for oxidizing ethanol by this organism (45).
The smaller (2.9 Mb) genome of Methylobacillus flagellatus, a favorite model for studying conventional methylotrophy by Betaproteobacteria, was published a few months later (19), demonstrating that the two organisms have little in common when it comes to methylotrophy. The M. flagellatus genome encodes the classic MDH, the classic MADH, and the classic RuMP cycle. Unlike M. capsulatus, M. extorquens, and M. petroleiphilum, this bacterium does lack the complete tricarboxylic acid cycle, as no genes for α-ketoglutarate, malate, or succinate dehydrogenases could be identified. Thus, interpretation of the obligate nature of this organism’s methylotrophic lifestyle is more straightforward. The only methylotrophy module common to both M. flagellatus and M. petroleiphilum is the H4MPT-linked formaldehyde oxidation pathway. The genes for this pathway in M. flagellatus are related phylogenetically more to the genes of M. capsulatus and other gammaproteobacterial methylotrophs than to the genes in M. petroleiphilum, suggesting that methylotrophy in Betaproteobacteria is of polyphyletic origin.
The other available genome of a Methylophilaceae representative is the small genome of strain HTCC2181, which belongs to the abundant coastal ocean clade OM43 (38). In fact, this is the smallest genome so far enabling a free-living lifestyle for a bacterium (38). The organism was originally cultivated using the dilution-to-extinction technique in medium made from seawater (25). In the lab, it appears to respond positively to the addition of methanol or formaldehyde in terms of cell yield. However, the doubling time during exponential growth was independent of the addition of substrates (38). The genome of strain HTCC2181 is a minimal genome, only encoding the functions necessary for autonomous existence, including the methylotrophy functions. Comparisons with the M. flagellatus genome reveal extensive synteny (38). However, some methylotrophy pathways typically present in Methylophilacea are entirely missing, such as the H4MPT pathway and the mxa (or mdh2) and mau gene clusters. It is concluded that strain HTCC2181 diverged from its relatives by reducing the size of its genome (38). The lack of known MDH-encoding genes questions the ability of this strain to effectively utilize methanol, but suggests the intriguing possibilities that OM43 contains yet another unknown MDH or that the primary substrate for the OM43 clade-type bacteria is a different methylated compound commonly present in coastal ocean waters.
Genome of Verrucomicrobia Strain V4
The fast progress from isolating methanotrophic Verrucomicrobia to sequencing the genome of strain V4, “Candidatus Methylacidiphilum infernorum” (44), has been exciting. This is also the first Verrucomicrobia genome to be formally analyzed. The single chromosome genome is 2.3 Mb in size, which again agrees with the obligate methylotrophic lifestyle of the organism (other publicly available Verrucomicrobia genomes are of considerably larger size). As mentioned above, strain V4 appears to encode bona fide methane monooxygenases, possessing three gene clusters highly similar to the pmoCAB clusters in Proteobacteria (up to 60% amino acid identity). However, there are no recognizable genes for MDH; instead, the genome contains only the xox cluster of PQQ-linked dehydrogenases, which so far has not been shown to encode an MDH (53). These results suggest either another novel MDH or a different type of an enzyme system for utilizing methanol. There are also no recognizable genes for any of the major systems for formaldehyde oxidation (i.e., the H4MPT-linked pathway, the glutathione-linked pathway, the oxidative branch of the RuMP cycle, or the specific NAD-linked formaldehyde dehydrogenase) (107). For the potential C1 assimilation pathways, only the Calvin Benson Basham (CBB) cycle appears to be encoded. The heavy energetic requirement for assimilation via the CBB cycle makes it a dubious candidate for supporting growth on methane, and if it were the assimilatory pathway, it would be unique among known methanotrophs. Clearly, experiments are required to assess the biochemistry of C1 metabolism downstream of methanol in this novel clade of methylotrophs, and that work promises interesting and novel results.
Genome-Based Predictions of Methylotrophy Capability
Methylotrophy has been previously discussed in terms of specific functional metabolic modules, as determined by mutational studies in model methylotrophs and as deduced from genomic sequences of methylotrophs (15, 18). It appears that for bona fide methylotrophy, a minimal set of methylotrophy modules is necessary that would allow for oxidation/demethylation of a primary C1 substrate, for oxidation/detoxification of the product of primary oxidation (formaldehyde or methyl-H4F), and for assimilation of C1 units. Thus, the presence of a complete set of signature methylotrophy modules in a genome may be used to predict methylotrophic capability for an organism. For example, the organism Granulibacter bethesdensis has been recently described as a causative agent in chronic granulomatous disease and has been classified as a member of the family Acetobacteraceae (39). The genomic sequence of this organism reveals the presence of an essential set of methylotrophy modules to enable growth on methanol; that is, it encodes the classic (mxaFI-type) MDH, the reactions of the H4MPT-linked formaldehyde oxidation pathway, the serine cycle reactions, and the glyoxylate shunt reactions (41). Indeed, routine tests for growth on methanol were positive (40).
Another example is the recently sequenced genome of Burkholderia phymatum, a highly effective nitrogen-fixing plant symbiont (32, 77). Its genome revealed the presence of genes for trimethylamine oxidation via trimethylamine dehydrogenase, methylamine oxidation via the NMG pathway, formaldehyde oxidation via the H4MPT-linked pathway, and the serine cycle for C1 assimilation (L. Chistoserdova, unpublished observations). Indeed, robust growth of the organism was observed on methylated amines (L. Chistoserdova, unpublished observations).
The presence of the complete set of genes for the serine cycle, as well as for the ethylmalonyl-CoA pathway in the genome of Silicibacter pomeroyi (76), an organism important in environmental degradation of dimethylsulfoniopropionate (12), suggests that this organism is likely to use these pathways for assimilation of methyl groups resulting from demethylation reactions (93). Although no known genes for primary oxidation of C1 compounds are recognizable in the genome of this organism, other organisms within the family Rhodobacteraceae are known for bona fide methylotrophy (95, 96). Two of the signature methylotrophy modules, the hexulosephosphate synthase/isomerase pair and the H4MPT-linked formaldehyde oxidation pathway, play a role in formaldehyde detoxification in heterotrophic bacteria (13, 71, 75, 114), so their presence does not necessarily imply methylotrophy unless other functional methylotrophy modules are present. We refer the reader to Table 1 for comparative analysis of physical characteristics of the genomes discussed above and major metabolic features deduced from these genomes.
Table 1.
Summary of methylotroph genomes discussed in this review
| Organism | Phylogenetic group | Genome size (Mb) | Plasmids | Methane oxidation | Methanol oxidation | Methylamine oxidation | H4MPT pathway | C1 assimilation pathway | TCA cycle |
|---|---|---|---|---|---|---|---|---|---|
| Methylobacterium extorquens AM1 | Alphaproteobacteria | 6.9 | Yes | No | MxaFI | MADH | Yes | Serine | Complete |
| Methylobacterium extorquens DM4 | Alphaproteobacteria | 6.1 | Yes | No | MxaFI | Unknown | Yes | Serine | Complete |
| Silicibacter pomeroyi DSS-3 | Alphaproteobacteria | 4.6 | Yes | No | None | None | No | Serine | Complete |
| Granulibacter bethesdensis NIH1.1 | Alphaproteobacteria | 2.7 | No | No | MxaFI | None | Yes | Serine | Complete |
| Burkholderia phymatum STM815 | Betaproteobacteria | 7.7 | Yes | No | None | NMG | Yes | Serine | Complete |
| Methylibium petroleiphilum PM1 | Betaproteobacteria | 4.6 | Yes | No | Mdh2 | None | Yes | Serine | Complete |
| Methylobacillus flagellatus KT | Betaproteobacteria | 2.9 | No | No | MxaFI | MADH | Yes | RuMP | Incomplete |
| Strain HTCC2181 | Betaproteobacteria | 1.3 | No | No | None | None | No | RuMP | Incomplete |
| Methylococcus capsulatus Bath | Gammaproteobacteria | 3.3 | No | Yes | MxaFI | None | Yes | RuMP | Complete |
| Isolate V4 | Verrucomicrobia | 2.3 | No | Yes | None | None | No | Unknown | Complete |
Abbreviations: H4MPT, tetrahydromethanopterin; MADH, methylamine dehydrogenase; NMG, N-methylglutamate; RuMP, ribulosemonophosphate; TCA, tricarboxylic acid cycle.
NEW APPROACHES IN ECOLOGY OF METHYLOTROPHS
The past decade in microbiology has been marked by the realization that the majority of microbes in this world may remain uncultured (92). One of the consequences of this realization has been a surge in culture-independent approaches, especially PCR-based detection of ribosomal RNA and a handful of functional genes (43, 46). However, despite the value of these technologies, a consensus has been emerging that a combination of approaches that would connect sequence to function, including more creative attempts at cultivation, would best serve the goal of understanding natural microbial communities (29, 84). When it comes to methylotrophs, the important question is, how well do the model methylotrophs studied in the lab reflect natural populations that carry out cycling of a variety of C1 compounds in the environment? Indeed, the discovery of entirely new methylotrophic phyla, such as Verrucomicrobia, and methylotrophs representing traditional groups but not possessing the traditional primary oxidation genes, such as Burkholderiales, provides a warning against overreliance on PCR-based approaches.
Methylotroph-Specific Probes
For environmental detection of methylotrophs, fragments of genes believed to be signatures of methylotrophs are PCR amplified using specific oligonucleotide primers. Some of these PCR approaches target ribosomal genes, thus detecting species belonging to established methylotrophic phyla (14), while others detect functional genes. Traditionally, pmoA and mmoX (encoding conserved subunits of the pMMO and sMMO, respectively) and mxaF are used to detect methane and methanol oxidizers, respectively (30, 101). Special probes have also been developed to detect specialized functions such as methylamine and methylhalide oxidation (73). The target enzymes listed above tend to be highly conserved in known methylotrophs, especially proteobacterial methylotrophs. Degenerate environmental primers have also been designed to target more divergent enzymes that are part of the H4MPT-linked formaldehyde oxidation pathway common to most methylotrophs (51, 55, 57). These primers allow a broader range of phylogenetic groups, including novel yet unidentified phyla, to be targeted (50, 80).
Redox Sensing
A method has been recently developed by our group that allows detection of a specific response to C1 substrate stimuli by members of complex natural communities, as monitored by observing fluorescence of a redox-sensing dye (56). The responding subpopulations detected in real time can be separated by flow cytometry and cell sorting, followed by PCR-based gene profiling. The sorted cells can also be used in enrichments to select for specific functional types (56). A community inhabiting Lake Washington sediment has been used as a model community to demonstrate that specific populations are activated by methane, methanol, methylamine, or formaldehyde. Some members of these populations belonged to established methylotrophic phyla, whereas others represented phyla not known for methylotrophy (56).
More recently, the same redox-sensing dye was employed to develop a new approach called respiration response imaging (RRI) (M. Kanopka & M.G. Kalyuzhnaya, unpublished data). On the basis of the premise of the respiration rate being proportional to fluorescence intensity, RRI monitors the response of microbial cell populations to an environmental perturbation/stress in real time at the single cell level. Responses of pure cultures of methanotrophic bacteria were monitored following the addition of C1 substrates. Similar experiments were also conducted on environmental samples demonstrating the feasibility of RRI for detecting individual cells responding to C1 stimuli.
Stable Isotope Probing
One powerful technique that has been widely utilized for targeting actively metabolizing methylotrophs is stable isotope probing (DNA-SIP), pioneered by the Murrell laboratory (89). Natural samples are fed a substrate labeled with a heavy isotope (typically, 13C). The label is efficiently incorporated into biomass of microbes actively consuming this substrate, and biomass components, most prominently their DNA, can be analyzed and phylogenetic affiliations of species whose DNA has been labeled can be determined. Feeding the same microbial community different isotopically labeled C1 compounds provides a more complex picture of methylotrophy specialists versus generalists functional guilds.
The methodology of SIP has been a subject of scrutiny recently, because, as any culture-independent approach, it is prone to biases (81–83). While it is understood that SIP experiments should be conducted in conditions resembling the in situ conditions as closely as possible to obtain a true picture of the activities and processes native to a given environment, the success of SIP inherently depends on altering the in situ conditions to a degree. A higher than in situ concentration of the substrate in question is required for successful labeling, and prolonged incubations often are required, which may result in some enrichment of DNAs of fast-growing microbes. However, if a labeled substrate concentration is in the range of environmental concentrations, little label is incorporated into the DNA and additional steps are required such as whole-genome amplification. In addition, longer incubations increase the possibility of labeling other cells via cross-feeding. Although they possess the potential of uncovering unexpected and unsuspected participants in the C1 cycle, SIP results always need critical interpretation.
Metagenomics
Metagenomics or environmental genomics has recently become a powerful tool for collecting information on microbial communities, bypassing cultivation of individual species, and this approach continues to gain momentum (97). In cases of low-complexity microbial communities, genomic data of high quality can be obtained for individual species and almost complete genomic sequences can be assembled (105). However, in communities of high complexity, sequencing efforts typically result in short unassembled sequences (94, 103).
Methylotrophs are often members of such complex communities. To increase the resolution of metagenomics and to increase coverage of methylotroph genomes in a metagenomic dataset, our group has utilized a strategy that combined substrate-specific labeling of methylotroph DNA via SIP with metagenomic sequencing (54). Five different 13C-labeled substrates were employed, methane, methanol, methylamine, formaldehyde, and formate, resulting in five shotgun libraries that were sequenced, thus producing datasets of 26 to 59 Mb of sequence and totaling 255 Mb. On the basis of an analysis of 16S rRNA gene sequences, community complexity of the metagenomes was significantly reduced compared with the complexity of the nonenriched community estimated at over 5000 species (54), and shifted toward specific functional guilds that also included known methylotroph species. A nearly complete composite genome of one of the dominant methylotrophs, Methylotenera mobilis (49), could be extracted from the metagenome via compositional binning, allowing for whole-genome analysis and comparative genomics (54).
CONCLUDING REMARKS AND REMAINING QUESTIONS ON METHYLOTROPHY
The past few years in the methylotrophy field have been marked by a number of spectacular breakthroughs that have not only contributed to a better understanding of the metabolism, phylogenetic distribution, and ecological significance of C1 oxidation in nature, but also challenged a number of long-held dogmas in the field. Inevitably, moving to the next level of knowledge is accompanied by the emergence of new questions. The new challenges in methylotrophy will include uncovering the mechanisms for anaerobic methane oxidation coupled to denitrification and for aerobic denitrification with methanol. Another important challenge will be uncovering the enzymes and pathways enabling methylotrophy in Verrucomicrobia and other novel and poorly characterized methylotrophs, including the yet uncultured anaerobic denitrifying methane-utilizing bacteria. Deciphering the significance and the precise division of functions between key enzymes with overlapping specificities, such as MtdA, MtdB, and FolD, presents another tempting biochemical puzzle. A major ecological challenge will be in understanding the interconnection of methylotrophy with other biogeochemical processes and in uncovering the yet unknown important carbon cycle components whose conversion involves methylotrophy pathways. While the established and emerging -omics technologies, including next-generation sequencing technologies, are predicted to play an important role in solving the outstanding questions in methylotrophy, their combination with experimental approaches remains key to the success of the field. The increasing ability to analyze uncultured cells, both in bulk and at the single-cell level, is expected to be an important tool in the future for uncovering novel and surprising aspects of methylotrophy in the laboratory and in nature.
SUMMARY POINTS
Methylotrophy is more widespread in the bacterial domain of life than previously recognized and includes representatives of deeply branching phyla such as Verrucomicrobia.
Previously unknown enzyme systems for primary oxidation of C1 compounds have been discovered, including a low-affinity methane monooxygenase and a methanol dehydrogenase (Mdh2), that are distantly related to the previously studied enzymes. It is likely that more are present in yet uncultured strains of methylotrophs.
Genes responsible for transferring a methyl group from methylamine to glutamate, followed by oxidation to methylene-H4F, have been identified and are essential for metabolism of methylamine in some methylotrophs.
Formate, not formaldehyde, appears to be the branchpoint between assimilatory and dissimilatory metabolism in serine cycle methylotrophs.
The long-standing problem of conversion of acetyl-CoA into glyoxylate by the serine cycle methylotrophs lacking isocitrate lyase has been finally resolved, resulting in a new carbon balance for the serine cycle of 1 formaldehyde to 1 CO2.
Emerging genomics and metagenomics of methylotrophs support the modular nature of methylotrophy.
New means for environmental detection of methylotrophs have been developed, including probing for less-conserved, presumably ancient genes; SIP with a variety of C1 substrates; and redox-dye-based manipulations.
Outstanding questions for the future include understanding the biochemistry and physiology of methylotrophy in novel strains, as well as understanding the roles of familiar enzymes in model methylotrophs. In addition, the diversity, distribution, and role of methylotrophs in natural habitats remain important areas of study for the future.
- Methylotrophy
ability to use C1 compounds as substrates
- Obligate methylotrophs
microbes that can only use C1 substrates
- Facultative methylotrophs
microbes that can use both C1 and multicarbon compounds as substrates
- Serine cycle
a specialized pathway for incorporating methyl groups in the form of methylene H4F into cell constituents, used by some methylotrophs
- Ribulosemonophosphate (RuMP) cycle
a specialized pathway for incorporating methyl groups in the form of formaldehyde into cell constituents, used by some methylotrophs
- MDH
methanol dehydrogenase
- Methanotrophy
ability to use methane as a substrate
- H4MPT
tetrahydromethanopterin
- C1 compounds
reduced organic compounds containing no carbon-carbon bonds
- Pmo
particulate methane monooxygenase
- PQQ
pyrroloquinoline quinone
- H4F
tetrahydrofolate
- MtdA
methylene H4MPT-dehydrogenase/methylene H4F-dehydrogenase
- Fch
methenyl H4F-cyclohydrolase
- MtdB
methylene H4MPT-dehydrogenase
- FolD
methylene H4F-dehydrogenase/methenyl H4F-cyclohydrolase
- Ethylmalonyl-CoA pathway
the pathway used by some serine cycle methylotrophs to convert acetyl-CoA into glyoxylate
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alber BE, Spanheimer R, Ebenau-Jehle C, Fuchs G. Study of an alternate glyoxylate cycle for acetate assimilation by Rhodobacter sphaeroides. Mol Microbiol. 2006;61:297–309. doi: 10.1111/j.1365-2958.2006.05238.x. [DOI] [PubMed] [Google Scholar]
- 2.Anthony C. The Biochemistry of Methylotrophs. London: Academic; 1982. [Google Scholar]
- 3.Anthony C. The quinoprotein dehydrogenases for methanol and glucose. Arch Biochem Biophys. 2004;428:2–9. doi: 10.1016/j.abb.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 4.Baani M, Liesack W. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc Natl Acad Sci USA. 2008;105:10203–8. doi: 10.1073/pnas.0702643105. Shows that an organism of the Methylocystis genus is capable of low-affinity methane oxidation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker SC, Ferguson SJ, Ludwig B, Page MD, Richter OM, van Spanning RJ. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol Mol Biol Rev. 1998;62:1046–1078. doi: 10.1128/mmbr.62.4.1046-1078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender M, Conrad R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol Ecol. 1992;101:261–70. [Google Scholar]
- 7.Betlach MR. Evolution of bacterial denitrification and denitrifier diversity. Antonie van Leeuwenhoek. 1982;48:585–607. doi: 10.1007/BF00399543. [DOI] [PubMed] [Google Scholar]
- 8.Boden R, Thomas E, Savani P, Kelly DP, Wood AP. Novel methylotrophic bacteria isolated from the River Thames (London, UK) Environ Microbiol. 2008;10:3225–36. doi: 10.1111/j.1462-2920.2008.01711.x. [DOI] [PubMed] [Google Scholar]
- 9.Boetius A. Anaerobic methane oxidation. Annu Rev Microbiol. 2009;63:311–34. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 10.Bosch G, Wang T, Latypova E, Kalyuzhnaya MG, Hackett M, Chistoserdova L. Insights into the physiology of Methylotenera mobilis as revealed by metagenome-based shotgun proteomic analysis. Microbiology. 2009;155:1103–10. doi: 10.1099/mic.0.024968-0. [DOI] [PubMed] [Google Scholar]
- 11.Bull ID, Parekh NR, Hall GH, Ineson P, Evershed RP. Detection and classification of atmospheric methane oxidizing bacteria in soil. Nature. 2000;405:175–78. doi: 10.1038/35012061. [DOI] [PubMed] [Google Scholar]
- 12.Bürgmann H, Howard EC, Ye W, Sun F, Sun S, et al. Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP) Environ Microbiol. 2007;9:2742–55. doi: 10.1111/j.1462-2920.2007.01386.x. [DOI] [PubMed] [Google Scholar]
- 13.Chain PS, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA. 2006;103:15280–87. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Dumont MG, Cébron A, Murrell JC. Identification of active methanotrophs in a landfill cover soil through detection of expression of 16S rRNA and functional genes. Environ Microbiol. 2007;9:2855–69. doi: 10.1111/j.1462-2920.2007.01401.x. [DOI] [PubMed] [Google Scholar]
- 15.Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J Bacteriol. 2003;185:2980–87. doi: 10.1128/JB.185.10.2980-2987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chistoserdova L, Crowther GJ, Vorholt JA, Skovran E, Portais JC, Lidstrom ME. Identification of a fourth formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role of formate oxidation in methylotrophy. J Bacteriol. 2007;189:9076–9081. doi: 10.1128/JB.01229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chistoserdova L, Jenkins C, Kalyuzhnaya MG, Marx CJ, Lapidus A, et al. The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol Biol Evol. 2004;21:1234–41. doi: 10.1093/molbev/msh113. [DOI] [PubMed] [Google Scholar]
- 18.Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. C1 transfer modules: from genomics to ecology. ASM News. 2005;71:521–28. [Google Scholar]
- 19.Chistoserdova L, Lapidus A, Han C, Goodwin L, Saunders L, et al. Genome of Methylobacillus flagellatus, molecular basis for obligate methylotrophy, and polyphyletic origin of methylotrophy. J Bacteriol. 2007;189:4020–27. doi: 10.1128/JB.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chistoserdova L, Laukel M, Portais JC, Vorholt JA, Lidstrom ME. Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J Bacteriol. 2004;186:22–28. doi: 10.1128/JB.186.1.22-28.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chistoserdova LV, Lidstrom ME. Genetics of the serine cycle in Methylobacterium extorquens AM1: identification of sgaA and mtdA and sequences of sgaA, hprA, and mtdA. J Bacteriol. 1994;176:1957–68. doi: 10.1128/jb.176.7.1957-1968.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chistoserdova LV, Lidstrom ME. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquens AM1. Microbiology. 1996;142:1459–68. doi: 10.1099/13500872-142-6-1459. [DOI] [PubMed] [Google Scholar]
- 23.Chistoserdova L, Vorholt JA, Lidstrom ME. A genomic view of methane oxidation by aerobic bacteria and anaerobic archaea. Genome Biol. 2005;6:208. doi: 10.1186/gb-2005-6-2-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chistoserdova L, Vorholt JA, Thauer RK, Lidstrom ME. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 25.Connon SA, Giovannoni SJ. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol. 2002;68:3878–85. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crowther GJ, Kosály G, Lidstrom ME. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J Bacteriol. 2008;190:5057–62. doi: 10.1128/JB.00228-08. Demonstrates that the H4F-linked C1 transfer pathway involving the enzymes MtdA and Fch is the major pathway to supply C1 units into the serine cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson VL. Electron transfer in quinoproteins. Arch Biochem Biophys. 2004;428:32–40. doi: 10.1016/j.abb.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 28.Dedysh SN, Knief C, Dunfield PF. Methylocella species are facultatively methanotrophic. J Bacteriol. 2005;187:4665–70. doi: 10.1128/JB.187.13.4665-4670.2005. Demonstrates that some methane utilizers are facultatively methylotrophic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donachie SP, Foster JS, Brown MV. Culture clash: challenging the dogma of microbial diversity. ISME J. 2007;1:97–99. doi: 10.1038/ismej.2007.22. [DOI] [PubMed] [Google Scholar]
- 30.Dumont MG, Murrell JC. Community-level analysis: key genes of aerobic methane oxidation. Methods Enzymol. 2005;397:413–27. doi: 10.1016/S0076-6879(05)97025-0. [DOI] [PubMed] [Google Scholar]
- 31.Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, et al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature. 2007;450:879–82. doi: 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- 32.Elliott GN, Chen WM, Chou JH, Wang HC, Sheu SY, et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 2007;173:168–80. doi: 10.1111/j.1469-8137.2006.01894.x. [DOI] [PubMed] [Google Scholar]
- 33.Erb TJ, Rétey J, Fuchs G, Alber BE. Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J Biol Chem. 2008;283:32283–93. doi: 10.1074/jbc.M805527200. [DOI] [PubMed] [Google Scholar]
- 34.Erb TJ, Berg IA, Brecht V, Müller M, Fuchs G, Alber BE. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalyl-CoA pathway. Proc Natl Acad Sci USA. 2007;104:10631–36. doi: 10.1073/pnas.0702791104. Demonstrates that ccr encodes a novel type of enzyme, crotonyl-CoA reductase/carboxylase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ettwig KF, Shima S, van de Pas-Schoonen KT, Kahnt J, Medema MH, et al. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol. 2008;10:3164–73. doi: 10.1111/j.1462-2920.2008.01724.x. [DOI] [PubMed] [Google Scholar]
- 36.Gak ER, Tsygankov YD, Chistoserdov AY. Organization of methylamine utilization genes (mau) in ‘Methylobacillus flagellatum’ KT and analysis of mau mutants. Microbiology. 1997;143:1827–35. doi: 10.1099/00221287-143-6-1827. [DOI] [PubMed] [Google Scholar]
- 37.Ginige MP, Hugenholtz P, Daims H, Wagner M, Keller J, Blackall LL. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl Environ Microbiol. 2004;70:588–96. doi: 10.1128/AEM.70.1.588-596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, et al. The small genome of an abundant coastal ocean methylotroph. Environ Microbiol. 2008;10:1771–82. doi: 10.1111/j.1462-2920.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- 39.Greenberg DE, Ding L, Zelazny AM, Stock F, Wong A, et al. A novel bacterium associated with lymphadenitis in a patient with chronic granulomatous disease. PLoS Pathog. 2006;2:e28. doi: 10.1371/journal.ppat.0020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenberg DE, Porcella SF, Stock F, Wong A, Conville PS, et al. Granulibacter bethesdensis gen. nov., sp nov., a distinctive pathogenic acetic acid bacterium in the family Acetobacteraceae. Int J Syst Evol Microbiol. 2006;56:2609–16. doi: 10.1099/ijs.0.64412-0. [DOI] [PubMed] [Google Scholar]
- 41.Greenberg DE, Porcella SF, Zelazny AM, Virtaneva K, Sturdevant DE, et al. Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis. J Bacteriol. 2007;189:8727–36. doi: 10.1128/JB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–71. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, et al. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 2007;1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- 44.Hou S, Makarova KS, Saw JH, Senin P, Ly BV, et al. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol Direct. 2008;3:26. doi: 10.1186/1745-6150-3-26. Analyzes the first genome of a Verrucomicrobia representative, a novel thermophilic and acidophilic methanotroph, “Candidatus Methylacidiphilum infernorum.”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hristova KR, Schmidt R, Chakicherla AY, Legler TC, Wu J, et al. Comparative transcriptome analysis of Methylibium petroleiphilum PM1 exposed to the fuel oxygenates methyl tert-butyl ether and ethanol. Appl Environ Microbiol. 2007;73:7347–57. doi: 10.1128/AEM.01604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Islam T, Jensen S, Reigstad LJ, Larsen O, Birkeland NK. Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci USA. 2008;105:300–4. doi: 10.1073/pnas.0704162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jewell T, Huston SL, Nelson DC. Methylotrophy in freshwater Beggiatoa alba strains. Appl Environ Microbiol. 2008;74:5575–78. doi: 10.1128/AEM.00379-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalyuzhnaya MG, Bowerman S, Lara JC, Lidstrom ME, Chistoserdova L. Methylotenera mobilis gen. nov., sp nov., an obligately methylamine-utilizing bacterium within the family. Methylophilaceae Int J Syst Evol Microbiol. 2006;56:2819–23. doi: 10.1099/ijs.0.64191-0. [DOI] [PubMed] [Google Scholar]
- 50.Kalyuzhnaya MG, Bowerman S, Nercessian O, Lidstrom ME, Chistoserdova L. Highly divergent genes for methanopterin-linked C1 transfer reactions in Lake Washington, assessed via metagenomic analysis and mRNA detection. Appl Environ Microbiol. 2005;71:8846–54. doi: 10.1128/AEM.71.12.8846-8854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalyuzhnaya MG, Chistoserdova L. Community-level analysis: genes encoding methanopterin-dependent enzymes. Methods Enzymol. 2005;397:443–54. doi: 10.1016/S0076-6879(05)97027-4. [DOI] [PubMed] [Google Scholar]
- 52.Kalyuzhnaya MG, De Marco P, Bowerman S, Pacheco CC, Lara JC, et al. Methyloversatilis universalis gen. nov., sp nov., a novel taxon within the Betaproteobacteria represented by three methylotrophic isolates. Int J Syst Evol Microbiol. 2006;56:2517–22. doi: 10.1099/ijs.0.64422-0. [DOI] [PubMed] [Google Scholar]
- 53.Kalyuzhnaya MG, Hristova KR, Lidstrom ME, Chistoserdova L. Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: implications for environmental detection of methylotrophy and evidence for convergent evolution. J Bacteriol. 2008;190:3817–23. doi: 10.1128/JB.00180-08. Demonstrates that some methylotrophs utilize a novel type of MDH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, et al. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol. 2008;26:1029–1034. doi: 10.1038/nbt.1488. Applies a massive sequencing approach (metagenomics) to characterize natural methylotroph communities. [DOI] [PubMed] [Google Scholar]
- 55.Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Utility of environmental primers targeting ancient enzymes: methylotroph detection in Lake Washington. Microb Ecol. 2004;48:463–72. doi: 10.1007/s00248-004-0212-6. [DOI] [PubMed] [Google Scholar]
- 56.Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Real-time detection of actively metabolizing microbes by redox sensing as applied to methylotroph populations in Lake Washington. ISME J. 2008;2:696–706. doi: 10.1038/ismej.2008.32. [DOI] [PubMed] [Google Scholar]
- 57.Kalyuzhnaya MG, Nercessian O, Lidstrom ME, Chistoserdova L. Development and application of polymerase chain reaction primers based on fhcD for environmental detection of methanopterin-linked C1-metabolism in bacteria. Environ Microbiol. 2005;7:1269–74. doi: 10.1111/j.1462-2920.2004.00831.x. [DOI] [PubMed] [Google Scholar]
- 58.Kane SR, Chakicherla AY, Chain PS, Schmidt R, Shin MW, et al. Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1. J Bacteriol. 2007;189:1931–45. doi: 10.1128/JB.01259-06. Erratum. 2007 . J Bacteriol. 189:4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kao WC, Chen YR, Yi EC, Lee H, Tian Q, et al. Quantitative proteomic analysis of metabolic regulation by copper ions in Methylococcus capsulatus (Bath) J Biol Chem. 2004;279:51554–60. doi: 10.1074/jbc.M408013200. [DOI] [PubMed] [Google Scholar]
- 60.Kiefer P, Portais JC, Vorholt JA. Quantitative metabolome analysis using liquid chromatography-high-resolution mass spectrometry. Anal Biochem. 2008;382:94–100. doi: 10.1016/j.ab.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 61.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol. 2005;71:467–79. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korotkova N, Chistoserdova L, Kuksa V, Lidstrom ME. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J Bacteriol. 2002;184:1750–58. doi: 10.1128/JB.184.6.1750-1758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korotkova N, Lidstrom ME. Connection between poly-beta-hydroxybutyrate biosynthesis and growth on C(1) and C(2) compounds in the methylotroph Methylobacterium extorquens AM1. J Bacteriol. 2001;183:1038–1046. doi: 10.1128/JB.183.3.1038-1046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korotkova N, Lidstrom ME. MeaB is a component of the methylmalonyl-CoA mutase complex required for protection of the enzyme from inactivation. J Biol Chem. 2004;279:13652–58. doi: 10.1074/jbc.M312852200. [DOI] [PubMed] [Google Scholar]
- 65.Korotkova N, Lidstrom ME, Chistoserdova L. Identification of genes involved in the glyoxylate regeneration cycle in Methylobacterium extorquens AM1, including two new genes, meaC and meaD. J Bacteriol. 2005;187:1523–26. doi: 10.1128/JB.187.4.1523-1526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Labbé N, Laurin V, Juteau P, Parent S, Villemur R. Microbiological community structure of the biofilm of a methanol-fed, marine denitrification system, and identification of the methanol-utilizing microorganisms. Microb Ecol. 2007;53:621–30. doi: 10.1007/s00248-006-9168-z. [DOI] [PubMed] [Google Scholar]
- 67.Large PJ, Peel D, Quayle JR. Microbial growth on C1 compounds. II Synthesis of cell constituents by methanol- and formate-grown Pseudomonas AM 1, and methanol-grown Hyphomicrobium vulgare. Biochem J. 1961;81:470–80. doi: 10.1042/bj0810470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lidstrom ME. Aerobic methylotrophic procaryotes. In: Balows A, Truper HG, Dworkin M, Harder W, Schleifer K-H, editors. The Prokaryotes. New York: Springer-Verlag; 2006. pp. 618–34. [Google Scholar]
- 69.Lidstrom ME, Anthony C, Biville F, Gasser F, Goodwin P, et al. New unified nomenclature for genes involved in the oxidation of methanol in gram-negative bacteria. FEMS Microbiol Lett. 1994;117:103–6. doi: 10.1111/j.1574-6968.1994.tb06749.x. [DOI] [PubMed] [Google Scholar]
- 70.Marx CJ, Lidstrom ME. Development of an insertional expression vector system for Methylobacterium extorquens AM1 and generation of null mutants lacking mtdA and/or fch. Microbiology. 2004;150:9–19. doi: 10.1099/mic.0.26587-0. [DOI] [PubMed] [Google Scholar]
- 71.Marx CJ, Miller JA, Chistoserdova L, Lidstrom ME. Multiple formaldehyde oxidation/detoxification pathways in Burkholderia fungorum LB400. J Bacteriol. 2004;186:2173–78. doi: 10.1128/JB.186.7.2173-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marx CJ, Van Dien SJ, Lidstrom ME. Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol. 2005;3:e16. doi: 10.1371/journal.pbio.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McDonald IR, Warner KL, McAnulla C, Woodall CA, Oremland RS, Murrell JC. A review of bacterial methyl halide degradation: biochemistry, genetics and molecular ecology. Environ Microbiol. 2002;4:193–203. doi: 10.1046/j.1462-2920.2002.00290.x. [DOI] [PubMed] [Google Scholar]
- 74.Meister M, Saum S, Alber BE, Fuchs G. L-malyl-coenzyme A/beta-methylmalyl-coenzyme A lyase is involved in acetate assimilation of the isocitrate lyase-negative bacterium Rhodobacter capsulatus. J Bacteriol. 2005;187:1415–25. doi: 10.1128/JB.187.4.1415-1425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitsui R, Kusano Y, Yurimoto H, Sakai Y, Kato N, Tanaka M. Formaldehyde fixation contributes to detoxification for growth of a nonmethylotroph, Burkholderia cepacia TM1, on vanillic acid. Appl Environ Microbiol. 2003;69:6128–32. doi: 10.1128/AEM.69.10.6128-6132.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moran MA, Buchan A, González JM, Heidelberg JF, Whitman WB, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–13. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 77.Moulin L, Munive A, Dreyfus B, Boivin-Masson C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature. 2001;411:948–50. doi: 10.1038/35082070. Erratum. 2001. Nature. 412:926. [DOI] [PubMed] [Google Scholar]
- 78.Nakatsu CH, Hristova K, Hanada S, Meng XY, Hanson JR, et al. Methylibium petroleiphilum gen. nov., sp nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int J Syst Evol Microbiol. 2006;56:983–89. doi: 10.1099/ijs.0.63524-0. [DOI] [PubMed] [Google Scholar]
- 79.Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer KH. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–39. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nercessian O, Noyes E, Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl Environ Microbiol. 2005;71:6885–99. doi: 10.1128/AEM.71.11.6885-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Neufeld JD, Boden R, Moussard H, Schäfer H, Murrell JC. Substrate-specific clades of active marine methylotrophs associated with a phytoplankton bloom in a temperate coastal environment. Appl Environ Microbiol. 2008;74:7321–28. doi: 10.1128/AEM.01266-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neufeld JD, Dumont MG, Vohra J, Murrell JC. Methodological considerations for the use of stable isotope probing in microbial ecology. Microb Ecol. 2007;53:435–42. doi: 10.1007/s00248-006-9125-x. [DOI] [PubMed] [Google Scholar]
- 83.Neufeld JD, Wagner M, Murrell JC. Who eats what, where and when? Isotope-labeling experiments are coming of age. ISME J. 2007;1:103–10. doi: 10.1038/ismej.2007.30. [DOI] [PubMed] [Google Scholar]
- 84.Oremland RS, Capone DG, Stolz JF, Fuhrman J. Whither or wither geomicrobiology in the era of ‘community metagenomics’. Nat Rev Microbiol. 2005;3:572–78. doi: 10.1038/nrmicro1182. [DOI] [PubMed] [Google Scholar]
- 85.Peyraud R, Kiefer P, Christen P, Massou S, Portais J-C, Vorholt JA. Demonstration of the ethylmalonyl-CoA pathway using 13C metabolomics. Proc Natl Acad Sci USA. 2009;106:4846–51. doi: 10.1073/pnas.0810932106. Uses high-resolution mass spectrometry-based metabolomics to demonstrate the operation of the ethylmalonyl-CoA pathway in glyoxylate regeneration in Methylobacterium extorquens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MS, Op den Camp HJ. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature. 2007;450:874–78. doi: 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 87.Pomper BK, Saurel O, Milon A, Vorholt JA. Generation of formate by the formyltransferase/hydrolase complex (Fhc) from Methylobacterium extorquens AM1. FEBS Lett. 2002;523:133–37. doi: 10.1016/s0014-5793(02)02962-9. Demonstrates that formate and not formylmethanofuran is the product of the H4MPT-mediated formaldehyde oxidation. [DOI] [PubMed] [Google Scholar]
- 88.Pomper BK, Vorholt JA, Chistoserdova L, Lidstrom ME, Thauer RK. A methenyl tetrahydromethanopterin cyclohydrolase and a methenyl tetrahydrofolate cyclohydrolase in Methylobacterium extorquens AM1. Eur J Biochem. 1999;261:475–80. doi: 10.1046/j.1432-1327.1999.00291.x. [DOI] [PubMed] [Google Scholar]
- 89.Radajewski S, Ineson P, Parekh NR, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–49. doi: 10.1038/35001054. Describes a methodology for identifying functional types involved in utilization of C1 compounds followed by the analysis of phylogenetic markers in the 13C DNA. [DOI] [PubMed] [Google Scholar]
- 90.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, et al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–21. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 91.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta. 2008;1784:1873–98. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–94. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 93.Reisch CR, Moran MA, Whitman WB. Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J Bacteriol. 2008;190:8018–24. doi: 10.1128/JB.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schaefer JK, Goodwin KD, McDonald IR, Murrell JC, Oremland RS. Leisingera methylohalidivorans gen. nov., sp nov., a marine methylotroph that grows on methyl bromide. Int J Syst Evol Microbiol. 2002;52:851–59. doi: 10.1099/00207713-52-3-851. [DOI] [PubMed] [Google Scholar]
- 96.Schäfer H, McDonald IR, Nightingale PD, Murrell JC. Evidence for the presence of a CmuA methyltransferase pathway in novel marine methyl halide-oxidizing bacteria. Environ Microbiol. 2005;7:839–52. doi: 10.1111/j.1462-2920.2005.00757.x. [DOI] [PubMed] [Google Scholar]
- 97.Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6:229. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith LM, Meijer WG, Dijkhuizen L, Goodwin PM. A protein having similarity with methylmalonyl-CoA mutase is required for the assimilation of methanol and ethanol by Methylobacterium extorquens AM1. Microbiology. 1996;142:675–84. doi: 10.1099/13500872-142-3-675. [DOI] [PubMed] [Google Scholar]
- 99.Stoecker K, Bendinger B, Schöning B, Nielsen PH, Nielsen JL, et al. Cohn’s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc Natl Acad Sci USA. 2006;103:2363–67. doi: 10.1073/pnas.0506361103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Studer A, McAnulla C, Büchele R, Leisinger T, Vuilleumier S. Chloromethane-induced genes define a third C1 utilization pathway in Methylobacterium chloromethanicum CM4. J Bacteriol. 2002;184:3476–84. doi: 10.1128/JB.184.13.3476-3484.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tavormina PL, Ussler W, 3rd, Orphan VJ. Planktonic and sediment-associated aerobic methanotrophs in two seep systems along the North American margin. Appl Environ Microbiol. 2008;74:3985–95. doi: 10.1128/AEM.00069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Theisen AR, Ali MH, Radajewski S, Dumont MG, Dunfield PF, et al. Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol Microbiol. 2005;58:682–92. doi: 10.1111/j.1365-2958.2005.04861.x. [DOI] [PubMed] [Google Scholar]
- 103.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–57. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 104.Trotsenko YA, Murrell JC. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol. 2008;63:183–229. doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- 105.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 106.Vigliotta G, Nutricati E, Carata E, Tredici SM, De Stefano M, et al. Clonothrix fusca Roze 1896, a filamentous, sheathed, methanotrophic gamma-proteobacterium. Appl Environ Microbiol. 2007;73:3556–65. doi: 10.1128/AEM.02678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vorholt JA. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch Microbiol. 2002;178:239–49. doi: 10.1007/s00203-002-0450-2. [DOI] [PubMed] [Google Scholar]
- 108.Vorholt JA, Chistoserdova L, Lidstrom ME, Thauer RK. The NADP-dependent methylene tetrahydromethanopterin dehydrogenase in Methylobacterium extorquens AM1. J Bacteriol. 1998;180:5351–56. doi: 10.1128/jb.180.20.5351-5356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vuilleumier S, Chistoserdova L, Lee M-C, Bringel F, Lajus A, et al. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS ONE. 2009;4:e5584. doi: 10.1371/journal.pone.0005584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wagner M, Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol. 2006;17:241–49. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 111.Ward N, Larsen Ø, Sakwa J, Bruseth L, Khouri H, et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath) PLoS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamamoto S, Wakayama M, Tachiki T. Characterization of theanine-forming enzyme from Methylovorus mays no. 9 in respect to utilization of theanine production. Biosci Biotechnol Biochem. 2007;71:545–52. doi: 10.1271/bbb.60590. [DOI] [PubMed] [Google Scholar]
- 113.Yamamoto S, Wakayama M, Tachiki T. Cloning and expression of Methylovorus mays no. 9 gene encoding γ-glutamylmethylamide synthetase: an enzyme usable in theanine formation by coupling with the alcoholic fermentation system of baker’s yeast. Biosci Biotechnol Biochem. 2008;72:101–9. doi: 10.1271/bbb.70462. [DOI] [PubMed] [Google Scholar]
- 114.Yasueda H, Kawahara Y, Sugimoto S. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J Bacteriol. 1999;181:7154–60. doi: 10.1128/jb.181.23.7154-7160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zarzycki J, Schlichting A, Strychalsky N, Müller M, Alber BE, Fuchs G. Mesaconyl-coenzyme A hydratase, a new enzyme of two central carbon metabolic pathways in bacteria. J Bacteriol. 2008;190:1366–74. doi: 10.1128/JB.01621-07. [DOI] [PMC free article] [PubMed] [Google Scholar]