Abstract
Notch signaling regulates many aspects of metazoan development and tissue renewal. Accordingly, misregulation or loss of Notch signaling underlies multiple human disorders, from developmental syndromes to adult onset diseases and cancer. Notch receptor activation is irreversible as it involves proteolysis-mediated release of the Notch intracellular domain, translocation to the nucleus, and association with a DNA-bound protein. Even though each Notch molecule signals only once without amplification by secondary messenger cascades, Notch signaling is remarkably robust in most tissues. In this review, we highlight the recent studies that reveal new molecular details involved in regulating ligand-mediated activation, receptor proteolysis and target selection.
The evolutionary conserved Notch signaling pathway functions as a mediator of short-range cell-cell communication. Notch signals select among preexisting cellular potentials; in a context dependent manner they will either promote or suppress proliferation, cell death, acquisition of specific cell fates and activation of differentiation programs throughout development and during maintenance of self-renewing adult tissues. Because Notch plays a critical role in many fundamental processes and in a wide range of tissues, it is not surprising that aberrant gain or loss of Notch signaling components have been directly linked to multiple human disorders, from developmental syndromes (e.g., Alagille, Teratology of Fallot, Syndactyly, Spondylocostal dysostosis, Familial Aortic Valve Disease; (Garg et al., 2005; Gridley, 2003)) to adult onset diseases (e.g., Cancer, Alzheimer’s disease, CADASIL (Louvi et al., 2006)). Notch signaling has emerged as a specific target in T-ALL (Weng et al., 2003) and colon cancer (van Es et al., 2005), and as a potential therapeutic target in the effort to curb tumor angiogenesis (Noguera-Troise et al., 2006; Ridgway et al., 2006). In addition, any meaningful manipulation of embryonic or adult stem cells will require development of receptor-specific antagonists and agonists of Notch signaling. As a consequence, research into the finer mechanistic detail of Notch activation and nuclear activity is of growing clinical and commercial relevance.
A Growing Family Of Notch Pathway Core Components
Notch receptors are large single pass Type I transmembrane proteins (see Figure 1A for domain organization). Whereas the fly genome contains only one Notch receptor, and worms have two that act redundantly (Fitzgerald et al., 1993), mammals have four Notch paralogs (Table 1) that display both redundant (for example, see (Krebs et al., 2003)) and unique functions (for example, see (Cheng et al., 2007)). The extracellular domain of all Notch proteins contains 29–36 tandem Epidermal growth factor (EGF)-like repeats, some of which mediate interactions with ligand (Figure 1A). Productive interaction with ligand presented by neighboring cells (trans interactions) are mediated by repeats 11–12 (whereas inhibitory interaction with ligand co-expressed in the same cell (cis interactions) are mediated by repeats 24–29 (de Celis and Bray, 2000). Many EGF repeats bind calcium, which plays an important role in determining the structure and affinity of Notch to its ligands (Cordle et al., 2008b; Raya et al., 2004). The EGF repeats are followed by a unique negative regulatory region (NRR) composed of three cysteine-rich Lin12-Notch repeats (LNR) and a heterodimerization domain (HD) (Figure 1A). The NRR plays a critical role in preventing receptor activation in the absence of ligand, and a detailed description of its structure will follow. Most surface Notch proteins are cleaved by furin-like convertases at site 1 (S1) located within an unstructured loop protruding from the HD subdomain, thereby converting the Notch polypeptide into an NECD-NTMIC (Notch extracellular domain-Notch transmembrane and intracellular domain) heterodimer held together by non-covalent interactions between the N- and C-terminal halves of HD (Figure 1A). S1 cleavage likely occurs in the secretory pathway (Figure 2) as secreted NRR modules undergo S1 cleavage (Malecki et al., 2006). However, the presence of less stable, uncleaved Notch molecules at the cell surface (Blaumueller et al., 1997; Bush et al., 2001) is also consistent with convertase cleavage occurring after receptor recycling.
Figure 1.
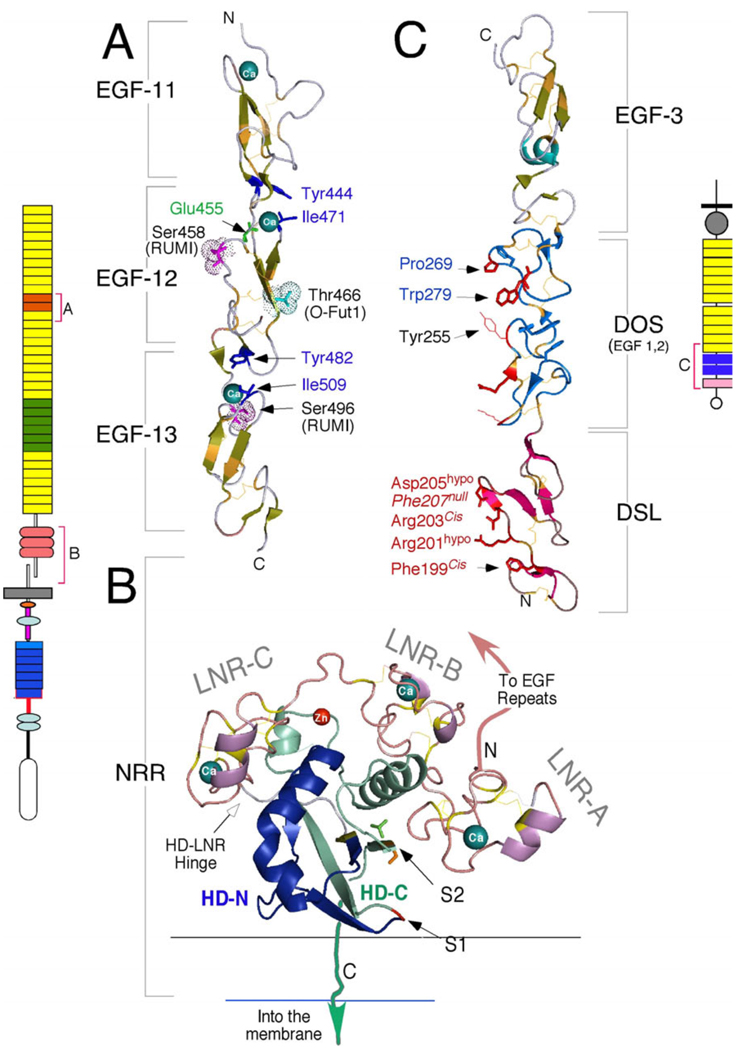
Domain organization of the Notch pathway receptors, ligands and co-ligands from fly, worm and mammals. A) Notch receptors are large Type I proteins that contain multiple extracellular EGF-like repeats. The single Drosophila (dNotch) and 4 mammalian Notch receptors (mNotch1–4) differ in the number of repeats (29–36) but all are much longer than the C. elegans Notch proteins (cLIN-12 and cGLP-1). Repeats 11–12 (orange) and 24–29 (green) mediate interactions with ligands. EGF repeats may contain consensus motifs for fucosylation by O-Fut1 and glycosylation by Rumi; the putative distribution of shared (green) and unique fucosylation (Cyan) and glycosylation (magenta) sites are shown for mNotch1 and mNotch2. Note that the ligand binding regions differ in their modification patterns. EGF repeats are followed by the Negative Regulatory Region (NRR), which is composed of three cysteine-rich Lin12-Notch repeats (LNR-A, B and C) and a heterodimerization domain (HD). In contrast to Drosophila Notch (dNotch), mammalian Notch proteins are cleaved by furin-like convertases at site 1 (S1). See text for more details on the intracellular domain. B) Ligands and potential ligands of Notch receptors can be divided into several groups based on their domain composition. Classical DSL ligands contain DSL, DOS and EGF motifs, and are not found in C. elegans (DSL/DOS/EGF ligands). C. elegans and mammalian DSL-only ligands lacking the DOS motif (DSL/EGF ligands) are a subtype of DSL ligands that may act alone (e.g., mDLL4) or in combination with DOS co-ligands (e.g., cDSL-1, and perhaps Dll3). This sub-family includes diffusible ligands. Functionally tested DOS co-ligands are marked with an asterisk; the role of mammalian DOS proteins is yet to be explored. Non-canonical ligands lack DSL and DOS domains but may act to facilitate the activation of Notch by DSL ligands and/or DOS co-ligands. Red brackets mark domains that have been crystallized alone or in combination with binding partners; some structural details will be addressed here but see (Blacklow upcoming review JCI). C) Details of the mouse Notch1 TMD (boxed) and flanking residues showing the cleavage sites and corresponding products. After ligand binding, Notch is cleaved at S2 by metalloproteases. γ-secretase can cleave multiple scissile bonds at S3 but only NICD molecules initiating at Val (V1744) evade N-end rule degradation (NICD-V). Cleavage then proceeds towards S4 until the short Nβ peptides can escape the lipid bilayer; most Nβ peptides are 21 amino acids long. The V1744G and K1749R amino acid substitutions (colorized) shift the S3 cleavage site (see text for details).
Table 1.
Core components and modifiers of the Notch pathway
| Component & Function | Drosophila |
Caenorhabditis elegans |
Mammals |
|---|---|---|---|
| Receptor | Notch | LIN-12, GLP-1 | Notch 1–4 |
| Ligand | |||
| DSL/DOS | Delta, Serrate | Dll1, Jagged1 and 2 | |
| DSL-only | APX-1, LAG-2, ARG-2, DSL1–7 | Dll3 and 4 | |
| DOS Co-ligands | DOS1–3, OSM7, 11 | DLK-1, DLK-2/EGFL9 | |
| Non-canonical | DNER, MAGP-1 and 2, F3/Contactin1, NB-3/Contactin6 | ||
| Nuclear Effectors | |||
| CSL DNA-binding transcription factor | Su(H) | LAG-1 | RBPjκ/CBF-1 |
| Transcriptional Co-activator | Mastermind | LAG-3 | MAML1–3 |
| Transcriptional Co-repressors | Hairless, SMRTR | Mint/Sharp/SPEN, NCoR/SMRT, KyoT2 | |
| Receptor Proteolysis | |||
| Furin convertase (S1 cleavage) | ? | ? | PC5/6, Furin |
| metalloprotease (S2 cleavage) | Kuzbanian, Kuzbanian-like, TACE | SUP-17/Kuzbanian, ADM-4/TACE | ADAM10/Kuzbanian, ADAM17/TACE |
| γ-secretase (S3/S4 cleavage) | Presenilin, Nicastrin, APH-1, PEN-2 | SEL-12, APH-1, APH-2, PEN-2 | Presenilin 1 and 2, Nicastrin, APH-1a–c, PEN-2 |
| Glycosyltransferase modifiers | |||
| O-fucosyl-transferase | OFUT-1 | OFUT-1 | POFUT-1 |
| O-glucosyl-transferase | RUMI | ||
| β1,3-GlcNAc-transferase | Fringe | Lunatic, Manic & Radical Fringe | |
| Endosomal Sorting/Membrane Trafficking Regulators | |||
| Ring Finger E3 Ubiquitin ligase (ligand endocytosis) | Mindbomb 1–2, Neuralized | Y47D3A.22 | Mindbomb, Skeletrophin, Neuralized 1–2 |
| Ring Finger E3 Ubiquitin ligase (receptor endocytosis) | Deltex | Deltex 1–4 | |
| HECT Domain E3 Ubiquitin ligase (receptor endocytosis) | Nedd4, Su(Dx) | WWP-1 | Nedd4, Itch/AIP4 |
| Negative regulator | Numb | Numb, Numb-like, ACBD3 | |
| Neuralized Inhibitors | Bearded, Tom, M4 | ||
| Other endocytic modifiers | sanpodo | ||
| NICD Degradation | |||
| F-Box Ubiquitin ligase | Archipelago | SEL-10 | Fbw-7/SEL-10 |
| Canonical Target bHLH Repressor Genes | E(spl) | REF-1 | HES/ESR/HEY |
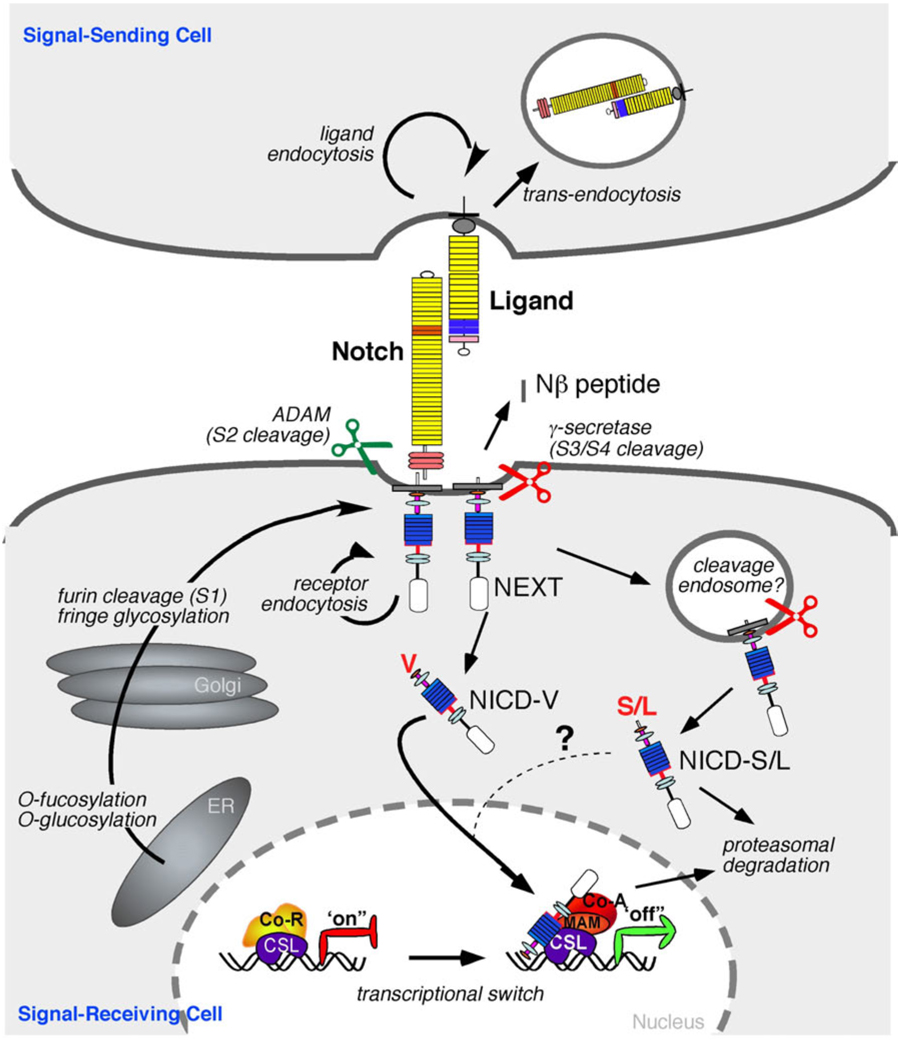
Figure 2.
The Core Notch Signaling Pathway is mediated by regulated proteolysis. Upon translation, the Notch protein is glycosylated by O-fut and Rumi, which are essential for the production of a functional receptor. The mature receptor is produced after proteolytic cleavage by PC5/furin at Site 1 (S1) and thereafter targeted to the cell surface as a heterodimer held together by non-covalent interactions. In cells expressing Fringe, the O-fucose is extended by the glycosyltransferase activity of Fringe, altering the ability of specific ligands to activate Notch. The Notch receptor is activated by binding to a ligand presented by a neighboring cell. Endocytosis and membrane trafficking regulate ligand and receptor availability at the cell surface. Ligand endocytosis is also thought to generate sufficient force to promote a conformational change that exposes Notch to cleavage at site S2 by ADAM metalloproteases (perhaps following heterodimer dissociation at S1). Juxtamembrane cleavage at S2 generates the membrane-anchored NEXT (Notch extracellular truncation) fragment, which is a subtrate for the γ-secretase complex. γ-secretase cleaves the Notch TMD progressively (from site S3 to S4; see Figure 1C) to release NICD (Notch intracellular domain) and Nβ peptides. γ-secretase cleavage can occur at the cell surface or in endosomal compartments however cleavage at the membrane favors the production of more stable form of NICD (see text for details). In the absence of NICD, the DNA-binding protein CSL associates with ubiquitous corepressor (Co-R) proteins and histone deacetylases (HDACs) to repress transcription of target genes. When NICD enters the nucleus, its binding to CSL may trigger an allosteric change that facilitates displacement of transcriptional repressors. Mastermind (MAM) then recognizes the NICD/CSL interface, and this tri-protein complex recruits coactivators (Co-A) to activate transcription.
The single transmembrane domain (TMD) is terminated by a ‘stop translocation’ signal comprised of 3–4 Arg/Lys residues. Intracellularly, the RAM (1RBPjκ association module) domain forms a high affinity binding module of 12–20 amino acids centered around a conserved WxP motif (Lubman et al., 2007). A long, unstructured linker containing one nuclear localizing sequence (NLS) links RAM to seven ankyrin repeats (ANK domain). Following the ANK domain are an additional bipartite NLS and a loosely defined and evolutionarily divergent transactivation domain (TAD). The very C-terminus contains conserved proline/glutamic acid/serine/threonine-rich motifs (PEST), containing degrons that regulate the stability of NICD. Drosophila Notch also contains the glutamine-rich OPA repeat (Figure 1A).
Our understanding of Notch ligands is rapidly evolving. Most Notch ligands are themselves Type I transmembrane proteins (Figure 1B), and recent studies have refined our understanding of their structure and function (Cordle et al., 2008a; Komatsu et al., 2008). The largest class of Notch ligands is characterized by three related structural motifs: an N-terminal DSL (Delta/Serrate/LAG-2) motif, specialized tandem EGF repeats called the DOS domain (Delta and OSM-11-like proteins, (Komatsu et al., 2008)), and EGF-like repeats (both calcium binding and non-calcium binding) (Figure 1B). As we will detail later, both the DSL and DOS domains are involved in receptor binding, with the DSL domain involved in both trans and cis interactions with Notch. DSL ligands can be classified based on the presence/absence of a cysteine-rich domain (Jagged/Serrate vs Delta, respectively) and a DOS domain (Komatsu et al., 2008)(Figure 1C). Positively acting DSL-only ligands (some of which are diffusible) have been described only in C. elegans (Chen and Greenwald, 2004; Komatsu et al., 2008). The activity of DSL-only ligands requires the presence of DOS-only co-ligands (Komatsu et al., 2008). In addition to the DSL/DOS and DSL-only ligands, and to the DOS-only co-ligands, the neural adhesion molecule F3/contactin (Hu et al., 2003), the related NB-3 protein (Cui et al., 2004), and the EGF repeat protein DNER (Eiraku et al., 2005; Saito and Takeshima, 2006) have been identified as potential Notch ligands in the CNS. The secreted microfibril associated proteins (MAGP-1 and 2) can activate Notch receptors in cultured cells (Miyamoto et al., 2006)(Figure 1B, Table 1), but a physiological function for these proteins in the Notch pathway is yet to be established.
It is now well established that Notch receptor activation is mediated by a sequence of proteolytic events (Figure 1C, Figure 2). Ligand binding leads to cleavage of Notch by ADAM metalloprotease(s) at site 2 (S2), which is located ~12 amino acids before the TMD and deeply buried within the NRR (Figure 1C, 3B and Supplemental movie 1). S2 cleavage is a key regulatory step in Notch activation but some ambiguity still exists regarding the enzyme(s) that mediate cleavage: only ADAM17/TACE is able to cleave Notch substrates in vitro (Brou et al., 2000), however, TACE null mice do not have a Notch phenotype (Peschon et al., 1998). In contrast, Kuzbanian/ADAM10/Sup-17 function is essential for Notch activity in all phyla (Hartmann et al., 2002; Lieber et al., 2002; Rooke and Xu, 1998; Sotillos et al., 1997; Wen et al., 1997).
Figure 3.
Atomic resolution details of the ligand-binding domain (EGF 11–13) from human Notch1 (PDB:2VJ3), the NRR from human Notch2 (PDB:2OO4) and the Notch-binding domain from human Jagged1 (PDB:2VJ2) generated with MacPymol (http://www.pymol.org). A) The ligand-binding domain of Notch1 (schematically depicted to the left) is centered on EGF repeat 12. The essential amino acids that coordinate Ca++ binding are shown in blue. O-glycosylation (Ser458, Ser496) and O-fucosylation (Thr466) sites are shown. Of note, the equivalent mutation to Glu455Val abolishes ligand binding in Drosophila, and this interface in hNotch1 was suggested to interact with hJagged1 DSL based on in silico docking models (Cordle et al., 2008a). However, glycosylation on Ser458 may block access to this site. Thr466 is essential for productive Notch activation in the mouse but not in the fly. B) The NRR domain folds to protect the S2 cleavage site (colorized side chains), which is located in a pocket protected by LNR-A, the HD-C helix, and the LNR-B/A linker. The furin cleavage site S1 lies within an unstructured loop that was removed to facilitate crystallization. LNR repeats bind calcium; chelation of these Ca++ atoms lead to NRR dissociation and Notch activation. See text and (Gordon et al., 2007) for further details. C) The crystal structure of the DSL, DOS and EGF repeat 3 of hJagged1 (schematically depicted to the right in trans binding orientation), highlighting the putative Notch binding interface (facing left)). The DSL fold is distinct from the EGF fold; amino acids in DSL that were shown to be required for interaction with Notch are labeled in red (see (Cordle et al., 2008a) for detail). Phe207Ala substitution generates a null protein whereas Arg203Ala and Phe199Ala substitutions ablate trans but not cis binding. Asp205Ala and Arg201Ala are hypomorphic. The DOS domain contains two conserved, atypical EGF repeats (defined by the presence of the conserved amino acids shown in blue (Komatsu et al., 2008)). Tyr255 is characteristic of Jagged DSL ligands and is replaced by a small hydrophobic amino acid in Delta-like proteins; this residue may be involved in defining sensitivity to Fringe.
The shedding of the Notch ectodomain creates a membrane-tethered intermediate (NEXT, for Notch extracellular truncation), which becomes a substrate for γ-secretase, a multi-component member of a growing family of Intramembrane cleaving proteases (I-CLiPs) (Wolfe and Kopan, 2004). γ-Secretase cleaves NEXT progressively within the TMD, most likely starting near the inner leaflet at S3 and ending near the middle of the TMD at S4 (Figure 1C). Only now is the Notch intracellular domain (NICD) free to translocate into the nucleus where it initially interacts with the DNA-binding protein CSL (CBF1/RBPjκ/Su(H)/Lag-1) via its RAM domain. The ANK domain then associates with CSL to help recruit the coactivator Mastermind/Lag-3. Mastermind recruits the MED8 mediator complex thereby leading to the upregulation of downstream target genes (reviewed in (Lubman et al., 2004)) (Figure 2). Additional proteins can modify the output from Notch receptors (listed in Table 1 and presented visually in (Ilagan and Kopan, 2007)). In the following sections, we will review in a step-wise fashion the recent significant developments in our understanding of the regulation of ligand-receptor recognition, receptor activation, its intramembrane proteolysis, and update the details of target activation by the Notch/CSL complexes.
I. Regulation of ligand-receptor interactions
Given that each Notch molecule undergoes proteolysis to generate a signal and thus can only signal once, regulation of either ligand or receptor availability at the cell surface are key to controlling Notch activation. A simple way of regulating availability is to restrict ligand and/or receptor expression spatially and temporally. Indeed, different ligands and receptors can have overlapping as well as distinct expression patterns during development and are subject to regulation by other signaling pathways ((Kolev et al., 2008) reviewed in (Tsuda et al., 2002; Wu and Bresnick, 2007)). While important, differential expression patterns of the ligands and receptors are not enough to explain the observed differences in signaling activity. Regulation of trafficking and post-translational modifications have emerged as important mechanisms controlling either ligand or receptor availability and/or productive ligand-receptor interactions. This has been discussed extensively in excellent recent reviews (Haines and Irvine, 2003; Le Borgne et al., 2005; Rampal et al., 2007; Stanley, 2007; Vodovar and Schweisguth, 2008); therefore, we will only briefly describe the key observations and highlight recent findings and controversies.
Ligand and receptor endocytosis and trafficking
Endocytic trafficking of the DSL ligands play a critical role in enhancing their signaling activity (Le Borgne, 2006; Nichols et al., 2007b). Ligand endocytosis is triggered by monoubiquitination mediated by the E3 ubiquitin ligases Neuralized, which preferentially recognizes Delta-type ligands, and Mindbomb, which preferentially recognizes Serrate/Jagged. Following endocytosis, a poorly characterized process produces a more active cell surface ligand. Current models explaining the nature of ligand modification include ligand clustering, post-translational modifications and/or recycling into specific membrane microdomains. Although the significance of the differences in ligand intracellular domains to Notch biology is unclear, interactions with intracellular scaffold proteins that associate with Delta but do not recognize Jagged proteins may send ligands to different membrane microdomains (Pfister et al., 2003). Interestingly, bearded family members, which negatively regulate Neuralized activity and thus, reduce the efficiency of Notch activation by Delta (Bardin and Schweisguth, 2006), are themselves Notch target genes (Lai et al., 2000) thereby forming a negative feedback loop. Fine-tuning this regulation are miRNAs that target Bearded and E(spl) proteins (Lai et al., 2005; Stark et al., 2003), thereby reducing their half life (Bray, 2006) and mitigating their impact on Neuralized. In addition to regulating endocytosis, miRNAs can also regulate Delta expression (Kwon et al., 2005). Whether endocytosis is also required for Notch activation by DNER, F3/Contactin1 or NB-3/Contactin6, and whether their concentration is regulated by miRNA, still have to be determined.
As with the DSL ligands, several mechanisms control the steady-state levels of the Notch receptors at the cell surface and therefore regulate their availability for ligand binding. For example, several E3 ubiquitin ligases - Deltex, Nedd4, Su(Dx)/Itch, Cbl - can control Notch receptor trafficking either towards lysosomal degradation or recycling, impacting its half life (reviewed in (Bray, 2006; Le Borgne et al., 2005)). Numb, in cooperation with the AP2 component α-adaptin and the AP2- or Numb-associated kinase (NAK), may promote Notch degradation in daughters of a asymmetric dividing cell. Numb is not active when lateral Notch signaling is occurring between resting cells (Le Borgne et al., 2005). Recent studies suggest that the restriction to dividing cells reflects a need for a partner that is trapped in the Golgi: the protein ACBD3 (Zhou et al., 2007). What makes this discovery fascinating is that physical separation between Numb (a cytosolic protein) and ACBD3 (a Golgi protein) prevents Numb from affecting Notch in resting cells. During mitosis, however, Golgi fragmentation allows the ACBD3/Numb complexes to form, allowing Numb to antagonize Notch activity via an unknown mechanism that is independent of Numb concentration (W.Z, personal communication) and thus may be catalytic. Notably, although mechanistic details of the inhibitory mechanism remain to be discovered, this mechanism can be activated in all cells with expression of a myristoylated form of ACBD3, which can constitutively associate with Numb. The Numb/ACBD3 complex inhibits lateral signaling even in cases where Notch activation results from contact between unrelated cells (Zhou et al., 2007). Thus, coupling the Golgi retention of ACBD3 with asymmetric segregation of Numb to one daughter cell is a unique way to take advantage of Golgi fragmentation and mitosis to assure that Notch activity is regulated in a precise spatial pattern (Zhou et al., 2007). As with Neuralized and Bearded, Notch can feed back and regulate Numb levels (Chapman et al., 2006), sustaining the signal in cells that attained high levels of Notch activation.
Receptor glycosylation
The role of glycosylation in Notch signaling is a growing field. It has been appreciated for some time that Notch receptors are large glycoproteins; many of their EGF repeats can be modified by two forms of O-glycosylation, O-fucose and O-glucose (reviewed in (Haines and Irvine, 2003; Rampal et al., 2007; Stanley, 2007; Visan et al., 2006b; Vodovar and Schweisguth, 2008)). Upon translation, the Notch protein is fucosylated on EGF repeats containing the consensus [(C2XXX(A/G/S)(S/T)C3] by the GDP fucose protein O-fucosyltransferase (O-fut1 in Drosophila/Pofut1 in mammals). This modification was previously thought to be essential for the production of a functional receptor (Lei et al., 2003; Okajima and Irvine, 2002; Okajima et al., 2003; Shi and Stanley, 2003) as loss of Ofut1 phenocopies Notch loss-of-function in flies and mice. However, later studies in Drosophila demonstrated that this was mainly due to the loss of the fucosylation-independent ER chaperone activity of Ofut; non-fucosylated Notch receptors still reach the cell surface, bind ligand and transduce signal (Okajima et al., 2008; Rampal et al., 2007; Stanley, 2007; Vodovar and Schweisguth, 2008).
Although it is still possible that O-fucosylation can facilitate proper Notch folding in the ER, fucosylation appears to be required only during Notch signaling events that depend on Fringe glycosyltransferase activity. Fringe extends O-fucose and this modification in the ligand binding domain can impact the ability of specific ligands to bind and activate Drosophila Notch: Fringe-mediated addition of a single N-acetylglucosamine on EGF repeat 12 is sufficient to enhance binding to Delta and reduce binding to Serrate in vivo and in vitro (Xu et al., 2007). Although glycosyltransferase enzymes have been conserved between flies and mice, the consequences of changes in Notch glycosylation and fucosylation in flies are not always mirrored in mammals and a role for Notch glycosylation is yet to be determined in C. elegans. Mice in which the fucosylation site on repeat 12 of Notch1 had been eliminated by a Thr466 substitution generated a hypomorphic allele that was unable to support T-cell differentiation in homozygoous animals, and was embryonic lethal in trans-heterozygotes over a null allele (Ge and Stanley, 2008). A similar allele in flies is hyperactive (Lei et al., 2003). Mammalian cells (Stahl et al., 2008) or animals (Ge and Stanley, 2008; Zhou et al., 2008) defective in fucosylation display a profound reduction in Notch signaling that extends beyond fringe-dependent processes. In contrast to Drosophila, surface Notch3 levels were not reduced in mouse Pofut1−/− ES cells relative to wild type controls (Stahl et al., 2008). Binding of monoclonal antibodies suggested that proper folding occurred, however, ligand binding was completely abolished, consistent with a subtle change in folding (Stahl et al., 2008). Indeed, ligand binding in Pofut1-deficient cells can be rescued by overexpressing an inactive, unrelated protein (α-glycosydase I), which refolds the Notch ECD and rescues ligand binding in the absence of Pofut1 (Stahl et al., 2008). This experiment confirms that fucose is not required for ligand binding and suggests that a global upregulation in chaperone activity, induced by over expression, and not the dedicated chaperone activity of Ofut/Pofut1, rescues Notch folding in mammals. Further support for ligand binding to “sugarless” Notch is provided by in vitro binding studies with bacterially produced, unmodified receptor and ligand interacting domains (Cordle et al., 2008a; Cordle et al., 2008b).
In mammals, full elucidation of the effects of Fringe on Notch activity is complicated by the presence of multiple receptors, ligands and Fringe proteins (Lunatic (Lfng), Manic (Mfng), and Radical (Rfng) fringe). Moreover, vertebrate glycosyltransferases appear to make a mechanistic contribution to Notch biology different to that in flies. Lunatic Fringe (Lfng) modifies Notch in T-cells in a manner that enhances Delta-to-Notch signaling and limits Jagged to Notch signals (Tsukumo et al., 2006; Visan et al., 2006a; Visan et al., 2006b), whereas it inhibits Delta-to-Notch signaling in the somite (Dale et al., 2003; Kageyama et al., 2007). In another puzzle, fringe-modified Notch2 retains its ability to respond to Jagged1 whereas fringe-modified Notch1 does not (Hicks et al., 2000). The distribution of consensus fucosylation and glycosylation sites on mouse Notch1 and Notch2 reveals a largely conserved glycosylation pattern (green, Figure 1A); however, distinct, paralog-specific distribution of glycosylation sites is apparent within the ligand-binding domain, which may contribute to some of the observed receptor-specific responses to ligands (Hicks et al., 2000). Further modifications by β1,4-galactosyltransferases (and perhaps sialyltransferases) may also play a modulatory role in mammalian cells in certain contexts (Chen et al., 2001).
RUMI, the glycosyltransferase that adds the O-glucose to serine residues in the consensus [(C1XSXPC2], was recently identified in Drosophila (Acar et al., 2008). Loss of Rumi leads to impaired Notch signaling in a variety of contexts, indicating that it is a general regulator of Notch signaling. Unlike Ofut1, Rumi’s function resides mainly in its glucosyltransferase activity. Rumi is required for productive ligand binding to occur and induce S2 and subsequent S3/S4 cleavages. It is possible that glycosylation contributes to the strength of ligand-receptor interaction (see below). Notably, O-glucose can be further extended, possibly by xylosyltransferases, but the existence of such modifications in Notch remains to be demonstrated (Haines and Irvine, 2003).
Delta and Serrate/Jagged ligands also contain consensus glycosylation sites and can be substrates for both O-fucosylation and Fringe modification (Panin et al., 2002). As Ofut1 and Rumi both appear to function cell-autonomously in signal-receiving cells (Acar et al., 2008; Okajima and Irvine, 2002; Sasamura et al., 2003), the biological significance of glycosylation in the Notch pathway is largely based on receptor modulation.
The generation of glycosylation-deficient Notch alleles in vertebrates coupled with the development of improved methods to detect glycosylation status of receptors in various in vivo contexts will undoubtedly continue to make important contributions to Notch biology. However, the exact mechanistic contribution of sugar to Notch biology remains a mystery. Glycans appear to play a minor part in the ligand/receptor recognition mechanism (Cordle et al., 2008a; Cordle et al., 2008b), but they may contribute to the mature conformation of the Notch extracellular domain, thereby modulating the receptor activation process. While a full molecular explanation for the differential effects of glycosylation may have to wait for the crystal structure of the respective receptor/ligand complexes, the data we summarized above is consistent with the hypothesis that fucosylation and glycosylation at critical residues define the strength of receptor-ligand interactions, altering the probability of activation and consequently, signal strength. Mammalian and fly Notch proteins lacking serine in their 12th EGF repeat may behave differently due to the different distribution of fucose and glucose on their surface.
II. Receptor Activation
The key to Notch activation is the regulation of ectodomain shedding
Ligand-induced, metalloprotease mediated cleavage serves as a key regulatory point in Notch signal transduction (Brou et al., 2000; Mumm et al., 2000). The S2 cleavage site resides within the NRR domain, which encompasses the LNR and HD regions. The NRR functions to prevent Notch proteolysis in the absence of ligand ((Greenwald, 1994; Kimble et al., 1998; Kopan et al., 1996) Figure 1A)); “leaky” signaling occurs when point mutations (Weng et al., 2004) or viral integration (Girard et al., 1996; Girard and Jolicoeur, 1998) disrupts the NRR, causing human T-ALL and lymphomas in mouse, respectively. Mutations in the linker between the LNR domains also result in activated Notch phenotypes in C. elegans (Levitan and Greenwald, 1995), underscoring the conserved nature of the mechanism that keeps Notch “off” in the absence of ligands.
How does the NRR region prevent an ADAM protease from cleaving Notch, and how can ligand reverse this block? Early models attempting to explain the function of the NRR postulated that receptor oligomers were resistant to proteolysis and that ligand binding generated monomeric Notch molecules (Kopan et al., 1996; Struhl and Adachi, 2000). However, biochemical measurement of the oligomeric state of wild type and mutation-activated Notch proteins at the cell surface revealed that the oligomerization status of Notch did not correlate with its activity (Vooijs et al., 2004). Importantly, Notch dimerization was mediated by the EGF repeats and not by the NRR, leaving the NRR function unexplained (Vooijs et al., 2004). While it is unlikely that oligomerization plays a major role in NRR function (see below), changes in oligomerization status may still be important for optimal ligand-receptor binding, the stoichiometry of which remains to be determined. Ligand and/or receptor oligomerization plays an important role in several other signaling pathways (e.g., GPCRs, RTKs, integrins) and it will be interesting to see whether this will also apply to Notch. Hints that oligomerization could be involved emerge from (1) the ability of receptors and ligands to form homodimers via their EGF repeats (Vooijs et al., 2004), (2) the clustering of cell surface Notch receptors at sites of contact with Delta-expressing cells (Bardot et al., 2005; Nichols et al., 2007a), and (3) the requirement of soluble DSL ligands to be pre-clustered before they can activate Notch receptors on the cell surface (Hicks et al., 2002; Varnum-Finney et al., 2000). Receptor and ligand oligomerization may enhance binding and could explain the strong adhesion forces between Delta and Notch-expressing cells as determined by atomic force microscopy (Ahimou et al., 2004).
An alternative mechanism for Notch activation was inspired by the observation that in Drosophila, the Notch ectodomain was trans-endocytosed by ligand-presenting/signal-sending cells while the Notch intracellular domain was localized to signal-receiving cells (Parks et al., 2000) (Figure 2). A genetic link between endocytosis and Notch signaling was strengthened by the characterization of the Dynamin homolog in Drosophila, shibire (shi). Dynamin, a pleckstrin homology repeat containing GTPase, is necessary for pinching-off clathrin coated pits from the plasma membrane during endocytosis. Shi mutants show strikingly similar phenotypes to Notch loss-of-function alleles during several developmental processes in Drosophila (Parks et al., 2000; Seugnet et al., 1997). Genetic analyses during peripheral nervous system development indicated that NEXT-like molecules lacking NRR are properly processed at the S3 site in shi mutants (Struhl and Adachi, 2000), indicating that endocytosis was only required for NRR-containing molecules. Parks et. al. proposed that the mechanical strain generated by receptor trans-endocytosis somehow exposed the S2 site for cleavage (see also (Le Borgne and Schweisguth, 2003)). While unable to explain the details of how the NRR worked to block S2 cleavage, the conformational-change model was born to explain how trans-endocytosis would nullify the NRR.
Given that Notch molecules engaged by ligand have already been cleaved at S1 either en route to the plasma membrane or during receptor recycling, the mechanical force generated by trans-endocytosis could simply be facilitating NECD/NTMIC heterodimer dissociation and subsequent exposure of the S2 cleavage site. Observations supporting this model came from the demonstration that calcium chelation resulted in dissociation (NECD shedding), permitting subsequent S3 cleavage (Rand et al., 2000). Urea unfolding analysis of HD domains carrying T-ALL-associated mutations established that most activating mutations had destabilizing effects, thereby enhancing dissociation around the pre-existing S1 site (Malecki et al., 2006). Subsequently, it was demonstrated that ligand binding dissociated NECD/NTMIC heterodimers in the presence of metalloprotease inhibitors (Nichols et al., 2007a), indicating dissociation occurs prior to, and independent of S2 cleavage. Notably, S2 cleavage was still necessary to gain full Notch activity even in dissociated molecules (Nichols et al., 2007a). Because most Drosophila Notch proteins may not be cleaved by protein convertases (Kidd and Lieber, 2002), non-enzymatic dissociation alone may not be sufficient to explain the conserved receptor activation mechanism. Moreover, evidences in support of dissociation are also consistent with a conformational change model: Notch activation via calcium chelation or mutations in NRR could have altered the domain structure to permit S2 cleavage, and furin cleavage site deletions may have altered Notch structure to increase its stability and alter its transport and/or maturation. Nonetheless, these experiments underscore the need for force to bypass the NRR and shed the NECD.
Seeing is believing: the NRR forms a fortress around S2
A recent high-resolution structure of the NRR domain has provided molecular details regarding NRR function (Figure 3B) (Gordon et al., 2007). The HD domain (HD-N and HD-C) forms a globular folded domain that makes extensive contacts with the three calcium-binding LNR modules (LNR-A, B and C, Figure 3B). S1 is located within an unstructured loop that does not contribute to the stability of the HD domain (Gordon et al., 2007; Malecki et al., 2006). Conversely, S2 is located in a β-strand buried within an inaccessible pocket (Figure 3B, Supplemental movie 1). Direct steric occlusion (by the LNR-AB linker) and global domain stabilization (by interactions between LNR-B and the HD-C helix) both prevent premature cleavage of the receptor in the absence of ligand. Thus, LNR-A, the LNR-AB linker, and LNR-B must all be removed to produce molecules with constitutive signaling activity. Given the deep active site pocket in ADAM17/TACE (Ingram et al., 2006; Maskos et al., 1998; Wasserman et al., 2003), Gordon et al. speculate that not only does the receptor activation mechanism forcibly lift at least two of the three LNR repeats, but the process must also disengage the stabilizing helix within the HD domain from the S2 containing strand (perhaps by partially unfolding the helix) to permit access to the scissile bond at S2 (a model the authors dub “lift and cut”). The NRR crystal structure reveals that Ca++ chelation will most likely unfold the LNR repeats and not just the HD domain (Figure 3B), suggesting that “lift” could simultaneously induce complete receptor dissociation. Given the dissimilarity between TACE knockouts and Notch mutants, an alternative possibility that must be considered is that another enzyme, with a shallower active site, mediates cleavage of S2 with minimal conformational changes in the HD domain.
The NRR structure clearly defines the "off" state of the receptor, confirms that auto-inhibition is intrinsic to monomeric Notch molecules, and delineates the domain shown genetically to keep the receptor inactive. The structure also provides a molecular logic for requiring a large-scale conformational movement, supporting the idea first suggested by Parks et. al. that mechanical forces will be needed to expose the metalloprotease cleavage site. However, the precise mechanism involved in transferring tensile force along the extracellular domain remains obscure, awaiting crystallization of ligand/receptor complexes and/or measurement of the force within individual units of receptor-ligand interactions in living cells. In addition, it remains to be determined whether differential glycosylation regulates the adhesion strength between Notch and its ligands. If we assume that maximal adhesion (read, force) develops only between glycosylated receptors and their ligands, the need to unfold the NRR could explain how Rumi and Fringe contribute to Notch signaling without affecting binding (Acar et al., 2008; Hicks et al., 2000).
Further support for the importance of the NRR region and S2 accessibility was recently provided by a consortium effort to develop functional agonistic and antagonistic antibodies against Notch3 (Li et al., 2008). Two high affinity antagonists and one agonist were identified and found to bind to adjacent epitopes within the Notch3 NRR (Li et al., 2008). The agonist increased S2 cleavage and ectodomain shedding in a receptor-specific and metalloprotease-sensitive manner, whereas the antagonists blocked NEXT production in response to ligand. Interestingly, the antagonists formed a “lock” by binding to an epitope comprised by amino acids in both LNR-A and HD-C of Notch3, likely increasing the energy required for “lift”. In contrast, the agonist bound to an epitope in LNR-A and most likely interfered with the LNR-A/HD interaction. That binding of an antibody to LNR-A can result in NRR dissociation suggests that the NRR structure is dynamic, alternating between a “closed” (crystal structure) and a hypothetical “open” state. This dynamic structure could potentially even allow access to proteases at some low probability, providing a possible explanation for the recent report of ligand-independent cleavage of full-length (i.e., NRR-containing) Notch receptors by ADAMs (Delwig and Rand, 2008).
Collectively, these results are consistent with the view that limiting accessibility to S2 is the key function of the NRR, that force is likely involved, and that cancer-causing mutations in HD shift the equilibrium to an “open” state. Similarly, agonistic antibodies and high ADAM concentrations may be trapping or exploiting the “open” conformation to activate Notch in a ligand-independent manner. While turning Notch signaling “off” pharmacologically via γ-secretase inhibition has become a common experimental tool, a deeper understanding of the S2 control switch now makes it possible to transiently turn endogenous Notch signaling “on” whenever needed for therapeutic or tissue-engineering purpose. The agonistic antibody can thus be viewed as the first truly soluble ligand, binding to the NRR instead of the EGF repeats.
May the Force be with you: Notch activation by diffusible ligands
Multiple lines of evidence support the idea that leverage is important in ligand-mediated activation of Notch and that soluble ligands act as dominant negative proteins (Hukriede and Fleming, 1997; Hukriede et al., 1997). However, this view of Notch activation has been challenged mainly by the fact that 5 out of the 10 C. elegans DSL ligands are soluble (Chen and Greenwald, 2004; Komatsu et al., 2008). If the Notch activation mechanism is conserved and requires unfolding/dissociation of the NRR in all species, how can diffusible DSL ligands like the C. elegans DSL1 activate Notch? The recent discovery and characterization of five C. elegans co-ligands have perhaps moved us significantly closer to solving this mystery (Cordle et al., 2008a; Komatsu et al., 2008). Hart and colleagues noticed that all C. elegans DSL ligands lacked a DOS domain that is present in most DSL ligands from other phyla (Figure 1B). The DOS domain encompasses the first 2 EGF repeats and has a consensus of C-X(3)-C-X(3,8)-C-X(2,5)-C-[KVER]-C-X(10,12)-C-X(1,3)-PX(6,9)-CX(1,4)-W-X(1,4)-C (X=any amino acid). Hart and colleagues provide genetic evidence that the soluble DOS protein OSM-11 cooperates with the DSL-only ligands and perhaps other DOS proteins to stimulate Notch activation in a subset of developmental contexts in C. elegans. These observations have led to a model suggesting that secreted and membrane-bound DOS proteins work with membrane-bound and secreted DSL ligands, respectively, to gain sufficient leverage for receptor activation. This bipartite ligand binding remains to be confirmed biochemically. However, the importance of the DOS domain for receptor binding is supported by positive interactions between LIN-12 and OSM-11 in yeast two hybrid assays (Komatsu et al., 2008), and by previous biochemical studies showing that the first two EGF repeats of Jagged1 were critical for high affinity binding to cell surface receptors (Shimizu et al., 1999). Interestingly, DOS domain-only proteins are also present in mammalian cells: Delta-like1 (DLK1) and DLK2/EGFL9. Mammalian DLK1 (and perhaps, DLK2) can compensate for the loss of OSM-11 (Komatsu et al., 2008), raising the possibility that DLK-1 and DLK-2 proteins could enhance Notch activation by mammalian DSL-only ligands (Dll3, Dll4) in certain physiological contexts while competing with DSL/DOS ligands (Dll1, Jag1 and Jag2) in others. Dll3 is unable to replace Dll1 in vivo (Geffers et al., 2007) and is unable to activate Notch in cellulo (Ladi et al., 2005); the in cellulo experiments may need to be repeated in the presence of DLK1 and/or DLK2 to rule out Dll3 as a Notch activator. An additional possibility for the mode of action of the secreted DOS proteins and non-canonical ligands is that they may interact with extracellular matrix proteins to provide sufficient leverage to unfold/dissociate the NRR and activate Notch.
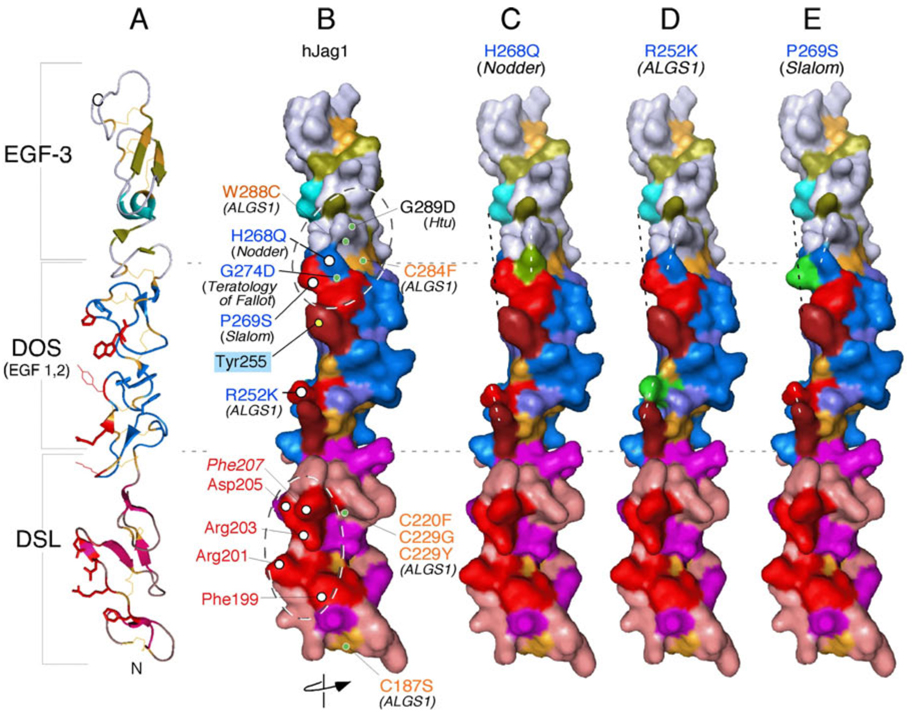
Further support for the importance of the DOS domain stems from the Jagged1 crystal structure (the first crystal structure of a mammalian Notch ligand fragment (Cordle et al., 2008a)) and from mapping known human and mouse mutations in Jagged1 onto the crystal structure (Figure 3C, Figure 4). The crystallized Jagged1 fragment, which contained the DSL domain and the first 3 EGF repeats, formed an extended rod-like structure. The DSL domain has a distinct organization that bears some structural similarities to an EGF repeat (Figure 3C). A positively charged cluster of highly conserved residues within the DSL domain identifies a Notch-binding surface, the importance of which was confirmed by mutagenesis, in vitro binding and in vivo functional assays (Figure 3C, 4)(Cordle et al., 2008a). The DOS domain encompasses EGF repeats 1 and 2 which exhibit atypical secondary structures while forming the classical EGF disulfide bond pattern, and therefore define a distinct functional domain. Mutations linked to Alagille syndrome and Teratology of Fallot in humans, and to autosomal dominant inner ear malformations in mice [headturner (Htu; (Kiernan et al., 2001), slalom (Tsai et al., 2001), and Nodder (Ingenium; (Russ et al., 2002))] cluster near a common DOS area (Figure 4). Altogether, these amino acids define another potential receptor-binding surface contiguous with the one identified in DSL. Mutations in headturner and Teratology of Fallot affected amino acids buried under the surface defined by slalom and nodder mutations and may impact the structure of the putative Notch binding site within DOS (Figure 4). Although many independent observations confirm that the 12th EGF repeat in Notch is critical for ligand binding, the interaction domains within Jagged1 span an area greater than repeat 12 alone (Figure 3A and C, all domains at the same scale). Elucidation of how the DOS and DSL domains engage the Notch receptor simultaneously will have to await the crystallization of the relevant ligand domains (DSL, DOS and EGF) with the appropriate interacting domain from Notch. Moreover, since both Rumi and Pofut1 sites are present on repeat 12 (Figure 3A), it will be interesting to see how sugar moieties affect the ligand binding sites.
Figure 4.
Mapping known autosomal dominant mutations on the surface of hJagged1 indicates that the DOS domain could form part of the Notch-binding interface. The hJagged1 ribbon structure (A) was surface rendered and rotated such that the Notch-binding interface is facing the reader (B–E). B) Structural model of the wild-type ligand showing biologically relevant residues. Alagille syndrome (ALGS)-associated missense mutations that are likely to affect disulfide bonding (and thus the structural integrity of these domains) are labeled in gold. Additional relevant DSL and DOS domain amino acids are labeled in red and blue, respectively, and further distinguished as being surface-exposed (white circles) or buried within the structure (green circles). A positively charged cluster of highly conserved surface-exposed residues within the DSL domain (labeled in red) identifies a putative Notch-binding surface (see text and Figure 3 legend). Interestingly, missense mutations associated with Teratology of Fallot in humans, and to autosomal dominant inner ear malformations in mice (i.e., headturner (Htu), slalom and Nodder) cluster near a common DOS region. Mutations in headturner and Teratology of Fallot affect amino acids buried under the surface defined by slalom and Nodder mutations and may impact the structure of the potential Notch binding site within DOS. Note that R252 (ALGS1) and Y255 (unique to Jagged; see Figure 3 legend) are also aligned with the putative Notch binding surfaces on the DSL and DOS domains. C–E) Modeling of specific substitutions of surface amino acids (highlighted in green) in the DOS domain results in realignment of the surface (e.g., Nodder and ALGS1; dashed white lines) or perturbations into space (e.g., slalom; dashed black line) that could potentially affect interactions with Notch. The exact topology of Notch/ligand interface remains to be explored by co-crystallization.
Altogether, the recent studies have further emphasized the importance of leverage in Notch activation. Therefore, significant concerns arise regarding the interpretation and physiological relevance of observations reliant on Notch activation mediated solely by diffusible ligand fragments, by synthetic DSL peptides or by bacterially produced DSL ligands. Additional studies should be done to establish whether they are exerting their apparent biological effects via the Notch pathway by using molecules harboring mutations in the Notch-binding DOS and DSL motifs defined by the studies summarized above.
What comes NEXT: Intramembrane proteolysis of Notch
Even though all membrane-tethered forms of Notch can interact with γ-secretase within the secretory pathway (Ray et al., 1999), only molecules with a free amino-terminus become substrates for intramembrane proteolysis by γ-secretase (Chavez-Gutierrez et al., 2008; Shah et al., 2005). The length of the extracellular domain is critical for efficient cleavage - longer regions are less efficiently cleaved (Mumm et al., 2000; Struhl and Adachi, 2000). This explains why, despite dissociation at S1 by ligand endocytosis (Nichols et al., 2007a) or addition of agonistic antibodies (Li et al., 2008), inhibition of metalloproteases still significantly reduced Notch S3 cleavage and target activation. This is also the likely explanation for the decreased cleavage of NEXT molecules containing extracellular dimerization domain fusions.
Once NEXT enters the active site, the TMD is sequentially cleaved, starting near the cytosolic leaflet (Qi-Takahara et al., 2005; Zhao et al., 2005). This initial cleavage at the S3 site releases NICD, while the last cleavage at S4 releases the Nβ peptide (so named after the Aβ peptide, which is released from another γ-secretase substrate Amyloid Precursor Protein (APP) and is associated with Alzheimer’s disease (Okochi et al., 2002)) (Figure 1C, 2). Immunoprecipitation and Edman sequencing of mouse Notch1 C-terminal fragments identified a single NICD species starting at Val1744 (Schroeter et al., 1998). More recently, mass spectrometric analysis of cleavage products from an in vitro assay using NEXT-like substrates identified NICD variants with diverse N-termini (NICD-V, NICD-L, NICD-S; Figure 1C) (Tagami et al., 2008). Quantification of the variants in a reconstituted system and in cells treated with proteasome inhibitors showed that the predominant scissile bond lies between L1746 and S1747, and not between G1745 and V1744, as previously thought. Importantly, these NICD variants were also produced from full-length Notch activated by co-culture with ligand-expressing cells and in embryonic and adult mouse tissues, suggesting that these occur in vivo. As expected from the N-end rule, NICD-S and NICD-L are rapidly degraded by the 26S proteasome degradation, making them extremely short-lived in cellulo (Blat et al., 2002; Tagami et al., 2008; Varshavsky, 1996). Although we cannot rule out some role for the short-lived products, NICD-V (starting at V1744) likely mediates the bulk of Notch1 signals due to its stability (Figure 2). The genetic evidence supporting this conclusion comes from re-analyzing mice homozygous for the Notch V1744G allele (Huppert et al., 2000), which was originally thought to be highly resistant to γ-secretase cleavage. Instead, this amino acid substitution shifted the cleavage site to generate more of the labile NICD-L(Tagami et al., 2008). The subsequent reduction in NICD stability proved detrimental to Notch signaling in vivo (Huppert et al., 2000), providing evidence that the labile NICD molecules are insufficient to compensate for loss of NICD-V.
Location, Location, Location
In addition to regulating receptor maturation and cell surface levels, endosomal sorting has an important role in preventing improper, ligand-independent, Notch receptor activation (Le Borgne et al., 2005). Mutations in ESCRT complex proteins vps25 or erupted/Tsg101/vps23 lead to accumulation of Notch in a late endosomal vesicle, which surprisingly permits ectopic activation of Notch via γ-secretase-dependent proteolysis (Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005; Vaccari et al., 2008). Another protein, Lethal (2) Giant Discs (LGD) is also required to maintain the “off” state of Notch; when LGD levels are altered by either loss or over-expression, ligand-independent activation is seen (Childress et al., 2006; Gallagher and Knoblich, 2006; Jaekel and Klein, 2006; Justice et al., 2003). It remains to be seen if ADAM and γ-secretase are involved in LGD-mediated activation. Since ligand also accumulates, it is unclear if the ectopic activation process represents cis-stimulation, shedding and intramembrane proteolysis, or shedding-independent activity of γ-secretase. Mis-trafficking of Notch may place it in a compartment where proteolysis is less constrained, perhaps because the NRR shifts to an “open” conformation at a lower pH. In summary, the ESCRT complexes and LGD are likely to be normally involved in Notch downregulation, indicating that endosomal sorting acts to restrict activation at or near the cell surface and may contribute to pathogenesis in different cellular contexts.
The identification of the subcellular location where Notch S3/S4 cleavage occurs during the normal ligand-activation process has been controversial. Dynamin/shi, Rab5 or the endocytic syntaxin avl, which are all involved in early endosome formation, are required in signal-receiving cells. Although it has been demonstrated that endocytosis is not required for NEXT cleavage in vivo (Struhl and Adachi, 2000), it was suggested that mono-ubiquitination and endocytosis of Notch are required to target the Notch/γ-secretase complex into an endocytic vesicle where efficient S3 cleavage will occur. The K1749R substitution in Notch simultaneously abolished mono-ubiquitination, endocytosis and NICD accumulation (Gupta-Rossi et al., 2004). However, an alternative explanation for this result emerged from analyzing the K1749R TMD mutant for scissile bond selection by γ-secretase. Like V1744G, the K1749R substitution caused a shift in scissile bond preference, producing labile NICD species instead of NICD-V and leading to a loss of Notch activity. In addition to TMD composition, scissile bond selection was also strongly influenced by the subcellular localization of the γ-secretase/substrate complex during cleavage (Figure 2) (Tagami et al., 2008). At the plasma membrane, the bond between G1743 and V1744 is preferentially cleaved, generating the stable NICD-V. However, in endosomes γ-secretase preferentially hydrolyzes the bond between L1746 and S1747, generating labile NICD-L and NICD-S perhaps due to lower pH (Fukumori et al., 2006). The idea that the stable NICD-V is generated at the plasma membrane or in the earliest vesicle to pinch off is consistent with the observation that non-cell permeable γ-secretase inhibitors can still block Notch proteolysis (Tarassishin et al., 2004). Therefore, although γ-secretase is active in many cellular membranes, and its proteolytic activity is independent of the composition of the TMD region, scissile bond selection and consequently, the stability of NICD, are highly dependent on both cellular location and TMD composition (Tagami et al., 2008). It is clear that from now on, efforts attempting to correlate Notch activity, endosomal location and proteolysis will have to take proteasome inhibition into consideration in order to properly assess the presence/absence of NICD/Notch activity. It is worth noting in this context that the apical polarity protein Crumbs was proposed to restrict the activity of γ-secretase and thus to limit the extent of Notch activation (Herranz et al., 2006). This too needs to be reevaluated as Crumbs may instead impact scissile bond selection.
So what is the role of endocytosis in Notch activation? The important observation that NICD-V is produced before/during budding of the endocytic vesicle led us to revisit the hypothesis that translocation into “cleavage endosome” is an important step in Notch activation. An alternative explanation for the phenotypes associated with the loss of dynamin/shi, Rab5 or syntaxin avl in signal-receiving cells (Vaccari et al., 2008) would be that these deficiencies lower the forces generated by trans-endocytosis thereby reducing ligand-induced NRR dissociation and subsequent activation. If so, expression of receptors containing NRR point mutations that affect domain folding but remain ligand-dependent should suppress Rab5, shi or avl mutations. Loss of hrs affects a later step and instead leads to Notch accumulation in an avl-positive, early endosomal compartment upstream of the ESCRT or LGD compartments. Accordingly, hrs loss does not lead to ectopic Notch activation (Childress et al., 2006; Jaekel and Klein, 2006; Vaccari et al., 2008) whereas the ESCRT and LGD mutations do.
Interestingly, it was discovered that loss of the aquaporin-related channel Big Brain (bib), one of the original neurogenic genes in Drosophila, not only affected endosomal maturation but also blocked all forms of Notch signaling, including ectopic activation in ESCRT mutants (Kanwar and Fortini, 2008). Surprisingly, Bib exerts its effects on Notch signaling not by preventing Notch from entering into a “cleavage endosome” but rather by acting downstream of S3 cleavage (Kanwar and Fortini, 2008). NEXT-like molecules were cleaved in bib−/− cells to form a NICD-like fragment, but ectopic Notch activity was not detected, indicating a defect in nuclear entry or in target activation (Kanwar and Fortini, 2008). However, NICD overexpression can bypass the requirement for Bib, establishing that loss of Bib does not compromise the NICD nuclear translocation machinery, CSL availability or target gene activation. In addition, the bib phenotypes do not indicated a general deficiency impeding nuclear entry of other cytosolic proteins (like SMAD, Armadillo). Instead, the generated NICD appears to remain associated with endosomes. Acidification of endosomes fails to occur in bib−/− cells and Bib proteins that lack ion channel activity mimic bib loss of function (Kanwar and Fortini, 2008). How release of NICD could be affected by the acidity of endocytic compartments remains to be elucidated. The authors proposed that the defect might reflect impaired association of the endosome with cytoskeletal or cytoplasmic transport factors. An alternative explanation for these observations is that in bib−/− cells, scissile bond selection by γ-secretase is altered, such that cleavage occurred closer to middle of the TMD, producing NICD molecules with longer, lipophilic amino termini that kept NICD anchored to the membrane. As the resolution of Western blots is not sufficient to compare the composition of NICD produced by wild type and bib−/− cells, testing this hypothesis will require mass spectrographic analysis of NICD produced in bib−/− endosomes.
γ-secretase function in Notch signaling: additional complexities
γ-Secretase cleavage of Notch had traditionally been thought of as a constitutive proteolytic event, with the critical regulatory steps occurring either upstream (i.e., ligand binding & ectodomain shedding) or downstream of intramembrane cleavage (i.e., NICD degradation, as discussed below). However, as γ-secretase activity and function has been further characterized, we have come to realize that intramembrane proteolysis can also be regulated by a variety of factors (Parks and Curtis, 2007). γ-Secretase in composed of 4 membrane proteins in a 1:1:1:1 stoichiometry (Sato et al., 2007): the catalytic component presenilin and three limiting cofactors NCT, Pen2 and Aph1, which are necessary and sufficient to reconstitute enzymatic activity. Because of the availability of 2 presenilin and 2 APH proteins (3 in mice), at least four different enzyme complexes exist in mammalian cells (Shirotani et al., 2007) that can have differing biochemical properties and protein interactions. Indeed, PS1 vs PS2 containing complexes exhibit different specific activities (Bentahir et al., 2006; Lai et al., 2003) and γ-secretase containing different Aph1 isoforms make specific contributions in vivo (Dejaegere et al., 2008; Serneels et al., 2005). Although the relevance of γ-secretase composition to Notch biology is still unexplored, but some studies suggest that different Aph1 complexes might contribute differentially to Notch signaling (Serneels 2005 PNAS). The remaining key questions are whether different γ-secretase complexes reside in different subcellular locations, have different optimal pH and membrane lipid composition and can provide a biochemical basis for scissile site selection in Notch.
Notably, like many Type I proteins, Notch ligands are also subject to extracellular cleavage by ADAM proteases followed by TMD cleavage by γ-secretase (Ikeuchi and Sisodia, 2003; LaVoie and Selkoe, 2003; Six et al., 2003). Ligand processing may be important to reduce its antagonistic function in cis (Mishra-Gorur et al., 2002), for limiting its availability, and/or for membrane clearance. Although it could, in principle, generate biologically active fragments, no physiological evidence has yet emerged to support bi-directional signaling by ligand ICDs.
III. Transcriptional regulation
Once NICD is released by γ-secretase, it translocates to the nucleus. The processes and proteins that regulate nuclear translocation are still unclear. In the nucleus, NICD is unable to bind DNA on its own, but it acts to affect transcription with the help of its partner (a CSL protein). CSL directs NICD to specific targets, the recognition of which appears to be independent of Notch (Kovall, 2007). NICD/CSL could also affect nuclear events by competing with other factors for MAM/MAML. The nuclear milieu that exists before NICD arrives in the nucleus will dictate which targets will be available to CSL and thus, activated by Notch (reviewed in (Bray, 2006)). Recent studies have begun to explore this regulation in greater detail.
CSL as a repressor
Studies in Drosophila indicate that in the absence of NICD, Su(H) actively represses its target promoters. Loss of Su(H) in a presenilin-deficient background leads to transient activation of Notch target genes (Koelzer and Klein, 2006). Su(H) mediates repression by recruiting SKIP (Zhou et al., 2000), hairless/CtBP (Morel et al., 2001) and Gro/TLE (Barolo et al., 2002; Nagel et al., 2005). In addition, Su(H) can silence transcription at multiple sites via recruitment of Asf1, a histone chaperone involved in nucleosome assembly (Goodfellow et al., 2007). Interestingly, modulating Asf1 levels does not impact targets of the Wnt, SHH, TGF or EGF pathways (Goodfellow et al., 2007) implying that it has a specific role in repressing Notch targets. In Drosophila S2-N cells, SKIP is a companion of Su(H) in the repressor complex. Importantly, whereas knockdown of Su(H) in S2-N cells de-repressed only the two transcripts regulated by Notch (M3 and Mβ; (Krejci and Bray, 2007)), knockdown of Asf1 de-repressed additional E(spl) transcripts that curiously, were all located centromeric to M3 (Goodfellow et al., 2007). Asf1 must remain associated with these promoters to maintain a stable nucleosome complex, and thus repression, in the absence of Su(H). By inference, Su(H)/SKIP/hairless/Asf1 complexes must transiently bind to such sites, delivering Asf1 to an unknown partner.
In the mammalian nucleus, although RBPjκ can form complexes with many ubiquitous co-repressor proteins such as CIR, FLH1C/KyoT2 and NCoR/SMRT (Bray, 2006), it is SHARP/MINT/SPEN (Kuroda et al., 2003; Oswald et al., 2002) that has emerged as the critical repressor of Notch target genes (Oswald et al., 2005; Tsuji et al., 2007). Interestingly, in human cell lines, RBPjκ is continually detected on the Hes1 promoter in the absence of NICD (Fryer et al., 2004), whereas recent studies in Drosophila cell lines demonstrated that occupancy of sites on the E(spl) complex is a dynamic process, with silenced sites being rarely occupied by Su(H) (Krejci and Bray, 2007).
It is important to note that target repression is not the rule: In C. elegans, in contrast to Drosophila, loss of the CSL protein LAG-1 does not result in phenotypes characteristic of a gain-of-Notch function. In addition, expression of ref-1, a target in many Notch-mediated decisions, is not elevated when CSL binding sites are mutated (Neves et al., 2007). Therefore, in nematodes, Notch targets are already poised for transcription and are not actively repressed by LAG-1. In mammalian skin, the phenotype of RBPjκ loss is not as severe as that seen when multiple Notch receptors or γ-secretase is lost, an observation that could be consistent with de-repression of Notch targets in RBPjκ null animals (Demehri et al., 2008). Surprisingly, removal of RBPjκ in Notch or presenilin mutants did not alleviate their phenotype, suggesting that in the skin, target repression does not play an important role and raising the possibility that Notch signals in a poorly-defined, RBPjκ-independent manner (Demehri et al., 2008). Similarly, target de-repression was not observed during the differentiation of T-helper cells (Ong et al., 2008).
Transcriptional Activation and Target Promoter Selection
Whether there is active repression or not, the binding of NICD to CSL mediates the “transcriptional switch” to activation of gene expression at the target promoter. The elucidation of several crystal structures of Notch, CSL, and the Notch/CSL/MAM nuclear complexes from multiple organisms provides detail at the atomic resolution (Kovall, 2007). They confirm that the activation complex forms in a step-wise manner (reviewed in (Lubman et al., 2004)) and provide insight with regards to the molecular changes likely to facilitate switching from repression to activation (reviewed in (Barrick and Kopan, 2006; Kovall, 2007)). The high affinity binding of the RAM domain to CSL increases the local concentration of ANK, thereby permitting it to bind to RBPjκ and promote dissociation of transcriptional repressors (Bertagna et al., 2008; Friedmann et al., 2008). The ANK/CSL interface is then recognized by Mastermind/LAG-3 protein (Nam et al., 2006; Petcherski and Kimble, 2000; Wilson and Kovall, 2006), and this ternary complex recruits histone acetyltansferases, chromatin remodeling factors and the mediator complex (Fryer et al., 2004) to assemble an active transcription complex on target promoters.
When the promoters contain optimally spaced head to head sites (SPS; Su(H) paired sites), cooperative binding is observed in vitro, mediated by ANK/ANK interactions between two CSL/RAMANK/MAM complexes (Nam et al., 2007). While cooperativity explains why site orientation is important (Cave et al., 2005; Ong et al., 2006), it is unclear whether such complexes are important in vivo and whether they also form on promoters in which spacing is suboptimal. Notably, the amino acids mediating these interactions are conserved on all 4 vertebrate Notch paralogs, allowing one to speculate that heterotypic interactions among different Notch paralogs may refine the regulation of transcription by Notch proteins (Nam et al., 2007). Interestingly, these specific amino acids are not conserved in C. elegans Notch proteins and consequently, neither are SPS sites in the genomes of different nematode species (Neves and Priess, 2005). It is however important to note that in vertebrates, multimerization of CSL binding sites is sufficient to elicit Notch-dependent activation in vivo in some but not all sites where Notch signaling is active (Mizutani et al., 2007; Souilhol et al., 2006).
It is tempting to describe SPS-containing genes as high affinity Notch targets, however it is clear that even Hes1, the archetypical SPS-containing Notch target, is not always responsive to Notch1 (Lee et al., 2007) and that many genes that contain SPS in their promoters do not respond to Notch signaling (our unpublished observations and (Neves and Priess, 2005)). Moreover, many characterized Notch-responsive enhancers are combinatorial. In Drosophila, Notch interacts with the bHLH protein daughterless (Cave et al., 2005) or the LIM domain protein grainyhead (Furriols and Bray, 2001) to activate SPS-containing promoters of the E(spl) complex genes. In C. elegans, Notch (LIN-12) interacts with a GATA related protein to regulate endodermal expression of ref-1 and with an NK-class factor to drive its mesodermal expression (Neves et al., 2007). It is conceivable that tissue-specific target expression will be controlled by the ability of different Notch paralogs to interact with diverse transcription factors bound on neighboring enhancers. Evidence for a qualitative difference among Notch paralogs was recently shown in vitro, where Notch3 seemed best equipped to interact with a nearby Zn-finger protein (Ong et al., 2006).
Additional partners in the nucleus?
Several studies have shown that when overexpressed, NICD can interact with different transcriptional cofactors from multiple signaling pathways (e.g., SMADs, NFκB and HIF1α) to impact transcription on their target promoters. If these reports do not reflect interactions between adjacent enhancer-bound complexes, NICD molecules would have to be distributed among putative partners according to their affinity and the local concentrations of such putative partners. As detailed earlier, interaction with CSL is mediated through a conserved WxP motif in the RAM domain with a small contribution from the ANK domain (Lubman et al., 2007; Tamura et al., 1995; Tani et al., 2001). Despite some sequence divergence, all 4 mammalian RAM domains interact with RBPjκ with similar affinity (~200 nM (Del Bianco et al., 2008; Friedmann et al., 2008; Lubman et al., 2007)). This affinity is not high enough to exclude the possibility that NICD associates with other proteins. However, it is important to note that free NICD is not detected at equilibrium in vitro when RBPjκ is in stoichiometric excess (Lubman et al., 2007). Therefore under physiological conditions, wherein a high level of RBPjk is coupled with low nuclear concentration of NICD, it is unlikely that a significant number of NICD molecules will associate with other partners. Notably, RBPjκ can associate with at least one partner other than Notch - the bHLH protein p48/PTF1a (Beres et al., 2006; Masui et al., 2007). Thus, it cannot be ruled out that when RBPjκ concentration is limiting, some NICD molecules could associate with other factors such as SMAD, HIF1α or NFκB. This remains to be demonstrated with physiological concentrations of NICD.
As is the case with RBPjκ, MAM proteins can also associate with factors other than Notch/CSL complexes (Kankel et al., 2007), such as β-catenin (Alves-Guerra et al., 2007), Mef2c (Shen et al., 2006) and p53 (Zhao et al., 2007). A competition between NICD and Mef2c for MAML1 offers a long sought after mechanism for myogenic inhibition by Notch: even truncated ANK domains that cannot activate Notch targets (Shawber et al., 1996) could still associate with RBPjκ when overexpressed (Del Bianco et al., 2008; Kato et al., 1997) and thus titrate MAM away from Mef2C (Shen et al., 2006). In summary, it appears that both co-activators and co-repressors acting in the Notch signaling pathway are shared with additional pathways, providing an alternative explanation for why overexpression of NICD could impact transcription driven by other factors.
All good things must pass: Signal downregulation
Activation of Notch receptors releases a quantum of signal in the form of NICD. Most Notch-mediated processes require a transient pulse of activity for instance, in developmental contexts where iterative activation of the Notch pathway is required. In some tissues, this could last a fraction of the cell cycle (Ambros, 1999; Bessho and Kageyama, 2003; Hirata et al., 2004). Even the few processes that require prolonged activation still seem to be sensitive to the activation “strength”, a yet to be defined aspect of Notch signaling. Given what we know about Notch biology, sustained Notch activation can be deleterious. Thus, in addition to the above-mentioned mechanisms that regulate NICD production, optimal signal strength is regulated in most cells by ensuring that NICD half-life is short. During the transcriptional activation process, NICD is phosphorylated on its PEST domain by the CDK8 kinase and targeted for proteasomal degradation by the E3 ubiquitin ligase Sel10/Fbw7 (Fryer et al., 2004; O'Neil et al., 2007; Thompson et al., 2007; Tsunematsu et al., 2004). This eliminates NICD, disassembles the ternary complex and resets the cell for the next round of signaling. Highlighting the importance of NICD turnover is the observation that C-terminal or PEST domain deletions or mutations that stabilize NICD can cause T-ALL in humans (Weng et al., 2004). Indeed, further studies on the T-ALL associated deletions identified additional conserved regulatory phosphorylation sites in Notch1 (Chiang et al., 2006). The kinase(s), phosphatase(s) and/or Ubiquitin ligases that target these sites remain to be identified. It is also unclear whether their regulatory mechanisms are coupled to transcriptional activation similar to CDK8 and Fbw7.
Conclusions and Perspectives
The efforts to understand the role developmental pathways in adult homeostasis and disease requires detailed knowledge of how the “on” and “off” states of such pathways are brought about, what mechanisms ensure their robustness, what vulnerabilities lead to disease and how to control or restore the balance to achieve a desired biological outcome. In the three decades following the cloning of Notch, a significant body of work has provided detailed mechanistic understanding of Notch activation and signal transduction. These efforts provided new tools with which to inhibit or activate Notch signals, and atomic level resolution of key structural elements involved in receptor activation and transcription complex assembly. Next, we await the structural analysis of ligand-receptor complexes as well as direct measurements of forces involved in Notch activation to help bridge the major gaps in our understanding of ligand-mediated receptor activation. The growing array of combinatorial possibilities of DOS protein-DSL ligand interactions could enable fine control over forces exerted by ligands on Notch (and hence, activation probability). CSL-corepressor complexes also need to be examined at the atomic level to better understand the transcriptional switch. The efforts to identify and characterize cellular activities that enable other signaling pathways to control the output from Notch proteins are ongoing (Hurlbut et al., 2007; Poellinger and Lendahl, 2008), offering promise of research tools to better our understanding of the pleiotropic effects of Notch signaling in development and disease as well as additional potential therapeutic avenues.
Recent studies have also led to a new appreciation of the underlying complexities of Notch proteolytic activation. Scissile bond selection, the impact of N-end rule degradation and the dependence of both on the subcellular location of cleavage will necessitate redesigning of experiments aiming to measure NICD production. Further developments in mass spectrometry may one day enable analysis of peptides isolated from small biological samples, improving investigation into the function of the various endocytic trafficking modulators of Notch. Improvements in ChIP technology will uncover new nuclear partners and help complete the story of target selection by different Notch paralogs in different cellular contexts and under physiological levels of NICD, which are too low to currently allow such investigation.
Another major hurdle yet to be addressed relates to the issue of redundancy. In which processes do Notch paralogs have specific or redundant functions? Do heterotypic NICD interactions occur at target promoters, and do they have a biological function? Developmental syndromes associated with Notch loss will benefit from receptor-specific agonists, or activation of paralog-specific targets, if present. Receptor-specific antagonists (e.g., Notch1 inhibition in T-ALL) are predicted to work better than γ-secretase inhibitors if redundancy with other Notch paralogs will alleviate toxicity associated for general pathway inhibition. Related to this issue are mechanistic questions differentiating qualitative and quantitative models for target selection and activation by different Notch paralogs and by different concentration of NICD. Despite some progress in tools for monitoring Notch pathway activity (for example, see (Ohtsuka et al., 2006; Souilhol et al., 2006; Vooijs et al., 2007), the field will benefit greatly from improved tools (e.g., antibodies, reporter strains, non-invasive imaging approaches) to identify cells engaged in Notch signaling, to quantify the levels of all four NICD proteins, and to monitor target activation and record its biological consequences.
Finally, despite genetic confirmation that a non-canonical, γ-secretase-dependent but RBPjκ-independent Notch signaling occurs in mammals and in flies, its mechanism remains as obscure as ever, presenting an interesting challenge to the field.
Supplementary Material
Acknowledgements
We apologize to our colleagues whose important contributions had not been mentioned due to the limited space. We are grateful to Dr. Craig Micchelli, Dr. Tim Schedl Dr. Jim Skeath, and Dr. Marc Vooijs for their critical reading of this review. We also thank Dr Anne Hart, Weimin Zhong. and members of the Kopan lab for helpful discussions. Our research work on Notch signaling is supported by grants from the National Institutes of Health GM55479 (to RK) and R21-NS06168001 (to MI).
Footnotes
RAM was identified in a yeast two-hybrid screen and originally named “RBP-jκ associated molecule”. The acronym has been retained to reflect function.
Contributor Information
Raphael Kopan, Email: kopan@wustl.edu.
Ma. Xenia G. Ilagan, Email: ilaganmg@wustl.edu.
References
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahimou F, Mok LP, Bardot B, Wesley C. The adhesion force of Notch with Delta and the rate of Notch signaling. J Cell Biol. 2004;167:1217–1229. doi: 10.1083/jcb.200407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Guerra MC, Ronchini C, Capobianco AJ. Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 2007;67:8690–8698. doi: 10.1158/0008-5472.CAN-07-1720. [DOI] [PubMed] [Google Scholar]
- Ambros V. Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development. 1999;126:1947–1956. doi: 10.1242/dev.126.9.1947. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Schweisguth F. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev Cell. 2006;10:245–255. doi: 10.1016/j.devcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bardot B, Mok LP, Thayer T, Ahimou F, Wesley C. The Notch amino terminus regulates protein levels and Delta-induced clustering of Drosophila Notch receptors. Exp Cell Res. 2005;304:202–223. doi: 10.1016/j.yexcr.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Barolo S, Stone T, Bang AG, Posakony JW. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002;16:1964–1976. doi: 10.1101/gad.987402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick D, Kopan R. The Notch transcription activation complex makes its move. Cell. 2006;124:883–885. doi: 10.1016/j.cell.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Bentahir M, Nyabi O, Verhamme J, Tolia A, Horre K, Wiltfang J, Esselmann H, De Strooper B. Presenilin clinical mutations can affect gamma-secretase activity by different mechanisms. J Neurochem. 2006;96:732–742. doi: 10.1111/j.1471-4159.2005.03578.x. [DOI] [PubMed] [Google Scholar]
- Beres TM, Masui T, Swift GH, Shi L, Henke RM, MacDonald RJ. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertagna A, Toptygin D, Brand L, Barrick D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochem Soc Trans. 2008;36:157–166. doi: 10.1042/BST0360157. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Kageyama R. Oscillations, clocks and segmentation. Curr Opin Genet Dev. 2003;13:379–384. doi: 10.1016/s0959-437x(03)00083-2. [DOI] [PubMed] [Google Scholar]
- Blat Y, Meredith JE, Wang Q, Bradley JD, Thompson LA, Olson RE, Stern AM, Seiffert D. Mutations at the P1(') position of Notch1 decrease intracellular domain stability rather than cleavage by gamma-secretase. Biochem Biophys Res Commun. 2002;299:569–573. doi: 10.1016/s0006-291x(02)02705-5. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Qi HL, Zagouras P, Artavanistsakonas S. Intracellular Cleavage Of Notch Leads to a Heterodimeric Receptor On the Plasma Membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A Novel Proteolytic Cleavage Involved in Notch Signaling: The Role of the Disintegrin-Metalloprotease TACE. Molecular Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Bush G, diSibio G, Miyamoto A, Denault JB, Leduc R, Weinmaster G. Ligand-induced signaling in the absence of furin processing of Notch1. Developmental Biology. 2001;229:494–502. doi: 10.1006/dbio.2000.9992. [DOI] [PubMed] [Google Scholar]
- Cave JW, Loh F, Surpris JW, Xia L, Caudy MA. A DNA transcription code for cell-specific gene activation by notch signaling. Curr Biol. 2005;15:94–104. doi: 10.1016/j.cub.2004.12.070. [DOI] [PubMed] [Google Scholar]
- Chapman G, Liu L, Sahlgren C, Dahlqvist C, Lendahl U. High levels of Notch signaling down-regulate Numb and Numblike. J Cell Biol. 2006;175:535–540. doi: 10.1083/jcb.200602009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Gutierrez L, Tolia A, Maes E, Li T, Wong PC, de Strooper B. Glu(332) in the Nicastrin ectodomain is essential for gamma-secretase complex maturation but not for its activity. J Biol Chem. 2008;283:20096–20105. doi: 10.1074/jbc.M803040200. [DOI] [PubMed] [Google Scholar]
- Chen JH, Moloney DJ, Stanley P. Fringe modulation of Jagged1-induced Notch signaling requires the action of beta 4galactosyltransferase-1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13716–13721. doi: 10.1073/pnas.241398098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Greenwald I. The Lateral Signal for LIN-12/Notch in C. elegans Vulval Development Comprises Redundant Secreted and Transmembrane DSL Proteins. Dev Cell. 2004;6:183–192. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MY, Xu ML, Histen G, Shestova O, Roy M, Nam Y, Blacklow SC, Sacks DB, Pear WS, Aster JC. Identification of a conserved negative regulatory sequence that influences the leukemogenic activity of NOTCH1. Mol Cell Biol. 2006;26:6261–6271. doi: 10.1128/MCB.02478-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress JL, Acar M, Tao C, Halder G. Lethal giant discs, a novel C2-domain protein, restricts notch activation during endocytosis. Curr Biol. 2006;16:2228–2233. doi: 10.1016/j.cub.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008a;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordle J, Redfieldz C, Stacey M, van der Merwe PA, Willis AC, Champion BR, Hambleton S, Handford PA. Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J Biol Chem. 2008b;283:11785–11793. doi: 10.1074/jbc.M708424200. [DOI] [PubMed] [Google Scholar]
- Cui XY, Hu QD, Tekaya M, Shimoda Y, Ang BT, Nie DY, Sun L, Hu WP, Karsak M, Duka T, et al. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J Biol Chem. 2004 doi: 10.1074/jbc.M313505200. [DOI] [PubMed] [Google Scholar]
- Dale JK, Maroto M, Dequeant ML, Malapert P, McGrew M, Pourquie O. Periodic Notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature. 2003;421:275–278. doi: 10.1038/nature01244. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Bray SJ. The Abruptex domain of Notch regulates negative interactions between Notch, its ligands and Fringe. Development. 2000;127:1291–1302. doi: 10.1242/dev.127.6.1291. [DOI] [PubMed] [Google Scholar]
- Dejaegere T, Serneels L, Schafer MK, Van Biervliet J, Horre K, Depboylu C, Alvarez-Fischer D, Herreman A, Willem M, Haass C, et al. Deficiency of Aph1B/C-gamma-secretase disturbs Nrg1 cleavage and sensorimotor gating that can be reversed with antipsychotic treatment. Proc Natl Acad Sci U S A. 2008;105:9775–9780. doi: 10.1073/pnas.0800507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch 1 transcription complexes. J Mol Biol. 2008;376:131–140. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwig A, Rand MD. Kuz and TACE can activate Notch independent of ligand. Cell Mol Life Sci. 2008;65:2232–2243. doi: 10.1007/s00018-008-8127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, Grigsby PW, Miner JH, Farr AG, Kopan R. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, Kengaku M. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8:873–880. doi: 10.1038/nn1492. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K, Wilkinson HA, Greenwald I. glp-1 can substitute for lin-12 in specifying cell fate decisions in Caenorhabditis elegans. Development. 1993;119:1019–1027. doi: 10.1242/dev.119.4.1019. [DOI] [PubMed] [Google Scholar]
- Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008;283:14781–14791. doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer CJ, White JB, Jones KA. Mastermind Recruits CycC:CDK8 to Phosphorylate the Notch ICD and Coordinate Activation with Turnover. Mol Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Fukumori A, Okochi M, Tagami S, Jiang J, Itoh N, Nakayama T, Yanagida K, Ishizuka-Katsura Y, Morihara T, Kamino K, et al. Presenilin-dependent gamma-secretase on plasma membrane and endosomes is functionally distinct. Biochemistry. 2006;45:4907–4914. doi: 10.1021/bi052412w. [DOI] [PubMed] [Google Scholar]
- Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless–dependent molecular switch. Current Biology. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- Gallagher CM, Knoblich JA. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Dev Cell. 2006;11:641–653. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;21:180–184. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Ge C, Stanley P. The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development. Proc Natl Acad Sci U S A. 2008;105:1539–1544. doi: 10.1073/pnas.0702846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffers I, Serth K, Chapman G, Jaekel R, Schuster-Gossler K, Cordes R, Sparrow DB, Kremmer E, Dunwoodie SL, Klein T, et al. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J Cell Biol. 2007;178:465–476. doi: 10.1083/jcb.200702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, Jolicoeur P. Frequent Provirus Insertional Mutagenesis Of Notch1 In Thymomas Of Mmtv(D)/Myc Transgenic Mice Suggests a Collaboration Of C-Myc and Notch1 For Oncogenesis. Genes & Development. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- Girard L, Jolicoeur P. A Full-Length Notch1 Allele Is Dispensable For Transformation Associated With a Provirally Activated Truncated Notch1 Allele In Moloney Mulv-Infected Mmtvd/Myc Transgenic Mice. Oncogene. 1998;16:517–522. doi: 10.1038/sj.onc.1201562. [DOI] [PubMed] [Google Scholar]
- Goodfellow H, Krejci A, Moshkin Y, Verrijzer CP, Karch F, Bray SJ. Gene-specific targeting of the histone chaperone asf1 to mediate silencing. Dev Cell. 2007;13:593–600. doi: 10.1016/j.devcel.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Greenwald I. Structure/function studies of lin-12/Notch proteins. [Review] Current Opinion in Genetics & Development. 1994;4:556–562. doi: 10.1016/0959-437x(94)90072-b. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12 Suppl 1:R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Six E, laBail O, Lugaet F, Chastagner P, Olry A, Israel A, Brou C. Monoubiquitination and endocytosis direct secretase cleavage of activated Notch receptor. J Cell Biol. 2004;166:73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- Herranz H, Stamataki E, Feiguin F, Milan M. Self-refinement of Notch activity through the transmembrane protein Crumbs: modulation of gamma-Secretase activity. EMBO Rep. 2006 doi: 10.1038/sj.embor.7400617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nature Cell Biology. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- Hicks C, Ladi E, Lindsell C, Hsieh JJD, Hayward SD, Collazo A, Weinmaster G. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. Journal of Neuroscience Research. 2002;68:655–667. doi: 10.1002/jnr.10263. [DOI] [PubMed] [Google Scholar]
- Hirata H, Bessho Y, Kokubu H, Masamizu Y, Yamada S, Lewis J, Kageyama R. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004 doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y, Chia W, Sankar N, Ng YK, et al. F3/Contactin Acts as a Functional Ligand for Notch during Oligodendrocyte Maturation. Cell. 2003;115:163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Hukriede NA, Fleming RJ. Beaded Of Goldschmidt, an Antimorphic Allele Of Serrate, Encodes a Protein Lacking Transmembrane and Intracellular Domains. Genetics. 1997;145:359–374. doi: 10.1093/genetics/145.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukriede NA, Gu Y, Fleming RJ. A dominant-negative form of Serrate acts as a general antagonist of Notch activation. Development. 1997;124:3427–3437. doi: 10.1242/dev.124.17.3427. [DOI] [PubMed] [Google Scholar]
- Huppert S, Le A, Schroeter EH, Mumm JS, Saxena MT, Milner LA, Kopan R. Embryonic Lethality in Mice Homozygous for a Processing Deficient Allele of Notch1. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007 doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Ikeuchi T, Sisodia SS. The notch ligands, delta1 and jagged2, are substrates for presenilin-dependent "gamma-secretase" cleavage. J Biol Chem. 2003 doi: 10.1074/jbc.C200711200. [DOI] [PubMed] [Google Scholar]
- Ilagan MX, Kopan R. SnapShot: Notch Signaling Pathway. Cell. 2007;128:1246. doi: 10.1016/j.cell.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Ingram RN, Orth P, Strickland CL, Le HV, Madison V, Beyer BM. Stabilization of the autoproteolysis of TNF-alpha converting enzyme (TACE) results in a novel crystal form suitable for structure-based drug design studies. Protein Eng Des Sel. 2006;19:155–161. doi: 10.1093/protein/gzj014. [DOI] [PubMed] [Google Scholar]
- Jaekel R, Klein T. The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev Cell. 2006;11:655–669. doi: 10.1016/j.devcel.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Justice N, Roegiers F, Jan LY, Jan YN. Lethal giant larvae acts together with numb in notch inhibition and cell fate specification in the Drosophila adult sensory organ precursor lineage. Curr Biol. 2003;13:778–783. doi: 10.1016/s0960-9822(03)00288-4. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Masamizu Y, Niwa Y. Oscillator mechanism of Notch pathway in the segmentation clock. Dev Dyn. 2007;236:1403–1409. doi: 10.1002/dvdy.21114. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Hurlbut GD, Upadhyay G, Yajnik V, Yedvobnick B, Artavanis-Tsakonas S. Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics. 2007;177:2493–2505. doi: 10.1534/genetics.107.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar R, Fortini ME. The big brain aquaporin is required for endosome maturation and notch receptor trafficking. Cell. 2008;133:852–863. doi: 10.1016/j.cell.2008.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kidd S, Lieber T. Furin cleavage is not a requirement for Drosophila Notch function. Mech Dev. 2002;115:41–51. doi: 10.1016/s0925-4773(02)00120-x. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Ahituv N, Fuchs H, Balling R, Avraham KB, Steel KP, de Angelis MH. The Notch ligand Jagged1 is required for inner ear sensory development. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3873–3878. doi: 10.1073/pnas.071496998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Henderson S, Crittenden S. Notch/Lin-12 Signaling - Transduction By Regulated Protein Slicing. Trends in Biochemical Sciences. 1998;23:353–357. doi: 10.1016/s0968-0004(98)01263-8. [DOI] [PubMed] [Google Scholar]
- Koelzer S, Klein T. Regulation of expression of Vg and establishment of the dorsoventral compartment boundary in the wing imaginal disc by Suppressor of Hairless. Dev Biol. 2006;289:77–90. doi: 10.1016/j.ydbio.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Kolev V, Mandinova A, Guinea-Viniegra J, Hu B, Lefort K, Lambertini C, Neel V, Dummer R, Wagner EF, Dotto GP. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008 doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, Tucey T, Dionne HM, White JQ, Wani K, Boxem M, et al. OSM-11 facilitates LIN-12 Notch signaling during C. elegans vulval development. PLoS Biol. 2008 doi: 10.1371/journal.pbio.0060196. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA. Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr Opin Struct Biol. 2007;17:117–127. doi: 10.1016/j.sbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Iwai N, Nonaka S, Welsh IC, Lan Y, Jiang R, Saijoh Y, O'Brien TP, Hamada H, Gridley T. Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 2003 doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, et al. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170:983–992. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Bodner R, Posakony JW. The Enhancer of split Complex of Drosophila includes four Notch-regulated members of the Bearded gene family. Development. 2000;127:3441–3455. doi: 10.1242/dev.127.16.3441. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MT, Chen E, Crouthamel MC, DiMuzio-Mower J, Xu M, Huang Q, Price E, Register RB, Shi XP, Donoviel DB, et al. Presenilin-1 and presenilin-2 exhibit distinct yet overlapping gamma-secretase activities. J Biol Chem. 2003;278:22475–22481. doi: 10.1074/jbc.M300974200. [DOI] [PubMed] [Google Scholar]
- LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem. 2003;278:34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- Le Borgne R. Regulation of Notch signalling by endocytosis and endosomal sorting. Curr Opin Cell Biol. 2006;18:213–222. doi: 10.1016/j.ceb.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Schweisguth F. Notch signaling: endocytosis makes delta signal better. Curr Biol. 2003;13:R273–R275. doi: 10.1016/s0960-9822(03)00199-4. [DOI] [PubMed] [Google Scholar]
- Lee J, Basak JM, Demehri S, Kopan R. Bi-compartmental communication contributes to the opposite proliferative behavior of Notch1-deficient hair follicle and epidermal keratinocytes. Development. 2007;134:2795–2806. doi: 10.1242/dev.02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Xu A, Panin VM, Irvine KD. An O-fucose site in the ligand binding domain inhibits Notch activation. Development. 2003;130:6411–6421. doi: 10.1242/dev.00883. [DOI] [PubMed] [Google Scholar]
- Levitan D, Greenwald I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature. 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M, Scoggin S, Fu T, Vien L, Histen G, et al. Modulation of notch signaling by antibodies specific for the extracellular negative regulatory region of Notch3. J Biol Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Young MW. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Arboleda-Velasquez JF, Artavanis-Tsakonas S. CADASIL: a critical look at a Notch disease. Dev Neurosci. 2006;28:5–12. doi: 10.1159/000090748. [DOI] [PubMed] [Google Scholar]
- Lubman OY, Ilagan MX, Kopan R, Barrick D. Quantitative Dissection of the Notch:CSL Interaction: Insights into the Notch-mediated Transcriptional Switch. J Mol Biol. 2007;365:577–589. doi: 10.1016/j.jmb.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubman OY, Korolev SV, Kopan R. Anchoring notch genetics and biochemistry; structural analysis of the ankyrin domain sheds light on existing data. Mol Cell. 2004;13:619–626. doi: 10.1016/s1097-2765(04)00120-0. [DOI] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskos K, Fernandez-Catalan C, Huber R, Bourenkov GP, Bartunik H, Ellestad GA, Reddy P, Wolfson MF, Rauch CT, Castner BJ, et al. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc Natl Acad Sci U S A. 1998;95:3408–3412. doi: 10.1073/pnas.95.7.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, Macdonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra-Gorur K, Rand MD, Perez-Villamil B, Artavanis-Tsakonas S. Downregulation of Delta by proteolytic processing. J Cell Biol. 2002;159:313–324. doi: 10.1083/jcb.200203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto A, Lau R, Hein PW, Shipley JM, Weinmaster G. Microfibrillar proteins MAGP-1 and MAGP-2 induce notch1 extracellular domain dissociation and receptor activation. J Biol Chem. 2006 doi: 10.1074/jbc.M600298200. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila Ortholog of Mammalian Tumor Susceptibility Gene 101, Elicit Non-Cell-Autonomous Overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F. Transcriptional repression by Suppressor of Hairless involves the binding of a Hairless-dCtBP complex in Drosophila. Current Biology. 2001;11:789–792. doi: 10.1016/s0960-9822(01)00224-x. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Nagel AC, Krejci A, Tenin G, Bravo-Patino A, Bray S, Maier D, Preiss A. Hairless-Mediated Repression of Notch Target Genes Requires the Combined Activity of Groucho and CtBP Corepressors. Mol Cell Biol. 2005;25:10433–10441. doi: 10.1128/MCB.25.23.10433-10441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Neves A, English K, Priess JR. Notch-GATA synergy promotes endoderm-specific expression of ref-1 in C. elegans. Development. 2007;134:4459–4468. doi: 10.1242/dev.008680. [DOI] [PubMed] [Google Scholar]
- Neves A, Priess JR. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev Cell. 2005;8:867–879. doi: 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Olsen SL, D'Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007a;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Weinmaster G. Notch signaling--constantly on the move. Traffic. 2007b;8:959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Imayoshi I, Shimojo H, Nishi E, Kageyama R, McConnell SK. Visualization of embryonic neural stem cells using Hes promoters in transgenic mice. Mol Cell Neurosci. 2006;31:109–122. doi: 10.1016/j.mcn.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Okajima T, Reddy B, Matsuda T, Irvine KD. Contributions of chaperone and glycosyltransferase activities of O-fucosyltransferase 1 to Notch signaling. BMC Biol. 2008;6:1. doi: 10.1186/1741-7007-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J Biol Chem. 2003;278:42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- Okochi M, Steiner H, Fukumori A, Tanii H, Tomita T, Tanaka T, Iwatsubo T, Kudo T, Takeda M, Haass C. Presenilins mediate a dual intramembranous {gamma}-secretase cleavage of Notch-1. EMBO J. 2002;21:5408–5416. doi: 10.1093/emboj/cdf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C, Cheng H, Chang LW, Ohtsuka T, Kageyama R, Stormo DG, Kopan R. Target selectivity of vertebrate Notch proteins: collaboration between discrete domains and CSL binding site architecture determine activation probability. J Biol Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Ong CT, Sedy JR, Murphy KM, Kopan R. Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PLoS ONE. 2008;3:e2823. doi: 10.1371/journal.pone.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knochel W, et al. SHARP is a novel component of the Notch/RBP-J{kappa} signalling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-J{kappa}/SHARP Recruits CtIP/CtBP Corepressors To Silence Notch Target Genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panin VM, Shao L, Lei L, Moloney DJ, Irvine KD, Haltiwanger RS. Notch Ligands are substrates for protein O-fucosyltransferase-1 and fringe. Journal of Biological Chemistry. 2002;277:29945–29952. doi: 10.1074/jbc.M204445200. [DOI] [PubMed] [Google Scholar]
- Parks AL, Curtis D. Presenilin diversifies its portfolio. Trends Genet. 2007 doi: 10.1016/j.tig.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. An Essential Role For Ectodomain Shedding in Mammalian Development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Petcherski AG, Kimble J. LAG-3 a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- Pfister S, Przemeck GK, Gerber JK, Beckers J, Adamski J, de Angelis MH. Interaction of the MAGUK Family Member Acvrinp1 and the Cytoplasmic Domain of the Notch Ligand Delta1. J Mol Biol. 2003;333:229–235. doi: 10.1016/j.jmb.2003.08.043. [DOI] [PubMed] [Google Scholar]
- Poellinger L, Lendahl U. Modulating Notch signaling by pathway-intrinsic and pathway-extrinsic mechanisms. Curr Opin Genet Dev. 2008 doi: 10.1016/j.gde.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Qi-Takahara y, Morishima-Kawashima M, Tanimura Y, Dolios G, Hirotani N, Horikoshi Y, Kametani F, Maeda M, C, T, Rong W, et al. Longer Forms of Amyloid {beta} Protein: Implications for the Mechanism of Intramembrane Cleavage by {gamma}-Secretase. J Neurosci. 2005;25:436–445. doi: 10.1523/JNEUROSCI.1575-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Luther KB, Haltiwanger RS. Notch signaling in normal and disease States: possible therapies related to glycosylation. Curr Mol Med. 2007;7:427–445. doi: 10.2174/156652407780831593. [DOI] [PubMed] [Google Scholar]
- Rand DM, Grimm MLM, Artavanis-Tsakonas S, Patriub V, Blacklow CS, Sklar CJ, Aster CJ. Calcium depletion dissociates and activates heterodimeric Notch receptors. Molecular and Cellular Biology. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray WJ, Yao M, Mumm J, Schroeter EH, Saftig P, Wolfe M, Selkoe DJ, Kopan R, Goate AM. Cell surface presenilin-1 participates in the gamma secretase-like proteolysis of notch. Journal of Biological Chemistry. 1999;274:36801–36807. doi: 10.1074/jbc.274.51.36801. [DOI] [PubMed] [Google Scholar]
- Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M, Rasskin-Gutman D, Rodriguez-Leon J, Buscher D, Feijo JA, Izpisua Belmonte JC. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature. 2004;427:121–128. doi: 10.1038/nature02190. [DOI] [PubMed] [Google Scholar]
- Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- Rooke JE, Xu T. Positive And Negative Signals Between Interacting Cells For Establishing Neural Fate [Review] Bioessays. 1998;20:209–214. doi: 10.1002/(SICI)1521-1878(199803)20:3<209::AID-BIES4>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Russ A, Stumm G, Augustin M, Sedlmeier R, Wattler S, Nehls M. Random mutagenesis in the mouse as a tool in drug discovery. Drug Discov Today. 2002;7:1175–1183. doi: 10.1016/s1359-6446(02)02515-1. [DOI] [PubMed] [Google Scholar]
- Saito SY, Takeshima H. DNER as key molecule for cerebellar maturation. Cerebellum. 2006;5:227–231. doi: 10.1080/14734220600632564. [DOI] [PubMed] [Google Scholar]
- Sasamura T, Sasaki N, Miyashita F, Nakao S, Ishikawa HO, Ito M, Kitagawa M, Harigaya K, Spana E, Bilder D, et al. neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development. 2003;130:4785–4795. doi: 10.1242/dev.00679. [DOI] [PubMed] [Google Scholar]
- Sato T, Diehl TS, Narayanan S, Funamoto S, Ihara Y, De Strooper B, Steiner H, Haass C, Wolfe MS. Active gamma-secretase complexes contain only one of each component. J Biol Chem. 2007;282:33985–33993. doi: 10.1074/jbc.M705248200. [DOI] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Serneels L, Dejaegere T, Craessaerts K, Horre K, Jorissen E, Tousseyn T, Hebert S, Coolen M, Martens G, Zwijsen A, et al. Differential contribution of the three Aph1 genes to gamma-secretase activity in vivo. Proc Natl Acad Sci U S A. 2005;102:1719–1724. doi: 10.1073/pnas.0408901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L, Simpson P, Haenlin M. Requirement For Dynamin During Notch Signaling In Drosophila Neurogenesis. Developmental Biology. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, Laplant Q, Ball H, Dann CE, 3rd, Sudhof T, Yu G. Nicastrin Functions as a gamma-Secretase-Substrate Receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Shawber C, Nofziger D, Hsieh JJD, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- Shen H, McElhinny AS, Cao Y, Gao P, Liu J, Bronson R, Griffin JD, Wu L. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci U S A. 2003 doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, Hamada Y, Yazaki Y, Hirai H. Mouse Jagged1 physically interacts with Notch2 and other Notch receptors - Assessment by quantitative methods. Journal of Biological Chemistry. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- Shirotani K, Tomioka M, Kremmer E, Haass C, Steiner H. Pathological activity of familial Alzheimer's disease-associated mutant presenilin can be executed by six different gamma-secretase complexes. Neurobiol Dis. 2007;27:102–107. doi: 10.1016/j.nbd.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, Logeat F. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and gamma-secretase. Proc Natl Acad Sci U S A. 2003;100:7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillos S, Roch F, Campuzano S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development. 1997;124:4769–4779. doi: 10.1242/dev.124.23.4769. [DOI] [PubMed] [Google Scholar]
- Souilhol C, Cormier S, Monet M, Vandormael-Pournin S, Joutel A, Babinet C, Cohen-Tannoudji M. Nas transgenic mouse line allows visualization of Notch pathway activity in vivo. Genesis. 2006;44:277–286. doi: 10.1002/dvg.20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M, Uemura K, Ge C, Shi S, Tashima Y, Stanley P. Roles of pofut1 and o-fucose in Mammalian notch signaling. J Biol Chem. 2008;283:13638–13651. doi: 10.1074/jbc.M802027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P. Regulation of Notch signaling by glycosylation. Curr Opin Struct Biol. 2007;17:530–535. doi: 10.1016/j.sbi.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Requirements for Presenilin-Dependent Cleavage of Notch and Other Transmembrane Proteins. Molecular Cell. 2000;6:625–663. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- Tagami S, Okochi M, Yanagida K, Ikuta A, Fukumori A, Matsumoto N, Ishizuka-Katsura Y, Nakayama T, Itoh N, Jiang J, et al. Regulation of Notch Signaling by Dynamic Changes in the Precision in S3 Cleavage of Notch-1. Mol Cell Biol. 2008;28:165–176. doi: 10.1128/MCB.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical Interaction Between a Novel Domain Of the Receptor Notch and the Transcription Factor Rbp-J-Kappa/Su(H) Current Biology. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Tani S, Kurooka H, Aoki T, Hashimoto N, Honjo T. The N-and C-terminal regions of RBP-J interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic Acids Research. 2001;29:1373–1380. doi: 10.1093/nar/29.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassishin L, Yin YI, Bassit B, Li YM. Processing of Notch and amyloid precursor protein by gamma-secretase is spatially distinct. Proc Natl Acad Sci U S A. 2004;101:17050–17055. doi: 10.1073/pnas.0408007101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, Cohen SM. Tumor Suppressor Properties of the ESCRT-II Complex Component Vps25 in Drosophila. Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Tsai H, Hardisty RE, Rhodes C, Kiernan AE, Roby P, Tymowska-Lalanne Z, Mburu P, Rastan S, Hunter AJ, Brown SDM, et al. The mouse slalom mutant demonstrates a role for Jagged1 in neuroepithelial patterning in the organ of Corti. Human Molecular Genetics. 2001;10:507–512. doi: 10.1093/hmg/10.5.507. [DOI] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipursky S, Banerjee U. An EGFR/Ebi/Sno Pathway Promotes Delta Expression by Inactivating Su(H)/SMRTER Repression during Inductive Notch Signaling. Cell. 2002;110:625. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc Natl Acad Sci U S A. 2007;104:1610–1615. doi: 10.1073/pnas.0610520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukumo S, Hirose K, Maekawa Y, Kishihara K, Yasutomo K. Lunatic fringe controls T cell differentiation through modulating notch signaling. J Immunol. 2006;177:8365–8371. doi: 10.4049/jimmunol.177.12.8365. [DOI] [PubMed] [Google Scholar]
- Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- Vaccari T, Bilder D. The Drosophila Tumor Suppressor vps25 Prevents Nonautonomous Overproliferation by Regulating Notch Trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol. 2008;180:755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. Immobilization of Notch ligand, Delta-1, is required for induction of Notch signaling. J Cell Sci. 2000;113:4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci U S A. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visan I, Tan JB, Yuan JS, Harper JA, Koch U, Guidos CJ. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat Immunol. 2006a;7:634–643. doi: 10.1038/ni1345. [DOI] [PubMed] [Google Scholar]
- Visan I, Yuan JS, Tan JB, Cretegny K, Guidos CJ. Regulation of intrathymic T-cell development by Lunatic Fringe-Notch1 interactions. Immunol Rev. 2006b;209:76–94. doi: 10.1111/j.0105-2896.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- Vodovar N, Schweisguth F. Functions of O-fucosyltransferase in Notch trafficking and signaling: towards the end of a controversy? J Biol. 2008;7:7. doi: 10.1186/jbiol68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, Korving J, van den Born M, Stappenbeck T, Wu Y, Clevers H, et al. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134:535–544. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M, Schroeter EH, Pan Y, Blandford M, Kopan R. Ectodomain shedding and intramembrane cleavage of mammalian notch proteins is not regulated through oligomerization. J Biol Chem. 2004;279:50864–50873. doi: 10.1074/jbc.M409430200. [DOI] [PubMed] [Google Scholar]
- Wasserman ZR, Duan JJ, Voss ME, Xue CB, Cherney RJ, Nelson DJ, Hardman KD, Decicco CP. Identification of a selectivity determinant for inhibition of tumor necrosis factor-alpha converting enzyme by comparative modeling. Chem Biol. 2003;10:215–223. doi: 10.1016/s1074-5521(03)00044-9. [DOI] [PubMed] [Google Scholar]
- Wen C, Metzstein MM, Greenwald I. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development. 1997;124:4759–4767. doi: 10.1242/dev.124.23.4759. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, IV, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating Mutations of NOTCH1 in Human T Cell Acute Lymphoblastic Leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, Aster JC. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Kopan R. Intramembrane Proteolysis: Theme and Variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- Wu J, Bresnick EH. Bare rudiments of notch signaling: how receptor levels are regulated. Trends Biochem Sci. 2007;32:477–485. doi: 10.1016/j.tibs.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Xu A, Haines N, Dlugosz M, Rana NA, Takeuchi H, Haltiwanger RS, Irvine KD. In vitro reconstitution of the modulation of Drosophila notch-ligand binding by fringe. J Biol Chem. 2007 doi: 10.1074/jbc.M707040200. [DOI] [PubMed] [Google Scholar]
- Zhao G, Cui MZ, Mao G, Dong Y, Tan J, Sun L, Xu X. gamma-Cleavage is dependent on zeta-cleavage during the proteolytic processing of amyloid precursor protein within its transmembrane domain. J Biol Chem. 2005;280:37689–37697. doi: 10.1074/jbc.M507993200. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Katzman RB, Delmolino LM, Bhat I, Zhang Y, Germaniuk-Kurowska A, Reddi HV, Solomon A, Zeng MS, Kung A, et al. The notch regulator maml1 interacts with p53 and functions as a coactivator. J Biol Chem. 2007 doi: 10.1074/jbc.M608974200. [DOI] [PubMed] [Google Scholar]
- Zhou L, Li LW, Yan Q, Petryniak B, Man Y, Su C, Shim J, Chervin S, Lowe JB. Notch-dependent control of myelopoiesis is regulated by fucosylation. Blood. 2008;112:308–319. doi: 10.1182/blood-2007-11-115204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G, Hayward SD. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Atkins JB, Rompani SB, Bancescu DL, Petersen PH, Tang H, Zou K, Stewart SB, Zhong W. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 2007;129:163–178. doi: 10.1016/j.cell.2007.02.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.